Impact of Biochar on Rhizosphere Bacterial Diversity Restoration Following Chloropicrin Fumigation of Planted Soil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Used Soil in This Study
2.2. Glasshouse Pot Experiments
2.3. Sampling and Determination of Plant Growth Responses
2.4. Illumina-Based 16S rRNA Sequencing and Sequence Data analysis
3. Results
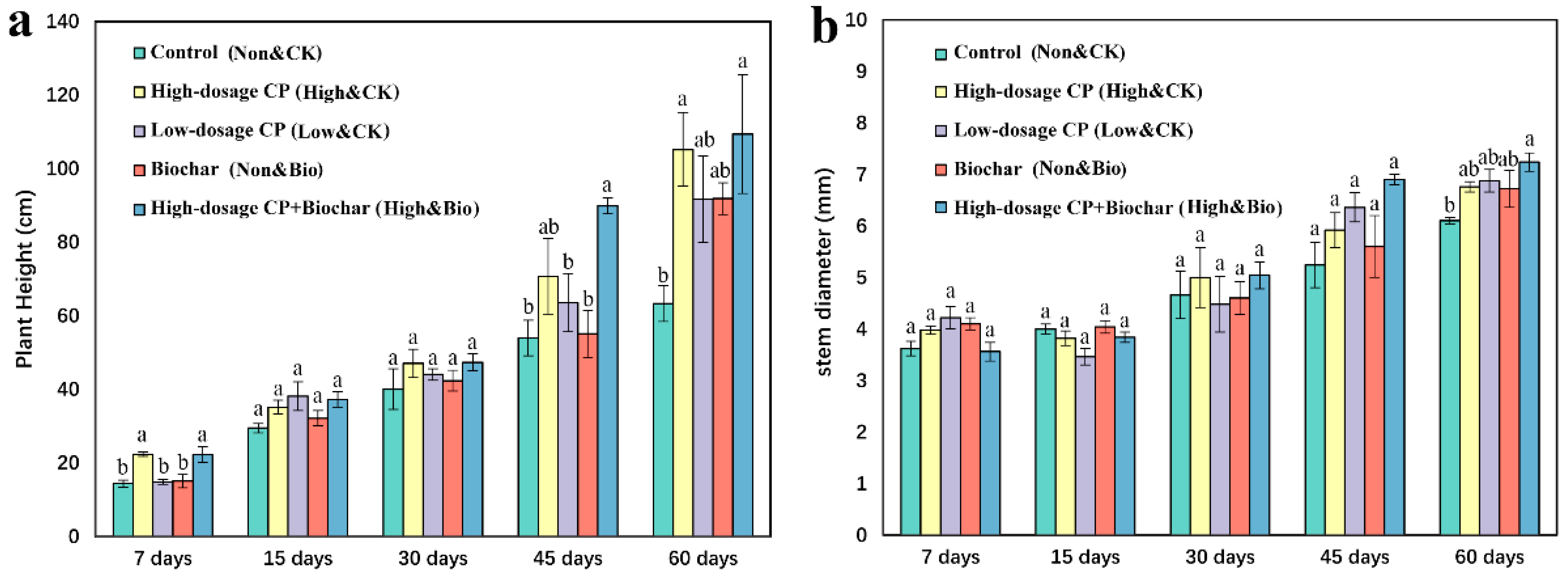
3.1. Effects of CP and Biochar on Plant Growth
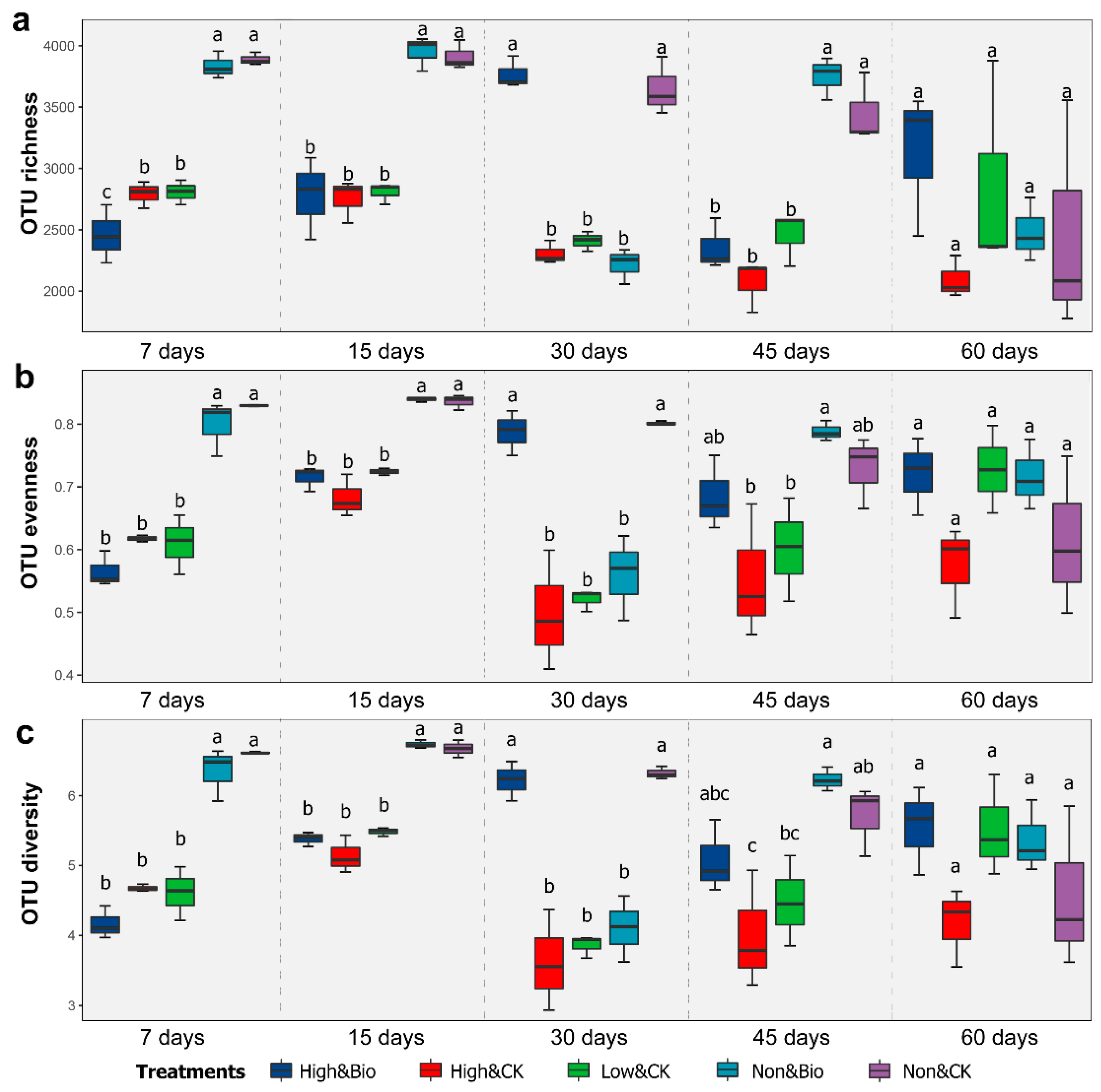
3.2. Effects of CP Fumigation and Biochar Addition on Rhizobacterial Alpha Diversity
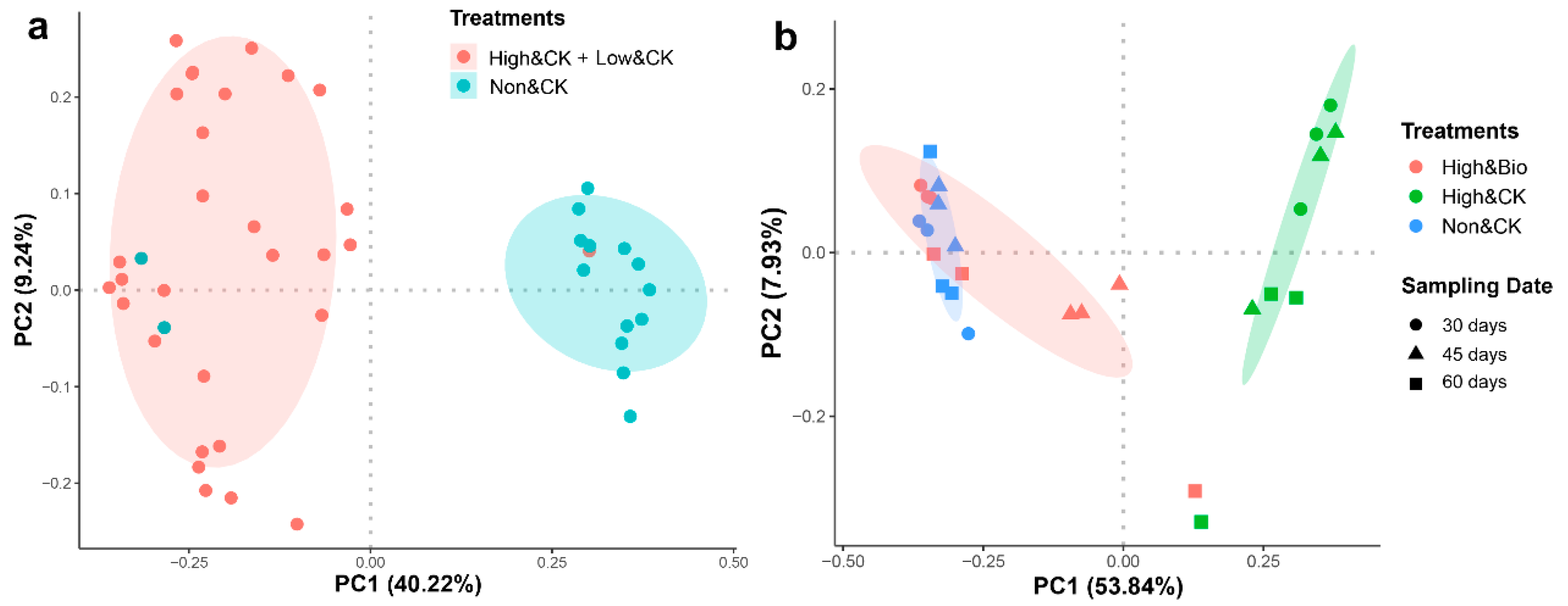
3.3. Effects of CP Fumigation and Biochar Addition on the Structural Composition of Rhizobacterial Communities
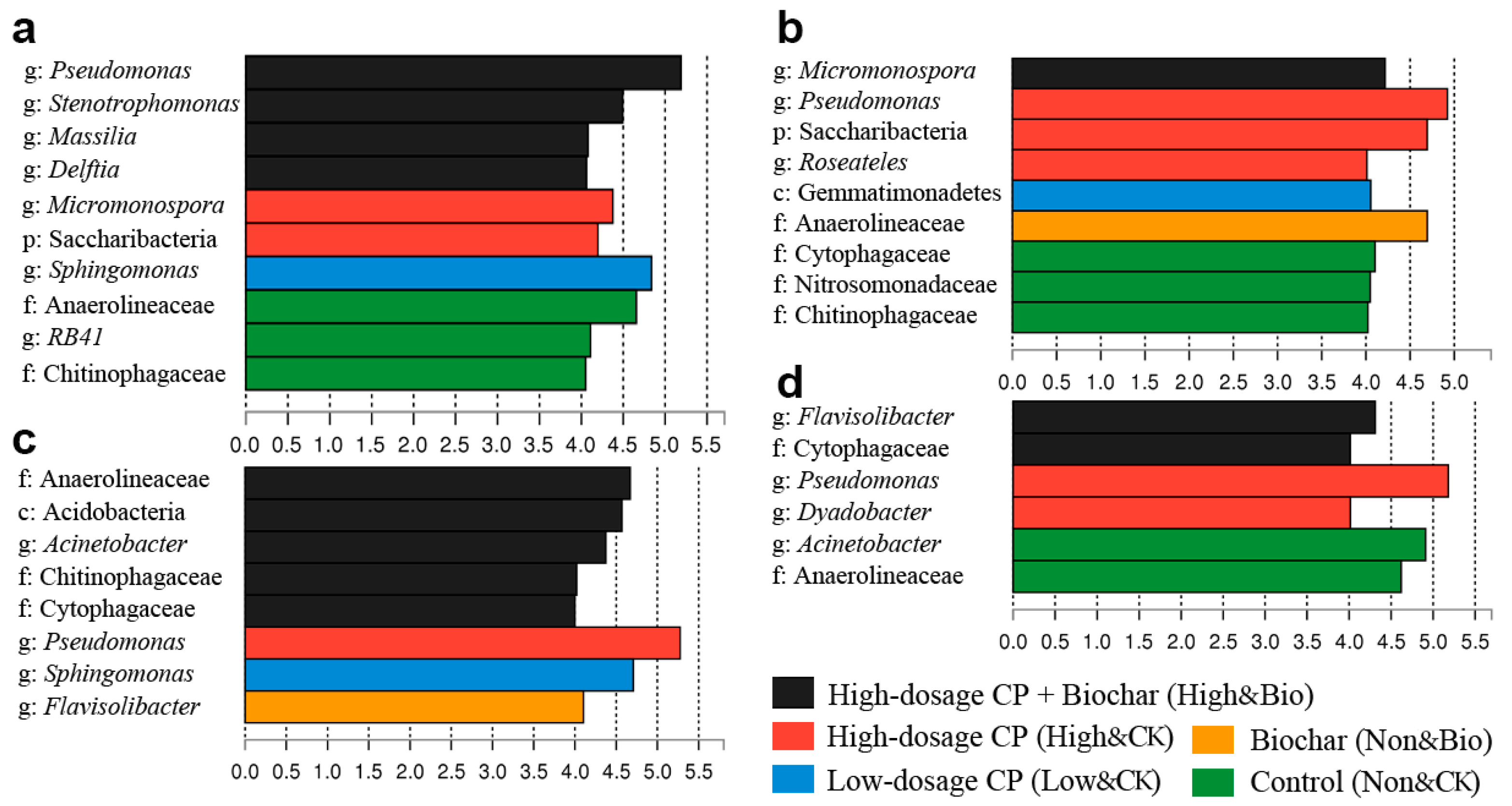
3.4. Shifts in the Predominant Rhizobacterial Populations Following CP Fumigation and Biochar Addition
4. Discussion
4.1. Effects of CP and Biochar on the Growth of Cucumber Seedlings
4.2. Different Changes between the Bulk Soil Bacterial Communities and Rhizosphere Soil Bacterial Communities after CP Fumigation
4.3. Biochar Accelerated the Reconstruction of Rhizobacterial Communities after CP Fumigation
4.4. New Predominant Rhizobacterial Populations after Biochar Amendments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bell, C.H. Fumigation in the 21st century. Crop Prot. 2000, 19, 563–569. [Google Scholar] [CrossRef]
- United Nations Environment Programme. Phasing-Out Methyl Bromide in Developing Countries: A Success Story and Its Challenges; UNEP: Nairobi, Kenya, 2014. [Google Scholar]
- Gullino, M.L.; Minuto, A.; Gilardi, G.; Garibaldi, A.; Ajwa, H.; Duafala, T. Efficacy of preplant soil fumigation with chloropicrin for tomato production in Italy. Crop Prot. 2002, 21, 741–749. [Google Scholar] [CrossRef]
- Mao, L.; Wang, Q.; Yan, D.; Ma, T.; Liu, P.; Shen, J.; Li, Y.; Ouyang, C.; Guo, M.; Cao, A. Evaluation of chloropicrin as a soil fumigant against Ralstonia solanacarum in ginger (Zingiber officinale Rosc.) production in China. PLoS ONE 2014, 9, e91767. [Google Scholar] [CrossRef]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The Role of Soil Microorganisms in Plant Mineral Nutrition—Current Knowledge and Future Directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, B.; Wang, Q.; Li, Y.; Fang, W.; Yan, D.; Guo, M.; Cao, A. Effect of fumigation with chloropicrin on soil bacterial communities and genes encoding key enzymes involved in nitrogen cycling. Environ. Pollut. 2017, 227, 534–542. [Google Scholar] [CrossRef]
- Dangi, S.R.; Gerik, J.S.; Tirado-Corbalá, R.; Ajwa, H. Soil Microbial Community Structure and Target Organisms under Different Fumigation Treatments. Appl. Environ. Soil Sci. 2015, 2015, 673264. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Huang, B.; Wang, Q.; Li, Y.; Fang, W.; Han, D.; Yan, D.; Guo, M.; Cao, A. Effects of fumigation with metam-sodium on soil microbial biomass, respiration, nitrogen transformation, bacterial community diversity and genes encoding key enzymes involved in nitrogen cycling. Sci. Total Environ. 2017, 598, 1027–1036. [Google Scholar] [CrossRef]
- Fang, W.; Wang, X.; Huang, B.; Zhang, D.; Liu, J.; Zhu, J.; Yan, D.; Wang, Q.; Cao, A.; Han, Q. Comparative analysis of the effects of five soil fumigants on the abundance of denitrifying microbes and changes in bacterial community composition. Ecotoxicol. Environ. Saf. 2020, 187, 109850. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; Bruijn, I.D.; Dekkers, E.; Voort, M.V.D.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Hoshino, Y.T.; Matsumoto, N. Changes in fungal community structure in bulk soil and spinach rhizosphere soil after chemical fumigation as revealed by 18S rDNA PCR-DGGE. Soil Sci. Plant Nutr. 2007, 53, 40–55. [Google Scholar] [CrossRef]
- Ibekwe, A.M.; Papiernik, S.K.; Yang, C.H. Influence of soil fumigation by methyl bromide and methyl iodide on rhizosphere and phyllosphere microbial community structure. J. Environ. Sci. Health Part B 2010, 45, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Zhang, N.; Shen, Z.; Zhu, C.; Li, R.; Salles, J.F.; Shen, Q. Rhizosphere bacteria assembly derived from fumigation and organic amendment triggers the direct and indirect suppression of tomato bacterial wilt disease. Appl. Soil Ecol. 2020, 147, 103364. [Google Scholar] [CrossRef]
- Lehmann, J.; Rillig, M.C.; Thies, J.; Masiello, C.A.; Hockaday, W.C.; Crowley, D. Biochar effects on soil biota—A review. Soil Biol. Biochem. 2011, 43, 1812–1836. [Google Scholar] [CrossRef]
- Fang, W.; Cao, A.; Yan, D.; Han, D.; Huang, B.; Li, J.; Liu, X.; Guo, M.; Wang, Q. The Effect of Two Types of Biochars on the Efficacy, Emission, Degradation, and Adsorption of the Fumigant Methyl Isothiocyanate. Energies 2017, 10, 16. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Wang, Q.; Yan, D.; Fang, W.; Mao, L.; Wang, D.; Li, Y.; Ouyang, C.; Guo, M.; Cao, A. Effects of Biochar Amendment on Chloropicrin Adsorption and Degradation in Soil. Energies 2016, 9, 869. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Lin, Y.; Tian, X.; Xu, Q.; Chen, Z.; Lin, W. Tobacco bacterial wilt suppression with biochar soil addition associates to improved soil physiochemical properties and increased rhizosphere bacteria abundance. Appl. Soil Ecol. 2017, 112, 90–96. [Google Scholar] [CrossRef]
- Sheng, Y.; Zhu, L. Biochar alters microbial community and carbon sequestration potential across different soil pH. Sci. Total Environ. 2018, 622–623, 1391–1399. [Google Scholar] [CrossRef]
- Fang, W.; Song, Z.; Tao, S.; Zhang, D.; Huang, B.; Ren, L.; Cheng, H.; Yan, D.; Li, Y.; Cao, A.; et al. Biochar mitigates the negative effect of chloropicrin fumigation on beneficial soil microorganisms. Sci. Total Environ. 2020, 738, 139880. [Google Scholar] [CrossRef]
- Gan, J.; Yates, S.R.; Ernst, F.F.; Jury, W.A. Degradation and Volatilization of the Fumigant Chloropicrin after Soil Treatment. J. Environ. Qual. 2000, 29, 1391–1397. [Google Scholar] [CrossRef]
- Yan, D.; Wang, Q.; Li, Y.; Ouyang, C.; Guo, M.; Cao, A. Analysis of the inhibitory effects of chloropicrin fumigation on nitrification in various soil types. Chemosphere 2017, 175, 459–464. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.S.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.A.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.L.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, 141–145. [Google Scholar] [CrossRef] [Green Version]
- Yan, D.; Wang, Q.; Mao, L.; Li, W.; Xie, H.; Guo, M.; Cao, A. Quantification of the effects of various soil fumigation treatments on nitrogen mineralization and nitrification in laboratory incubation and field studies. Chemosphere 2013, 90, 1210–1215. [Google Scholar] [CrossRef]
- Lamb, M.C.; Sorensen, R.B.; Butts, C.L. Crop response to biochar under differing irrigation levels in the southeastern USA. J. Crop Improv. 2018, 32, 305–317. [Google Scholar] [CrossRef]
- Gaskin, J.; Speir, R.; Harris, K.; Das, K.C.; Lee, R.; Morris, L.; Fisher, D. Effect of Peanut Hull and Pine Chip Biochar on Soil Nutrients, Corn Nutrient Status, and Yield. Agron. J. 2010, 102, 623–633. [Google Scholar] [CrossRef] [Green Version]
- Major, J.; Rondon, M.; Molina, D.; Riha, S.J.; Lehmann, J. Maize yield and nutrition during 4 years after biochar application to a Colombian savanna oxisol. Plant Soil 2010, 333, 117–128. [Google Scholar] [CrossRef]
- Van Zwieten, L.; Kimber, S.; Morris, S.; Chan, K.Y.; Downie, A.; Rust, J.; Joseph, S.; Cowie, A. Effects of biochar from slow pyrolysis of papermill waste on agronomic performance and soil fertility. Plant Soil 2010, 327, 235–246. [Google Scholar] [CrossRef]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P.; Monchy, S.; Cardinale, M.; Taghavi, S.; Crossman, L.; Avison, M.B.; Berg, G.; van der Lelie, D.; Dow, J.M. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 2009, 7, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.S.; Baruah, R.; Borah, M.; Singh, A.K.; Boruah, H.P.D.; Saikia, N.; Deka, M.; Dutta, N.; Bora, T.C. Bioremediation potential of native hydrocarbon degrading bacterial strains in crude oil contaminated soil under microcosm study. Int. Biodeterior. Biodegrad. 2014, 94, 79–89. [Google Scholar] [CrossRef]
- Volova, T.G.; Prudnikova, S.V.; Vinogradova, O.N.; Syrvacheva, D.A.; Shishatskaya, E.I. Microbial Degradation of Polyhydroxyalkanoates with Different Chemical Compositions and Their Biodegradability. Microb. Ecol. 2017, 73, 353–367. [Google Scholar] [CrossRef]
- Yan, D.; Wang, Q.; Mao, L.; Ma, T.; Li, Y.; Ouyang, C.; Guo, M.; Cao, A. Interaction between nitrification, denitrification and nitrous oxide production in fumigated soils. Atmos. Environ. 2015, 103, 82–86. [Google Scholar] [CrossRef]
- Zhang, D.; Cheng, H.; Hao, B.; Li, Q.; Wu, J.; Zhang, Y.; Fang, W.; Yan, D.; Li, Y.; Wang, Q.; et al. Fresh chicken manure fumigation reduces the inhibition time of chloropicrin on soil bacteria and fungi and increases beneficial microorganisms. Environ. Pollut. 2021, 286, 117460. [Google Scholar] [CrossRef]
- Jin, H. Characterization of Microbial Life Colonizing Biochar and Biochar-Amended Soils. Ph.D. Thesis, Cornell University, Ithaca, NY, USA, 2010. [Google Scholar]
- Han, D.; Yan, D.; Cao, A.; Fang, W.; Liu, P.; Li, Y.; Ouyang, C.; Wang, Q. Degradation of dimethyl disulphide in soil with or without biochar amendment. Pest Manag. Sci. 2017, 73, 1830–1836. [Google Scholar] [CrossRef]
- Nguyen, T.T.N.; Xu, C.; Tahmasbian, I.; Che, R.; Xu, Z.; Zhou, X.; Wallace, H.M.; Bai, S.H. Effects of biochar on soil available inorganic nitrogen: A review and meta-analysis. Geoderma 2017, 288, 79–96. [Google Scholar] [CrossRef] [Green Version]
- Yakabe, L.E.; Parker, S.R.; Kluepfel, D.A. Effect of pre-plant soil fumigants on Agrobacterium tumefaciens, pythiaceous species, and subsequent soil recolonization by A. tumefaciens. Crop Prot. 2010, 29, 583–590. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Chen, Y.; Qin, X.; Cao, A.; Lu, A. Impact of Biochar on Rhizosphere Bacterial Diversity Restoration Following Chloropicrin Fumigation of Planted Soil. Int. J. Environ. Res. Public Health 2022, 19, 2126. https://doi.org/10.3390/ijerph19042126
Li J, Chen Y, Qin X, Cao A, Lu A. Impact of Biochar on Rhizosphere Bacterial Diversity Restoration Following Chloropicrin Fumigation of Planted Soil. International Journal of Environmental Research and Public Health. 2022; 19(4):2126. https://doi.org/10.3390/ijerph19042126
Chicago/Turabian StyleLi, Jun, Yan Chen, Xiangyang Qin, Aocheng Cao, and Anxiang Lu. 2022. "Impact of Biochar on Rhizosphere Bacterial Diversity Restoration Following Chloropicrin Fumigation of Planted Soil" International Journal of Environmental Research and Public Health 19, no. 4: 2126. https://doi.org/10.3390/ijerph19042126
APA StyleLi, J., Chen, Y., Qin, X., Cao, A., & Lu, A. (2022). Impact of Biochar on Rhizosphere Bacterial Diversity Restoration Following Chloropicrin Fumigation of Planted Soil. International Journal of Environmental Research and Public Health, 19(4), 2126. https://doi.org/10.3390/ijerph19042126





