Identification of Candidate lncRNA and Pseudogene Biomarkers Associated with Carbon-Nanotube-Induced Malignant Transformation of Lung Cells and Prediction of Potential Preventive Drugs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Preprocessing
2.2. Weighted Correlation Network Analysis of lncRNAs and Pseudogenes
2.3. Differential Analysis of lncRNAs, Pseudogenes and mRNAs in GSE41178 Dataset
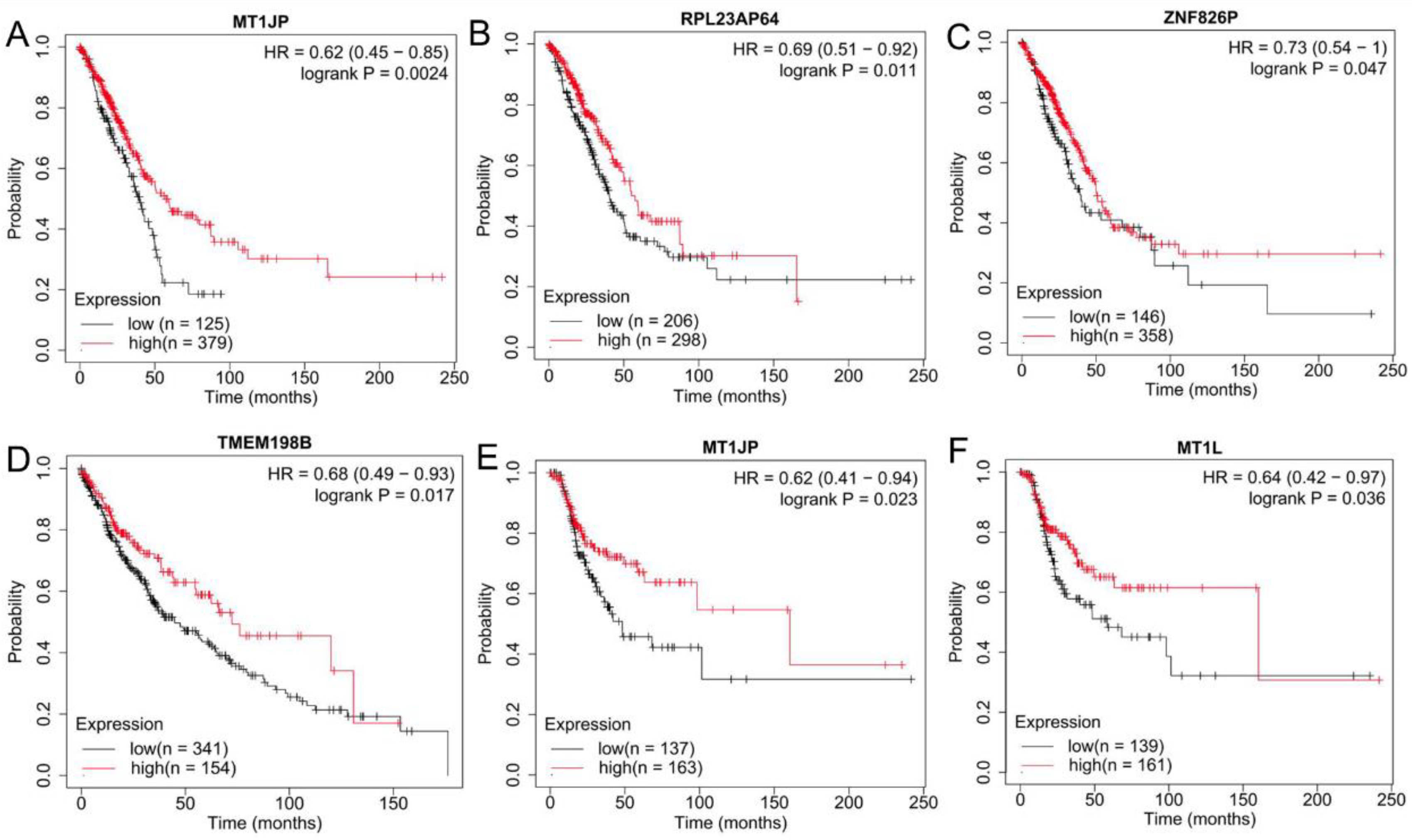
2.4. Validation of the Expression Levels and Prognosis of DE-lncRNAs and DE-Pseudogenes in Lung Cancer
2.5. Construction of lncRNA- and Pseudogene-Related ceRNA Networks
2.6. Construction of lncRNA- and Pseudogene-mRNA Co-Expression Networks
2.7. Construction of Protein–Protein Interaction (PPI) Networks
2.8. Validation of the Expression Levels and Survival Associations of DE-mRNAs in Lung Cancer
2.9. Validation of the Expression Levels and Survival Associations of miRNAs in Lung Cancer
2.10. Function Enrichment Analysis
2.11. Diagnostic Analysis of Hub lncRNAs and Pseudogenes
2.12. Prediction of Small-Molecule Drugs That Regulated Hub lncRNAs and Pseudogenes
3. Results
3.1. Identification of Key Modules Related to CNT-Induced Malignant Transformation of Lung Cells
3.2. Identification of Differential Genes in CNT-Induced Malignant Transformation of Lung Cells
3.3. Validation of the Expression Levels and Prognosis of lncRNAs and Pseudogenes in Tissues of Lung Cancer Patients
3.4. Establishment of lncRNA- and Pseudogene-Based ceRNA Regulatory Networks
3.5. Establishment of lncRNA- and Pseudogene-Based Co-Expression Networks
3.6. Establishment of PPI Networks for lncRNA- and Pseudogene-Related Target Genes
3.7. Validation of the Expression Levels and Prognosis of lncRNA- and Pseudogene-Related mRNAs and miRNAs in Lung Cancer
3.8. Function Enrichment Analysis
3.9. Diagnostic Values of Hub lncRNAs and Pseudogenes
3.10. Prediction of Small-Molecule Drugs That Regulated Hub lncRNAs and Pseudogenes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, X.; Liang, Y.; Weng, W.; Hu, Z.; Zhu, M. Polypyrrole-coated carbon nanotube/cotton hybrid fabric with high areal capacitance for flexible quasi-solid-state supercapacitors. Energy Storage Mater. 2020, 33, 11–17. [Google Scholar] [CrossRef]
- Luo, X.; Weng, W.; Liang, Y.; Hu, Z.; Zhang, Y.; Yang, J.; Yang, L.; Yang, S.; Zhu, M.; Cheng, H.M. Multifunctional fabrics of carbon nanotube fibers. J. Mater. Chem. A 2019, 7, 8790–8797. [Google Scholar] [CrossRef]
- Abazari, S.; Shamsipur, A.; Bakhsheshi-Rad, H.R.; Ismail, A.F.; Sharif, S.; Razzaghi, M. Carbon Nanotubes (CNTs)-Reinforced Magnesium-Based Matrix Composites: A Comprehensive Review. Materials 2020, 13, 4421. [Google Scholar] [CrossRef]
- Yin, Z.; Cui, C.; Chen, H.; Yu, X.; Qian, W. The Application of Carbon Nanotube/Graphene-Based Nanomaterials in Wastewater Treatment. Small 2020, 16, e1902301. [Google Scholar] [CrossRef] [PubMed]
- Negri, V.; Pacheco-Torres, J. Carbon Nanotube in Biomedicine. Top. Curr. Chem. 2020, 378, 15. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Saito, N. Biocompatibility and Carcinogenicity of Carbon Nanotubes as Biomaterials. Nanomaterials 2020, 10, 264. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Luanpitpong, S.; Castranova, V.; Tse, W.; Lu, Y.; Pongrakhananon, V.; Rojanasakul, Y. Carbon nanotubes induce malignant transformation and tumorigenesis of human lung epithelial cells. Nano. Lett. 2011, 11, 2796–2803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Stueckle, T.A.; Mishra, A.; Derk, R.; Meighan, T.; Castranova, V.; Rojanasakul, Y. Neoplastic-like transformation effect of single-walled and multi-walled carbon nanotubes compared to asbestos on human lung small airway epithelial cells. Nanotoxicology 2014, 8, 485–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luanpitpong, S.; Wang, L.; Castranova, V.; Dinu, C.Z.; Issaragrisil, S.; Chen, Y.C.; Rojanasakul, Y. Induction of cancer-associated fibroblast-like cells by carbon nanotubes dictates its tumorigenicity. Sci. Rep. 2016, 6, 39558. [Google Scholar] [CrossRef] [PubMed]
- Suzui, M.; Futakuchi, M.; Fukamachi, K.; Numano, T.; Abdelgied, M.; Takahashi, S.; Ohnishi, M.; Omori, T.; Tsuruoka, S.; Hirose, A.; et al. Multiwalled carbon nanotubes intratracheally instilled into the rat lung induce development of pleural malignant mesothelioma and lung tumors. Cancer Sci. 2016, 107, 924–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleh, D.M.; Alexander, W.T.; Numano, T.; Ahmed, O.H.M.; Gunasekaran, S.; Alexander, D.B. Comparative carcinogenicity study of a thick, straight-type and a thin, tangled-type multi-walled carbon nanotube administered by intra-tracheal instillation in the rat. Part. Fibre Toxicol. 2020, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Voronkova, M.A.; Luanpitpong, S. SOX9 Regulates Cancer Stem-Like Properties and Metastatic Potential of Single-Walled Carbon Nanotube-Exposed Cells. Sci. Rep. 2017, 7, 11653. [Google Scholar] [CrossRef]
- Polimeni, M.; Gulino, G.R.; Gazzano, E.; Kopecka, J.; Marucco, A.; Fenoglio, I.; Cesano, F.; Campagnolo, L.; Magrini, A.; Pietroiusti, A.; et al. Multi-walled carbon nanotubes directly induce epithelial-mesenchymal transition in human bronchial epithelial cells via the TGF-β-mediated Akt/GSK-3β/SNAIL-1 signalling pathway. Part. Fibre Toxicol. 2016, 13, 27. [Google Scholar] [CrossRef] [Green Version]
- Wang, P. Induction of Slug by Chronic Exposure to Single-Walled Carbon Nanotubes Promotes Tumor Formation and Metastasis. Chem. Res. Toxicol. 2017, 30, 1396–1405. [Google Scholar] [CrossRef]
- Pongrakhananon, V.; Luanpitpong, S.; Stueckle, T.A.; Wang, L.; Nimmannit, U.; Rojanasakul, Y. Carbon nanotubes induce apoptosis resistance of human lung epithelial cells through FLICE-inhibitory protein. Toxicol. Sci. 2015, 143, 499–511. [Google Scholar] [CrossRef] [Green Version]
- Luanpitpong, S.; Wang, L.; Stueckle, T.A.; Tse, W.; Chen, Y.C.; Rojanasakul, Y. Caveolin-1 regulates lung cancer stem-like cell induction and p53 inactivation in carbon nanotube-driven tumorigenesis. Oncotarget 2014, 5, 3541–3554. [Google Scholar] [CrossRef]
- Chen, D.; Stueckle, T.A.; Luanpitpong, S.; Rojanasakul, Y.; Lu, Y.; Wang, L. Gene expression profile of human lung epithelial cells chronically exposed to single-walled carbon nanotubes. Nanoscale Res. Lett. 2015, 10, 12. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Tian, X.; Zhang, J.; Tan, L.; Ouyang, N.; Jia, B.; Chen, C.; Ge, C.; Li, J. Postchronic Single-Walled Carbon Nanotube Exposure Causes Irreversible Malignant Transformation of Human Bronchial Epithelial Cells through DNA Methylation Changes. ACS Nano 2021, 15, 7094–7104. [Google Scholar] [CrossRef]
- He, X.; Despeaux, E.; Stueckle, T.A.; Chi, A.; Castranova, V.; Dinu, C.Z.; Wang, L.; Rojanasakul, Y. Role of mesothelin in carbon nanotube-induced carcinogenic transformation of human bronchial epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2016, 311, L538–L549. [Google Scholar] [CrossRef] [Green Version]
- Guo, N.L.; Wan, Y.W.; Denvir, J.; Porter, D.W.; Pacurari, M.; Wolfarth, M.G.; Castranova, V.; Qian, Y. Multiwalled carbon nanotube-induced gene signatures in the mouse lung: Potential predictive value for human lung cancer risk and prognosis. J. Toxicol. Environ. Health A 2012, 75, 1129–1153. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.J.; Tay, Y. Noncoding RNA: RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porter, D.W.; Hubbs, A.F.; Mercer, R.R.; Wu, N.; Wolfarth, M.G.; Sriram, K.; Leonard, S.; Battelli, L.; Schwegler-Berry, D.; Friend, S.; et al. Mouse pulmonary dose- and time course-responses induced by exposure to multi-walled carbon nanotubes. Toxicology 2010, 269, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Győrffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021, 19, 4101–4109. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015, 127, 67–72. [Google Scholar] [CrossRef]
- Ventura, C.; Vieira, L.; Silva, C.; Sousa-Uva, A.; Silva, M.J. Functional effects of differentially expressed microRNAs in A549 cells exposed to MWCNT-7 or crocidolite. Toxicol. Lett. 2020, 328, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, visualization, and Integrated Discovery. Genome. Biol. 2003, 4, P3. [Google Scholar] [CrossRef] [PubMed]
- Grondin, C.J.; Davis, A.P.; Wiegers, J.A.; Wiegers, T.C.; Sciaky, D.; Johnson, R.J.; Mattingly, C.J. Predicting molecular mechanisms, pathways, and health outcomes induced by Juul e-cigarette aerosol chemicals using the Comparative Toxicogenomics Database. Curr. Res. Toxicol. 2021, 2, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Hao, S.; Xie, L.; Xiang, G.; Hu, W.; Wu, Q.; Liu, Z.; Li, S. LncRNA NEAT1 contributes to the acquisition of a tumor like-phenotype induced by PM 2.5 in lung bronchial epithelial cells via HIF-1α activation. Environ. Sci. Pollut. Res. Int. 2021, 28, 43382–43393. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, B.; Yun, J.; Xu, J.; Meng, Q.; Li, X.; Chen, R. lncRNA SOX2-OT ceRNA network enhances the malignancy of long-term PM(2.5)-exposed human bronchial epithelia. Ecotoxicol. Environ. Saf. 2021, 217, 112242. [Google Scholar] [CrossRef]
- Qi, H.; Liu, Y.; Wang, N.; Xiao, C. Lentinan Attenuated the PM2.5 Exposure-Induced Inflammatory Response, Epithelial-Mesenchymal Transition and Migration by Inhibiting the PVT1/miR-199a-5p/caveolin1 Pathway in Lung Cancer. DNA Cell Biol. 2021, 40, 683–693. [Google Scholar] [CrossRef]
- Zhou, C.; Huang, C.; Wang, J.; Huang, H.; Li, J.; Xie, Q.; Liu, Y.; Zhu, J.; Li, Y.; Zhang, D.; et al. LncRNA MEG3 downregulation mediated by DNMT3b contributes to nickel malignant transformation of human bronchial epithelial cells via modulating PHLPP1 transcription and HIF-1α translation. Oncogene 2017, 36, 3878–3889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.P.; Rea, M.; Wang, Z.; Yang, C. Down-regulation of lncRNA MEG3 promotes chronic low dose cadmium exposure-induced cell transformation and cancer stem cell-like property. Toxicol. Appl. Pharmacol. 2021, 430, 115724. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.P.; Wang, Z.; Yang, C. LncRNA DUXAP10 up-regulation and the Hedgehog pathway activation are critically involved in chronic cadmium exposure-induced cancer stem cell-like property. Toxicol. Sci. 2021, 184, 33–45. [Google Scholar] [CrossRef]
- Ndika, J.; Karisola, P.; Kinaret, P. Profiling Non-Coding RNA Changes Associated with 16 Different Engineered Nanomaterials in a Mouse Airway Exposure Model. Cells 2021, 10, 1085. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zhang, X.; Gao, L.; Xue, H.; Liu, L.; Wang, S.; Chen, S.; Huang, L. LncRNA loc105377478 promotes NPs-Nd(2)O(3)-induced inflammation in human bronchial epithelial cells through the ADIPOR1/NF-κB axis. Ecotoxicol. Environ. Saf. 2021, 208, 111609. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Chang, X.; Wang, X.; Yang, M.; Gao, Q.; Liu, H.; Li, C.; Li, S.; Sun, Y. LncRNA MEG3 mediates nickel oxide nanoparticles-induced pulmonary fibrosis via suppressing TGF-β1 expression and epithelial-mesenchymal transition process. Environ. Toxicol. 2021, 36, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wang, Y.; Lv, W.; Wu, X.; Sheng, H.; He, C.; Hu, J. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019, 10, 383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, S.; Lu, J.; Ge, Y.; Wang, Q.; Ma, G.; Zhao, Q.; Wu, D.; Gong, W.; Du, M.; et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol. Cancer 2018, 17, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.H.; Xu, K.; Liu, J.H.; Du, L.L.; Li, X.S.; Su, Y.M.; Liu, J.C. LncRNA MT1JP inhibits the malignant progression of hepatocellular carcinoma through regulating AKT. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6647–6656. [Google Scholar] [PubMed]
- Ma, J.; Yan, H.; Zhang, J.; Tan, Y.; Gu, W. Long-Chain Non-Coding RNA (lncRNA) MT1JP Suppresses Biological Activities of Lung Cancer by Regulating miRNA-423-3p/Bim Axis. Med. Sci. Monit. 2019, 25, 5114–5126. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Liu, N.; Chen, M.; Xu, Y.; Fang, S.; Xiang, W.; Hua, X.; Chen, G.; Zhong, Y.; Yu, H. Overexpressed pseudogene MT1L associated with tumor immune infiltrates and indicates a worse prognosis in BLCA. World J. Surg. Oncol. 2021, 19, 133. [Google Scholar] [CrossRef] [PubMed]
- Sabbir Ahmed, C.M.; Paul, B.C.; Cui, Y.; Frie, A.L.; Burr, A.; Kamath, R.; Chen, J.Y.; Nordgren, T.M.; Bahreini, R.; Lin, Y.H. Integrative Analysis of lncRNA-mRNA Coexpression in Human Lung Epithelial Cells Exposed to Dimethyl Selenide-Derived Secondary Organic Aerosols. Chem. Res. Toxicol. 2021, 34, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.Y.; Qi, Q.L.; Hou, S.; Chen, Z.; Zhang, L.; Zhao, H.G.; Lin, C.H. LncRNA PVT1 Acts as a Tumor Promoter in Thyroid Cancer and Promotes Tumor Progression by Mediating miR-423-5p-PAK3. Cancer Manag. Res. 2020, 12, 13403–13413. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Qiu, T.; Xuan, Y.; Liu, A.; Sun, X.; Huang, Z.; Su, W.; Du, W.; Yun, T.; Wo, Y.; et al. circFBXW7 attenuates malignant progression in lung adenocarcinoma by sponging miR-942-5p. Transl. Lung Cancer Res. 2021, 10, 1457–1473. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, J.; Huang, H.; Jiang, Y.; Huang, Y.; Fang, H.; Zheng, G.; Zhou, X.; Wu, Y.; Lei, C.; et al. lncRNA LIFR-AS1 suppresses invasion and metastasis of non-small cell lung cancer via the miR-942-5p/ZNF471 axis. Cancer Cell Int. 2020, 20, 180. [Google Scholar] [CrossRef]
- Bi, C.; Wang, G. LINC00472 suppressed by ZEB1 regulates the miR-23a-3p/FOXO3/BID axis to inhibit the progression of pancreatic cancer. J. Cell Mol. Med. 2021, 25, 8312–8328. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, W.J.; Liu, H.T.; Que, Z.J.; Qian, F.F.; Liu, L.S.; Tian, J.H. TOB1-AS1 suppresses non-small cell lung cancer cell migration and invasion through a ceRNA network. Exp. Ther. Med. 2019, 18, 4249–4258. [Google Scholar] [CrossRef] [Green Version]
- Fan, L.; Qi, H.; Teng, J.; Su, B.; Chen, H.; Wang, C.; Xia, Q. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol. 2016, 37, 7777–7784. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Liu, G.; Jin, Y.; Yang, T.; Zhang, D.; Ding, L.; Zhou, F.; Pan, Y.; Wei, Y. miR-15b-5p Promotes Growth and Metastasis in Breast Cancer by Targeting HPSE2. Front. Oncol. 2020, 10, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Sheng, L. miR-15b-5p facilitates the tumorigenicity by targeting RECK and predicts tumour recurrence in prostate cancer. J. Cell Mol. Med. 2018, 22, 1855–1863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tordjman, J.; Majumder, M.; Amiri, M.; Hasan, A.; Hess, D.; Lala, P.K. Tumor suppressor role of cytoplasmic polyadenylation element binding protein 2 (CPEB2) in human mammary epithelial cells. BMC Cancer 2019, 19, 561. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Xu, Z.; Du, Z.; Wu, B. The Identification of Key Genes and Pathways in Glioma by Bioinformatics Analysis. J. Immunol. Res. 2017, 2017, 1278081. [Google Scholar] [CrossRef] [Green Version]
- Marwitz, S.; Depner, S.; Dvornikov, D.; Merkle, R.; Szczygieł, M.; Müller-Decker, K.; Lucarelli, P.; Wäsch, M.; Mairbäurl, H.; Rabe, K.F.; et al. Downregulation of the TGFβ Pseudoreceptor BAMBI in Non-Small Cell Lung Cancer Enhances TGFβ Signaling and Invasion. Cancer Res. 2016, 76, 3785–3801. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, M.; Hu, M.; Wei, P.; Zhu, W. BAMBI overexpression together with β-sitosterol ameliorates NSCLC via inhibiting autophagy and inactivating TGF-β/Smad2/3 pathway. Oncol. Rep. 2017, 37, 3046–3054. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Li, J.J.; Liang, X.L.; Wu, H.; Liang, Z. PSME3 Promotes TGFB1 Secretion by Pancreatic Cancer Cells to Induce Pancreatic Stellate Cell Proliferation. J. Cancer 2019, 10, 2128–2138. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Guo, C.; Chen, J.; Duan, S.; Hu, Y.; Zou, Y.; Chi, H.; Geng, J.; Zhou, J. Silencing PSME3 induces colorectal cancer radiosensitivity by downregulating the expression of cyclin B1 and CKD1. Exp. Biol. Med. 2019, 244, 1409–1418. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fan, Y.; Huang, X.; Luo, J.; Zhong, L.; Shu, X.S.; Lu, L.; Xiang, T.; Chan, A.T.C.; Yeo, W.; et al. Tumor Suppression of Ras GTPase-Activating Protein RASA5 through Antagonizing Ras Signaling Perturbation in Carcinomas. Iscience 2019, 21, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, J.M.; Gonzalez, A.; Massion, P.P.; Chen, H.; Olson, S.J.; Shyr, Y.; Diaz, R.; Lambright, E.S.; Sandler, A.; Carbone, D.P.; et al. Cytoplasmic clusterin expression is associated with longer survival in patients with resected non small cell lung cancer. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1845–1851. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Fan, Z.; Dou, X.; Zhou, Q.; Zeng, G.; Liu, L.; Chen, W.; Lan, R.; Liu, W.; Ru, G.; et al. Inactivation of tumor suppressor gene Clusterin leads to hyperactivation of TAK1-NF-κB signaling axis in lung cancer cells and denotes a therapeutic opportunity. Theranostics 2020, 10, 11520–11534. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.R.; Abdelsalam, R.; Ghosh, S.; Livingstone, J.; Palanisamy, N.; Boutros, P.C.; Yip, S.M.; Lees-Miller, S.P.; Bismar, T.A. Decreased ATM Protein Expression Is Substantiated with PTEN Loss in Defining Aggressive Phenotype of Prostate Cancer Associated with Lethal Disease. Eur. Urol. Open Sci. 2021, 29, 93–101. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, H.; Liu, L.; Zhu, K.; Yu, W.; Guo, J. Hsa_circ_0001546 acts as a miRNA-421 sponge to inhibit the chemoresistance of gastric cancer cells via ATM/Chk2/p53-dependent pathway. Biochem. Biophys. Res. Commun. 2020, 521, 303–309. [Google Scholar] [CrossRef]
- Xu, X.; Gao, H.; Qin, J.; He, L.; Liu, W. TMP21 modulates cell growth in papillary thyroid cancer cells by inducing autophagy through activation of the AMPK/mTOR pathway. Int. J. Clin. Exp. Pathol. 2015, 8, 10824–10831. [Google Scholar]
- Wang, X.; Duan, J.; Fu, W.; Yin, Z.; Sheng, J.; Lei, Z.; Wang, H. Decreased expression of NEDD4L contributes to NSCLC progression and metastasis. Biochem. Biophys. Res. Commun. 2019, 513, 398–404. [Google Scholar] [CrossRef]
- Bhushan, A.; Kumari, R.; Srivastava, T. Scouting for common genes in the heterogenous hypoxic tumor microenvironment and their validation in glioblastoma. 3 Biotech 2021, 11, 451. [Google Scholar] [CrossRef]
- Nalairndran, G.; Hassan Abdul Razack, A.; Mai, C.W.; Fei-Lei Chung, F.; Chan, K.K.; Hii, L.W.; Lim, W.M.; Chung, I.; Leong, C.O. Phosphoinositide-dependent Kinase-1 (PDPK1) regulates serum/glucocorticoid-regulated Kinase 3 (SGK3) for prostate cancer cell survival. J. Cell Mol. Med. 2020, 24, 12188–12198. [Google Scholar] [CrossRef]
- Liu, F.; Wu, X.; Jiang, X.; Qian, Y.; Gao, J. Prolonged inhibition of class I PI3K promotes liver cancer stem cell expansion by augmenting SGK3/GSK-3β/β-catenin signalling. J. Exp. Clin. Cancer Res. 2018, 37, 122. [Google Scholar] [CrossRef] [Green Version]
- Xie, D.; Luo, X. Identification of four methylation-driven genes as candidate biomarkers for monitoring single-walled carbon nanotube-induced malignant transformation of the lung. Toxicol. Appl. Pharmacol. 2021, 412, 115391. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, Y.; Lv, W.; Wu, X.; Sheng, H.; He, C.; Hu, J. Schizandrin A can inhibit non-small cell lung cancer cell proliferation by inducing cell cycle arrest, apoptosis and autophagy. Int. J. Mol. Med. 2021, 48, 214. [Google Scholar] [CrossRef]
- Bae, J.M. Serum Folate Levels and Lung Cancer Risk: A Meta-Epidemiological Study of Population-based Case-Control Studies. Asian Pac. J. Cancer Prev. 2020, 21, 1829–1833. [Google Scholar] [CrossRef]
- Charoenngam, N.; Ponvilawan, B.; Ungprasert, P. Higher zinc intake is associated with decreased risk of lung cancer. J. Evid. Based Med. 2021, 14, 185–187. [Google Scholar] [CrossRef]
- De Bravo, M.G.; Tournier, H.; Schinella, G.; Viaggi, M.; Quintans, C. Effect of dietary supplementation with gamma-linolenic acid on the growth of a human lung carcinoma implanted in nude mice. Medicina 1995, 55, 670–674. [Google Scholar]
- Wang, Y.; Shi, J.; Gong, L. Gamma linolenic acid suppresses hypoxia-induced proliferation and invasion of non-small cell lung cancer cells by inhibition of HIF1α. Genes Genom. 2020, 42, 927–935. [Google Scholar] [CrossRef]
- Barbarino, M.; Giordano, A. Assessment of the Carcinogenicity of Carbon Nanotubes in the Respiratory System. Cancers 2021, 13, 1318. [Google Scholar] [CrossRef]

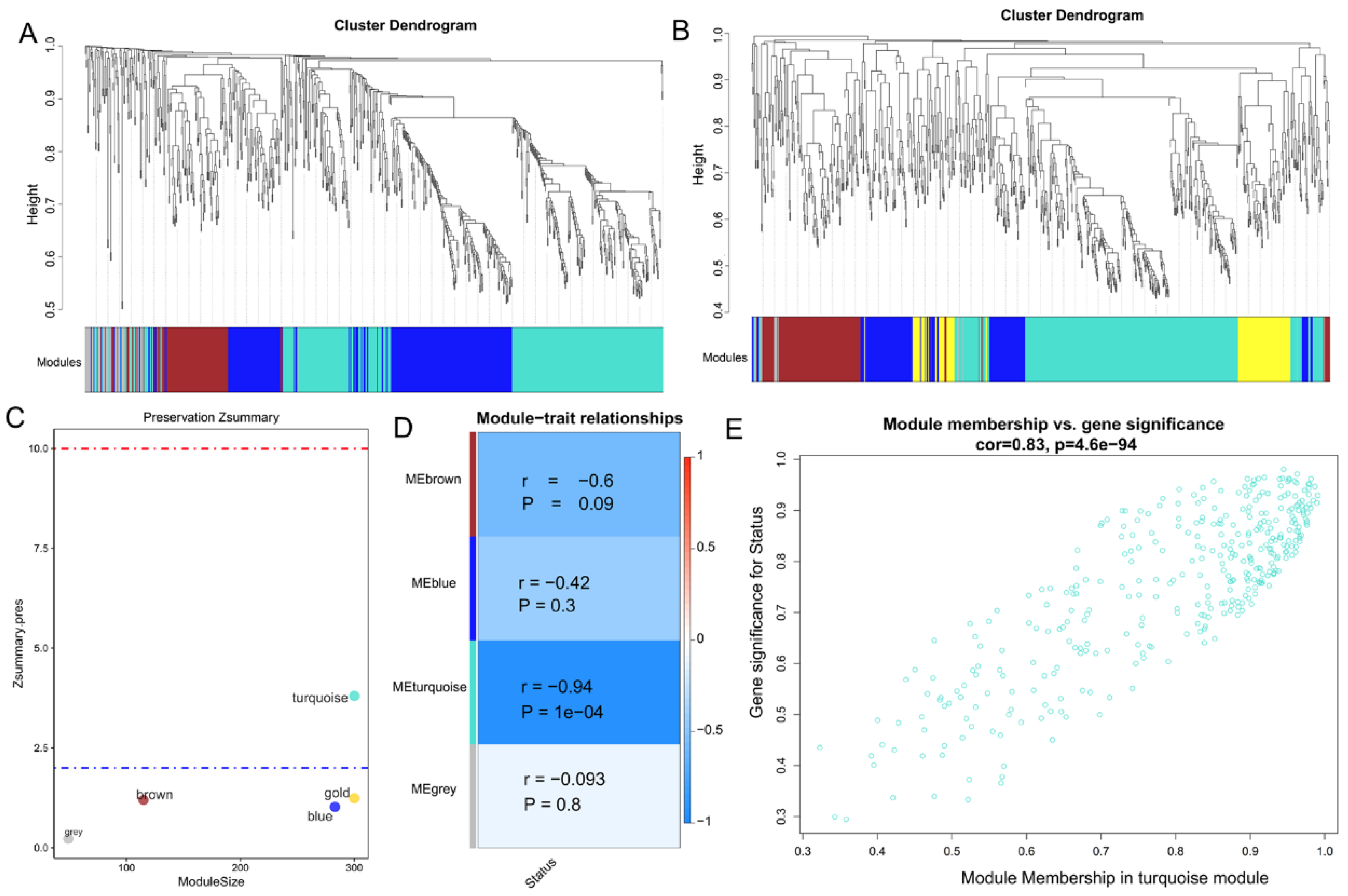
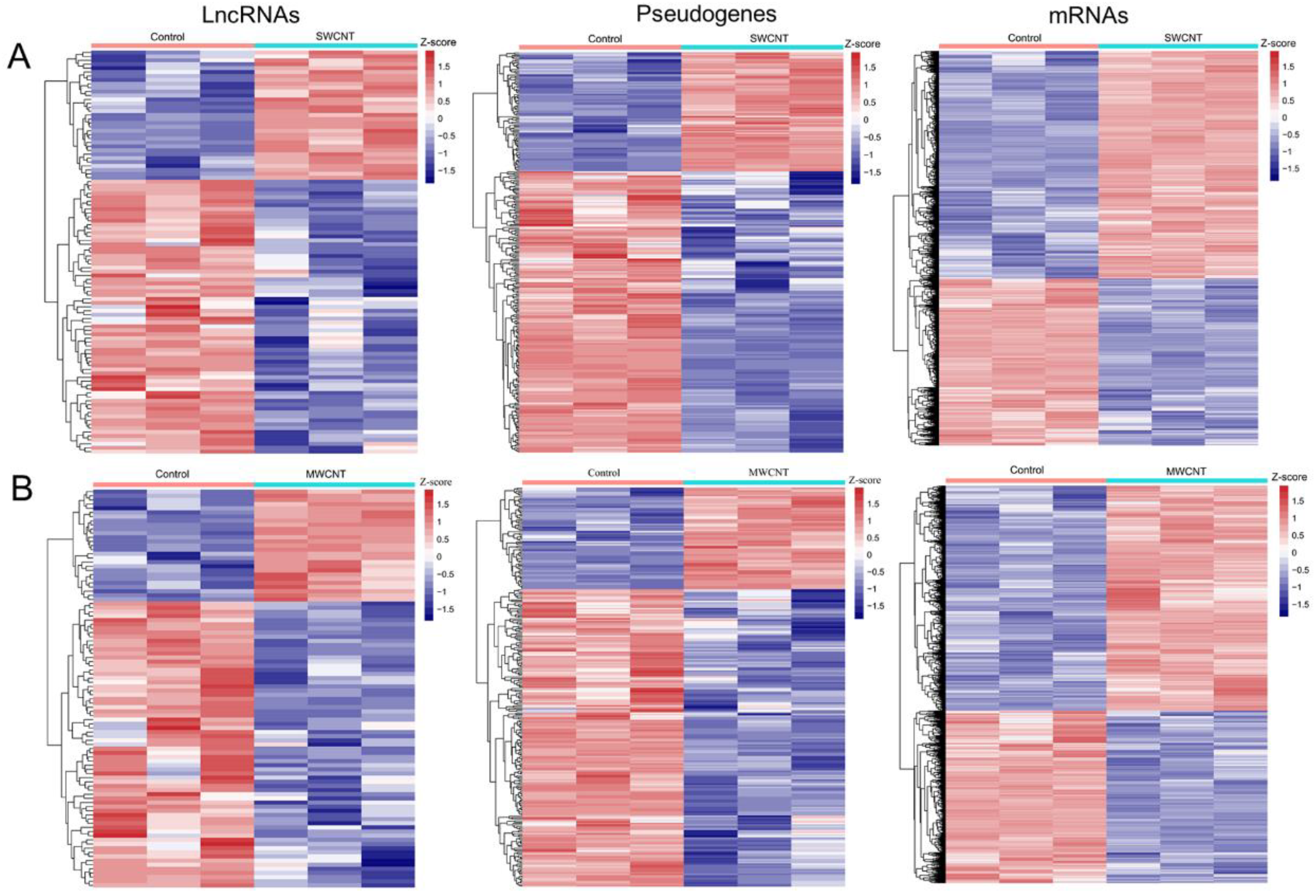
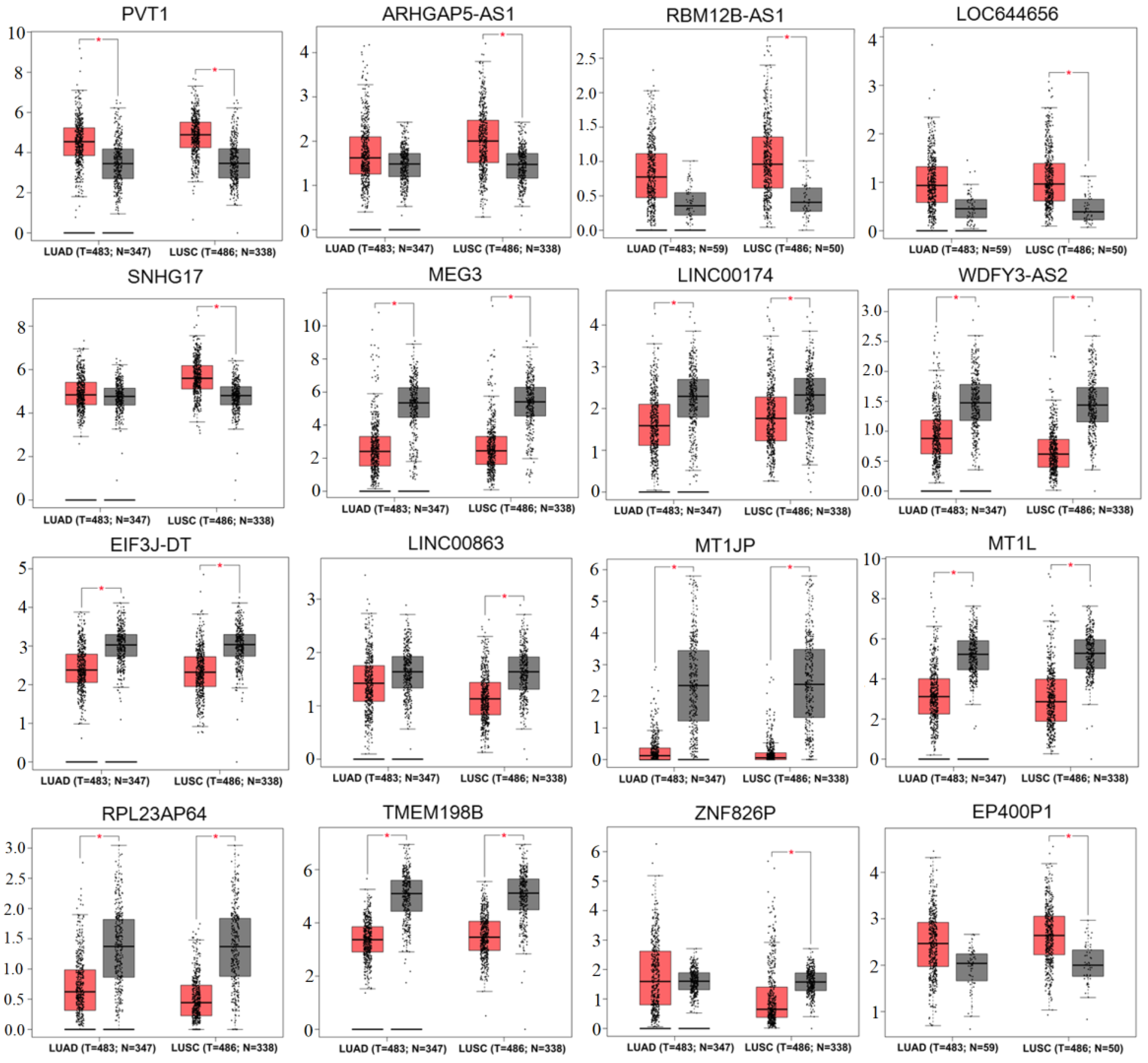
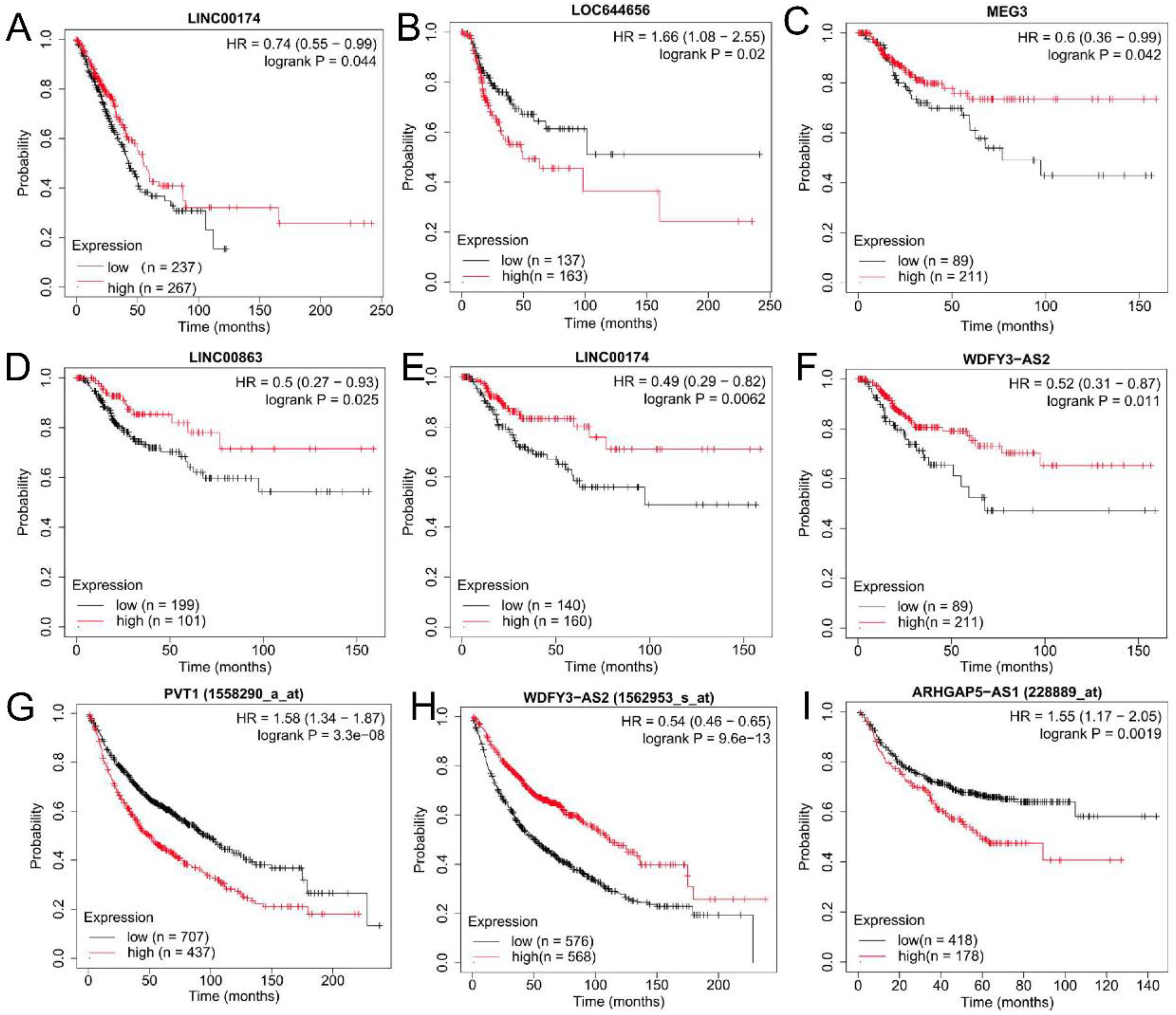


| GSE41178 (SWCNT vs. Control) | GSE41178 (MWCNT vs. Control) | ||||
|---|---|---|---|---|---|
| Log2FC | FDR | Log2FC | FDR | ||
| lncRNAs | ARHGAP5-AS1 | 1.46 | 3.91 × 104 | 1.46 | 5.35 × 104 |
| PVT1 | 0.7 | 1.04 × 103 | 1.14 | 2.86 × 104 | |
| LINC00174 | −1.23 | 3.31 × 104 | −1.01 | 2.14 × 103 | |
| MEG3 | −1.09 | 1.44 × 102 | −1.37 | 2.00 × 103 | |
| Pseudogenes | MT1L | −1.9 | 7.49 × 105 | −1.77 | 6.05 × 105 |
| ZNF826P | −1.19 | 6.06 × 103 | −1.24 | 3.38 × 103 | |
| MT1JP | −0.87 | 5.15 × 104 | −1.21 | 2.86 × 104 | |
| RPL23AP64 | −0.78 | 2.63 × 103 | −0.99 | 1.62 × 103 | |
| TMEM198B | −0.66 | 3.94 × 103 | −0.54 | 4.41 × 103 | |
| mRNAs | NIP7 | 0.66 | 3.56 × 103 | 0.75 | 8.43 × 104 |
| TMED10 | 0.61 | 4.28 × 103 | 0.61 | 6.93 × 103 | |
| PSME3 | 0.60 | 8.01 × 104 | 0.57 | 5.52 × 103 | |
| CPEB2 | −3.29 | 6.40× 105 | −3.25 | 1.63 × 104 | |
| CLU | −2.77 | 2.49 × 105 | −1.87 | 3.67 × 105 | |
| ATM | −2.26 | 3.58 × 104 | −1.7 | 3.58 × 104 | |
| SGK3 | −2.04 | 3.89× 105 | −2.08 | 3.19 × 105 | |
| SYNGAP1 | −1.45 | 6.28 × 104 | −1.08 | 1.50 × 103 | |
| BAMBI | −1.34 | 7.39 × 105 | −1.2 | 2.23 × 104 | |
| NEDD4L | −1.29 | 4.58 × 105 | −0.86 | 1.43 × 103 | |
| PHF21A | −1.15 | 9.71 × 105 | −0.79 | 6.65 × 103 | |
| Genes | DC | Genes | BC | Genes | CC | Genes | EC | Genes | LAC | Genes | IC | Genes | SC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MYC | 89 | MYC | 69325.03 | MYC | 0.046924 | MYC | 0.345324 | COL4A2 | 11.14286 | MYC | 4.519095 | MYC | 3.95 × 107 |
| ATM | 53 | CASP3 | 28914.29 | CASP3 | 0.046573 | HIST2H3PS2 | 0.232677 | COL6A1 | 10.95652 | ATM | 4.37312 | HIST2H3PS2 | 1.79 × 107 |
| HIST2H3PS2 | 49 | ATM | 26878.67 | ATM | 0.046346 | CASP3 | 0.218635 | COL11A1 | 10.88889 | HIST2H3PS2 | 4.345062 | CASP3 | 1.58 × 107 |
| CASP3 | 49 | PPARG | 21315.25 | HIST2H3PS2 | 0.046318 | ATM | 0.213316 | COL6A2 | 10.34783 | CASP3 | 4.345062 | ATM | 1.51 × 107 |
| PPARG | 40 | HIST2H3PS2 | 17839.02 | PPARG | 0.046209 | CCNB1 | 0.196486 | COL5A2 | 10.2 | PPARG | 4.263963 | CCNB1 | 1.28 × 107 |
| CCNB1 | 37 | NRAS | 16979.76 | NRAS | 0.046184 | CHEK1 | 0.175156 | COL1A1 | 9.67742 | CCNB1 | 4.229326 | CHEK1 | 1.02 × 107 |
| NRAS | 36 | PTK2 | 15259.33 | CCNB1 | 0.046091 | AURKA | 0.154292 | COL6A3 | 9.636364 | NRAS | 4.216698 | AURKA | 7896703 |
| CHEK1 | 34 | AURKA | 15109.43 | SMAD2 | 0.046085 | FOS | 0.145202 | NID1 | 8.923077 | CHEK1 | 4.189493 | FOS | 6983669 |
| AURKA | 33 | SMAD2 | 14891.42 | FOS | 0.046038 | CCND2 | 0.137003 | CCNB1 | 8.648648 | AURKA | 4.17482 | CCND2 | 6221439 |
| SMAD2 | 32 | HSPA9 | 12724.54 | SP1 | 0.04601 | NRAS | 0.13518 | LAMC1 | 8.6 | SMAD2 | 4.159399 | NRAS | 6054038 |
| FOS | 31 | TJP1 | 12550.71 | SOD2 | 0.045982 | CCNE2 | 0.135058 | CHEK1 | 8.588235 | COL1A1 | 4.14313 | CCNE2 | 6049281 |
| WDR5 | 31 | TGFBR1 | 11898.11 | PTK2 | 0.045967 | PPARG | 0.13326 | LAMA4 | 8.5 | FOS | 4.14313 | PPARG | 5887511 |
| COL1A1 | 31 | UBE2I | 11854.41 | CHEK1 | 0.045967 | RBBP4 | 0.12862 | HIST2H3PS2 | 8.408163 | WDR5 | 4.14313 | RBBP4 | 5483687 |
| RBBP4 | 30 | RERE | 11608.05 | RBBP4 | 0.045964 | LMNB1 | 0.124717 | MYC | 8.314607 | PTK2 | 4.125951 | LMNB1 | 5156968 |
| PTK2 | 30 | SOD2 | 11343.42 | UBE2I | 0.045952 | SMAD2 | 0.121389 | COL18A1 | 7.888889 | RBBP4 | 4.125951 | SMAD2 | 4895097 |
| HSPA9 | 30 | FOS | 10771.87 | AURKA | 0.045945 | SP1 | 0.117861 | LEPREL1 | 7.818182 | HSPA9 | 4.125951 | SP1 | 4601198 |
| UBE2I | 29 | TMED10 | 10569.32 | CCNE2 | 0.045884 | WDR5 | 0.112875 | COL8A1 | 7.5 | UBE2I | 4.107765 | WDR5 | 4226990 |
| AGO2 | 27 | RBBP4 | 10403.83 | TGFBR1 | 0.045859 | PLK4 | 0.111689 | FBN1 | 7.428571 | AGO2 | 4.068093 | PLK4 | 4140534 |
| CCNE2 | 26 | DMD | 10381.87 | SIRT7 | 0.045847 | RAD21 | 0.110684 | COL11A2 | 7.375 | CCNE2 | 4.046388 | RAD21 | 4063987 |
| NUP98 | 26 | WDR5 | 10283.43 | CCND2 | 0.045826 | MCL1 | 0.110118 | CCND2 | 7.363637 | NUP98 | 4.046388 | MCL1 | 4018060 |
| TGFBR1 | 25 | CHEK1 | 10165.99 | AGO2 | 0.045816 | NUP98 | 0.109068 | CASP3 | 6.979592 | TGFBR1 | 4.023259 | NUP98 | 3947886 |
| SP1 | 23 | TFRC | 9382.314 | TJP1 | 0.045813 | UBE2I | 0.10781 | CCNE2 | 6.923077 | COL6A1 | 3.972192 | UBE2I | 3852734 |
| SOD2 | 23 | COL1A1 | 9344.245 | RAD21 | 0.04581 | TOP1 | 0.106882 | NUP133 | 6.833334 | COL6A2 | 3.972192 | TOP1 | 3787653 |
| TJP1 | 23 | CCNB1 | 9142.68 | WDR5 | 0.045786 | AGO2 | 0.104175 | RAD51AP1 | 6.714286 | FH | 3.972192 | AGO2 | 3599771 |
| NCBP2 | 23 | TXNRD1 | 8277.166 | HSPA9 | 0.04578 | ATAD2 | 0.098454 | AURKA | 6.606061 | ACLY | 3.972192 | PTK2 | 3242236 |
| FH | 23 | PPP6C | 7909.077 | AKT3 | 0.04578 | PTK2 | 0.097769 | PLK4 | 6.454546 | SP1 | 3.972192 | ATAD2 | 3214702 |
| ACLY | 23 | SP1 | 7895.044 | FGFR2 | 0.045773 | DTL | 0.095207 | NDC1 | 6.428571 | SOD2 | 3.972192 | DTL | 3011290 |
| COL6A1 | 23 | FGFR2 | 7834.322 | NUP98 | 0.045773 | RNF2 | 0.091295 | ATM | 6.415094 | NCBP2 | 3.972192 | RNF2 | 2763911 |
| COL6A2 | 23 | CCT3 | 7686.299 | PGK1 | 0.045767 | CDC27 | 0.089433 | MCL1 | 6.375 | TJP1 | 3.972192 | CDC27 | 2653227 |
| CCND2 | 22 | CCNE2 | 7528.156 | TOP1 | 0.045758 | MYBL2 | 0.0881 | ATAD2 | 6.375 | COL6A3 | 3.943907 | MYBL2 | 2572502 |
| PLK4 | 22 | ITCH | 7308.306 | PRKCD | 0.045749 | KAT6A | 0.087586 | DTL | 6.363637 | CCND2 | 3.943907 | KAT6A | 2543617 |
| DTL | 22 | CSF2 | 7290.804 | LMNB1 | 0.045749 | AKT3 | 0.081991 | LEPREL2 | 6.285714 | PLK4 | 3.943907 | AKT3 | 2227275 |
| PGK1 | 22 | PINK1 | 7266.99 | MCL1 | 0.045734 | PAX6 | 0.080551 | LMNB1 | 6.210527 | DTL | 3.943907 | COL1A1 | 2210530 |
| COL6A3 | 22 | AKT3 | 7194.102 | RUNX1 | 0.045715 | SIRT7 | 0.080551 | ITGA7 | 6.166667 | PGK1 | 3.943907 | SIRT7 | 2151882 |
| CCT3 | 22 | AGO2 | 6931.627 | EPAS1 | 0.045691 | AGO1 | 0.080408 | NUP50 | 6.117647 | CCT3 | 3.943907 | PAX6 | 2149573 |
| TOP1 | 21 | UBQLN1 | 6730.845 | PAX6 | 0.045688 | RCOR1 | 0.07753 | NUP98 | 6.076923 | COL4A2 | 3.91349 | AGO1 | 2144101 |
| RNF2 | 21 | ACLY | 6575.635 | RNF2 | 0.045679 | PTCH1 | 0.077235 | HK2 | 6 | TOP1 | 3.91349 | RCOR1 | 1993021 |
| AGO1 | 21 | GOLPH3 | 6248.143 | AGO1 | 0.045664 | COL1A1 | 0.076574 | FH | 6 | RNF2 | 3.91349 | PTCH1 | 1976036 |
| COL4A2 | 21 | PLK4 | 6190.994 | PTCH1 | 0.045651 | SOD2 | 0.076143 | SERPINH1 | 5.888889 | AGO1 | 3.91349 | SOD2 | 1923597 |
| ABCE1 | 21 | SERPINH1 | 6138.03 | PAK2 | 0.045648 | TGFBR1 | 0.075727 | CS | 5.75 | ABCE1 | 3.91349 | TGFBR1 | 1910805 |
| RAD21 | 20 | MAPRE3 | 6080.12 | YES1 | 0.045648 | RUNX1 | 0.075226 | GPI | 5.733333 | COL5A2 | 3.880701 | RUNX1 | 1875071 |
| AKT3 | 20 | EPAS1 | 5907.968 | KAT6A | 0.045639 | RAD51AP1 | 0.073014 | PGK1 | 5.727273 | LAMC1 | 3.880701 | RAD51AP1 | 1771838 |
| EPAS1 | 20 | PAK2 | 5893.081 | COL1A1 | 0.045636 | CCNH | 0.072743 | MYBL2 | 5.571429 | RAD21 | 3.880701 | CCNH | 1754899 |
| ITCH | 20 | SRPR | 5857.721 | PLK4 | 0.045609 | FGFR2 | 0.070863 | ACLY | 5.565218 | DMD | 3.880701 | FGFR2 | 1663734 |
| LAMC1 | 20 | ABCE1 | 5784.403 | MAX | 0.045606 | PGK1 | 0.070127 | FOS | 5.548387 | TXNRD1 | 3.880701 | PGK1 | 1635160 |
| DMD | 20 | PPL | 5654.79 | CDC27 | 0.045603 | EPAS1 | 0.069382 | PTK2 | 5.533333 | AKT3 | 3.880701 | EPAS1 | 1595363 |
| COL5A2 | 20 | BCL2 | 5523.898 | ITCH | 0.045597 | TJP1 | 0.068124 | SEH1L | 5.466667 | EPAS1 | 3.880701 | TJP1 | 1540930 |
| TXNRD1 | 20 | RAD21 | 5511.951 | TFRC | 0.045591 | CSF2 | 0.068085 | SMAD2 | 5.4375 | ITCH | 3.880701 | CSF2 | 1535941 |
| TMED10 | 20 | PGK1 | 5500.145 | MYBL2 | 0.045579 | PRKCD | 0.066951 | AGO2 | 5.333334 | TMED10 | 3.880701 | PRKCD | 1485477 |
| LMNB1 | 19 | SLC25A33 | 5496.237 | PSME3 | 0.045572 | MAX | 0.065213 | NIP7 | 5.333334 | LMNB1 | 3.845279 | TGFB3 | 1411799 |
| FGFR2 | 19 | NEDD4L | 5484.699 | TGFB3 | 0.045566 | TGFB3 | 0.064884 | TRMT11 | 5.230769 | NEDD4L | 3.845279 | MAX | 1409118 |
| NEDD4L | 19 | CCNH | 5324.429 | ACLY | 0.04556 | NCBP2 | 0.064605 | SP1 | 5.217392 | FGFR2 | 3.845279 | NCBP2 | 1385477 |
| RUNX1 | 18 | F11R | 5165.416 | NCBP2 | 0.045551 | HSPA9 | 0.064468 | RCOR1 | 5.142857 | COL11A1 | 3.806871 | HSPA9 | 1381436 |
| CCNH | 18 | FH | 5138.533 | ATAD2 | 0.045548 | HK2 | 0.0622 | NRAS | 5.111111 | COL18A1 | 3.806871 | HK2 | 1286048 |
| DDX18 | 18 | AGO1 | 5105.469 | NEDD4L | 0.045542 | HIST1H3J | 0.061885 | RAD21 | 5.1 | SERPINH1 | 3.806871 | HIST1H3J | 1270916 |
| COL18A1 | 18 | DDX18 | 5075.202 | GOLPH3 | 0.045539 | NFYA | 0.06188 | PNO1 | 5.058824 | DDX18 | 3.806871 | NFYA | 1268920 |
| COL11A1 | 18 | SIRT7 | 5028.431 | HK2 | 0.045539 | NDC1 | 0.061369 | RBBP4 | 5 | CCNH | 3.806871 | NDC1 | 1251614 |
| SERPINH1 | 18 | NCBP2 | 4760.606 | CCNH | 0.045527 | MDM4 | 0.060536 | TGFB3 | 5 | RUNX1 | 3.806871 | MDM4 | 1214375 |
| CDC27 | 17 | TCEA1 | 4649.523 | CSF2 | 0.045521 | PAK2 | 0.06015 | NOC3L | 5 | NUP50 | 3.765087 | PAK2 | 1205645 |
| KAT6A | 17 | CCND2 | 4621.744 | RCOR1 | 0.045521 | POLE3 | 0.060104 | RBM19 | 5 | HK2 | 3.765087 | POLE3 | 1200180 |
| Genes | DC | Genes | BC | Genes | CC | Genes | EC | Genes | LAC | Genes | IC | Genes | SC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIST2H3PS2 | 24 | IGF1 | 12,657.02 | IGF1 | 0.020131 | HIST2H3PS2 | 0.336233 | HIST2H2AB | 8.666667 | HIST2H3PS2 | 2.791786 | HIST2H3PS2 | 15879.49 |
| ATM | 23 | ATM | 10,286.39 | ATM | 0.020117 | HIST1H4F | 0.293143 | HIST2H3D | 8.533334 | ATM | 2.778953 | HIST1H4F | 12090.06 |
| IGF1 | 21 | ICAM1 | 8398.269 | NFKBIA | 0.020094 | HIST3H3 | 0.282137 | HIST1H1E | 8.4 | IGF1 | 2.750203 | HIST3H3 | 11213.9 |
| HIST1H4F | 18 | NFKBIA | 6650.969 | HIST2H3PS2 | 0.020077 | HIST2H3D | 0.273608 | HIST1H3J | 8.307693 | HIST1H4F | 2.697331 | HIST2H3D | 10568.27 |
| CCL2 | 18 | HIST2H3PS2 | 5622.567 | ICAM1 | 0.020076 | HIST1H2AJ | 0.269709 | HIST3H3 | 8.125 | CCL2 | 2.697331 | HIST1H2AJ | 10266.41 |
| NFKBIA | 17 | CCL2 | 5539.708 | IKBKB | 0.020066 | HIST2H2AB | 0.2494 | HIST1H2AI | 8 | NFKBIA | 2.676373 | HIST2H2AB | 8764.285 |
| HIST3H3 | 16 | TUBB4A | 5141.43 | SOD2 | 0.020055 | HIST1H3J | 0.247382 | HIST1H2AJ | 7.625 | HIST3H3 | 2.65333 | HIST1H3J | 8645.107 |
| HIST1H2AJ | 16 | IKBKB | 5052.972 | CCL2 | 0.020028 | ATM | 0.233653 | HIST1H4F | 7.555555 | HIST1H2AJ | 2.65333 | ATM | 7783.067 |
| HIST2H3D | 15 | RCVRN | 4885.731 | PAX6 | 0.020013 | HIST1H2AI | 0.220319 | HIST1H1A | 7.4 | HIST2H3D | 2.627873 | HIST1H2AI | 6837.918 |
| POU5F1 | 14 | SOD2 | 4803.247 | POU5F1 | 0.020007 | HIST1H1E | 0.217499 | HIST2H3PS2 | 6.75 | POU5F1 | 2.599608 | HIST1H1E | 6684.745 |
| ICAM1 | 14 | AKT3 | 4681.91 | TUBB4A | 0.020004 | HIST1H1A | 0.209097 | HIST1H2AD | 5.333334 | ICAM1 | 2.599608 | HIST1H1A | 6168.202 |
| HIST1H3J | 13 | CCND2 | 4486.447 | AKT3 | 0.020001 | HIST1H2AD | 0.19763 | PHF21A | 5.333334 | HIST1H3J | 2.568042 | HIST1H2AD | 5525.728 |
| IKBKB | 13 | CD59 | 4309.046 | CSF2 | 0.019999 | PHF21A | 0.176246 | TNFAIP3 | 4.25 | IKBKB | 2.568042 | PHF21A | 4369.904 |
| HIST2H2AB | 12 | POU5F1 | 3915.681 | CCND2 | 0.019997 | WHSC1 | 0.160865 | WHSC1 | 4 | HIST2H2AB | 2.532561 | WHSC1 | 3640.597 |
| HIST1H2AD | 12 | WWC1 | 3888.129 | FGFR1 | 0.019978 | KAT6A | 0.150582 | IKBKB | 3.846154 | HIST1H2AD | 2.532561 | KAT6A | 3188.971 |
| PAX6 | 11 | CDK5 | 3827.028 | FGFR2 | 0.019959 | CBX6 | 0.137474 | ATM | 3.826087 | PAX6 | 2.492376 | CBX6 | 2673.861 |
| SOD2 | 11 | CSF2 | 3700.629 | WWC1 | 0.019931 | IGF1 | 0.112766 | KAT6A | 3.75 | SOD2 | 2.492376 | IGF1 | 2112.217 |
| CSF2 | 11 | KIF5C | 3555.034 | HIST1H4F | 0.01993 | POU5F1 | 0.102878 | CBX6 | 3.5 | CSF2 | 2.492376 | POU5F1 | 1541.507 |
| RCVRN | 11 | PAX6 | 3298.453 | RCVRN | 0.019922 | PAX6 | 0.100751 | CCL2 | 3.333333 | RCVRN | 2.492376 | PAX6 | 1454.034 |
| HIST1H2AI | 10 | WWOX | 2962.674 | TNFAIP3 | 0.019909 | AIRE | 0.096106 | TUBB8 | 3.2 | HIST1H2AI | 2.446504 | NFKBIA | 1431.716 |
| HIST1H1E | 10 | ZNF395 | 2778.826 | WWOX | 0.019906 | CCND2 | 0.089509 | NFKBIA | 3.058824 | HIST1H1E | 2.446504 | IKBKB | 1324.191 |
| HIST1H1A | 10 | TGM1 | 2707.593 | TUBA1A | 0.019905 | NFKBIA | 0.087309 | AIRE | 3 | HIST1H1A | 2.446504 | AIRE | 1304.235 |
| CCND2 | 10 | SYN3 | 2563.518 | CDK5 | 0.01989 | IKBKB | 0.084984 | SOD2 | 2.909091 | CCND2 | 2.446504 | CCND2 | 1161.27 |
| TUBB4A | 10 | GNG12 | 2428.181 | BCL2 | 0.019888 | CTBP1 | 0.074958 | ICAM1 | 2.857143 | TUBB4A | 2.446504 | CTBP1 | 804.1904 |
| PHF21A | 9 | FZR1 | 2387.046 | SIRPA | 0.01988 | TET3 | 0.06408 | TUBA1A | 2.857143 | PHF21A | 2.393643 | CCL2 | 775.7595 |
| WHSC1 | 9 | CAMK1G | 2352.95 | CTBP1 | 0.019877 | AKT3 | 0.059664 | TUBA4A | 2.666667 | WHSC1 | 2.393643 | SOD2 | 677.2321 |
| AKT3 | 9 | CAV3 | 2350.628 | KAT6A | 0.019877 | FGFR2 | 0.058941 | IGF1 | 2.571429 | AKT3 | 2.393643 | ICAM1 | 610.6983 |
| KAT6A | 8 | MYO5B | 2350.244 | ZC3H12A | 0.019873 | SOD2 | 0.056252 | TUBB4A | 2.4 | KAT6A | 2.332049 | TET3 | 582.8483 |
| CBX6 | 8 | XCL1 | 2339.667 | HIST3H3 | 0.019869 | EYA2 | 0.052028 | VAMP1 | 2.4 | CBX6 | 2.332049 | AKT3 | 560.5861 |
| FGFR2 | 8 | EXOC4 | 2334.192 | THPO | 0.019866 | WWC1 | 0.050389 | CSF2 | 2.363636 | FGFR2 | 2.332049 | FGFR2 | 529.6174 |
| FGFR1 | 8 | COPZ2 | 1880 | EYA2 | 0.019866 | TAL1 | 0.048814 | CCND2 | 2 | FGFR1 | 2.332049 | CSF2 | 405.3018 |
| TNFAIP3 | 8 | KCND3 | 1875.697 | WHSC1 | 0.019865 | CCL2 | 0.048485 | ZC3H12A | 2 | TNFAIP3 | 2.332049 | EYA2 | 384.0464 |
| CDK5 | 8 | CCK | 1872 | PHF21A | 0.019863 | FGFR1 | 0.047015 | SIRPA | 2 | CDK5 | 2.332049 | WWC1 | 368.5083 |
| KIF5C | 8 | LIN7C | 1762.686 | IGFBP3 | 0.019861 | TNRC6C | 0.046971 | NFKBIE | 2 | KIF5C | 2.332049 | FGFR1 | 354.2804 |
| VAMP8 | 8 | FGFR1 | 1600.013 | HIST2H2AB | 0.019858 | ICAM1 | 0.044627 | TNIP2 | 2 | VAMP8 | 2.332049 | TNFAIP3 | 343.6676 |
| CTBP1 | 7 | RAB11FIP5 | 1546.701 | HIST1H2AI | 0.019855 | ERCC6 | 0.044454 | SYT17 | 2 | CTBP1 | 2.259382 | TAL1 | 341.3784 |
| WWC1 | 7 | F11R | 1532.283 | HOXB7 | 0.019852 | TUBA1A | 0.03925 | LCE1D | 2 | WWC1 | 2.259382 | TNRC6C | 314.0037 |
| TUBA1A | 7 | F7 | 1457.162 | ADRBK1 | 0.019851 | CSF2 | 0.037267 | LCE1F | 2 | TUBA1A | 2.259382 | ERCC6 | 280.5435 |
| WWOX | 7 | UBE2K | 1414 | HIST1H2AJ | 0.019841 | TNFAIP3 | 0.032499 | POU5F1 | 1.857143 | WWOX | 2.259382 | TUBA1A | 252.0046 |
| CD59 | 7 | TCEAL6 | 1411 | CLU | 0.01984 | CDK5 | 0.030225 | FGFR2 | 1.75 | GNG12 | 2.259382 | ZC3H12A | 188.933 |
| GNG12 | 7 | WDFY2 | 1410 | ERCC6 | 0.019832 | WWOX | 0.02852 | FGFR1 | 1.75 | CD59 | 2.259382 | TUBB4A | 185.0031 |
| CAV3 | 7 | CREB3L3 | 1407.115 | HIST1H1A | 0.019829 | TUBB4A | 0.02729 | VAMP8 | 1.75 | CAV3 | 2.259382 | CDK5 | 147.8962 |
| ZC3H12A | 6 | BID | 1394.544 | TET3 | 0.019826 | ZC3H12A | 0.027068 | CTBP1 | 1.714286 | ZC3H12A | 2.172348 | WWOX | 136.4548 |
| BID | 6 | RGMA | 1341.624 | CD59 | 0.019821 | FOXP1 | 0.02444 | SYN3 | 1.666667 | BID | 2.172348 | SIRPA | 98.61929 |
| BCL2 | 6 | CACNA1E | 1306.522 | CADM1 | 0.019819 | TACC3 | 0.02356 | AKT3 | 1.555556 | BCL2 | 2.172348 | FOXP1 | 96.34972 |
| TUBA4A | 6 | SOD3 | 1285.36 | NRG1 | 0.019818 | BID | 0.0221 | TET3 | 1.5 | TUBA4A | 2.172348 | RCVRN | 90.32636 |
| SYN3 | 6 | VAMP8 | 1285.289 | HIST2H3D | 0.019818 | FGFRL1 | 0.01855 | IGFBP3 | 1.5 | EXOC4 | 2.172348 | TACC3 | 86.21384 |
| EXOC4 | 6 | TUBA1A | 1254.487 | CSH1 | 0.019812 | SIRPA | 0.01763 | KCND3 | 1.5 | SYN3 | 2.172348 | NFKBIE | 83.78503 |
| TGM1 | 6 | CLU | 1220.285 | HIST1H3J | 0.019812 | RCVRN | 0.017589 | PKP2 | 1.5 | TGM1 | 2.172348 | TNIP2 | 83.78503 |
| TAL1 | 5 | BCL2 | 1190.642 | BID | 0.019811 | BCL2 | 0.017578 | TUBA3C | 1.5 | FOXP1 | 2.066228 | BID | 81.1344 |
| FOXP1 | 5 | FNDC5 | 1174.097 | FNDC5 | 0.01981 | ZIC1 | 0.017441 | PPL | 1.5 | F7 | 2.066228 | IGFBP3 | 69.96769 |
| TUBB8 | 5 | SPTB | 1171.549 | SGK3 | 0.01981 | NFKBIE | 0.017346 | PAX6 | 1.454546 | VAMP1 | 2.066228 | BCL2 | 66.91744 |
| FZR1 | 5 | ZIC1 | 1159.743 | HAND2 | 0.019808 | TNIP2 | 0.017346 | RCVRN | 1.454546 | TAL1 | 2.066228 | FGFRL1 | 51.49969 |
| CLU | 5 | SYNGAP1 | 1106.794 | IGFBP5 | 0.019805 | HOXB7 | 0.016011 | BCL2 | 1.333333 | FZR1 | 2.066228 | THPO | 51.38118 |
| VAMP1 | 5 | PLAT | 1016.892 | PLAT | 0.019797 | IGFBP3 | 0.015366 | PTX3 | 1.333333 | CLU | 2.066228 | ZIC1 | 50.78407 |
| SYT11 | 5 | CTBP1 | 992.8071 | HIST1H2AD | 0.019796 | THPO | 0.013824 | RGMB | 1.333333 | TUBB8 | 2.066228 | CADM1 | 50.12931 |
| SCN1B | 5 | CABP4 | 974.4585 | LIN7C | 0.019793 | NRG1 | 0.013609 | TCEAL2 | 1.333333 | SYT11 | 2.066228 | HOXB7 | 46.41033 |
| F7 | 5 | SCN1B | 959.0853 | NFKBIE | 0.019793 | FZR1 | 0.012277 | FOXP1 | 1.2 | CACNA1E | 2.066228 | PTX3 | 45.3798 |
| CACNA1E | 5 | NLGN2 | 958.3921 | TNIP2 | 0.019793 | HAND2 | 0.011935 | SYT11 | 1.2 | SCN1B | 2.066228 | TUBA4A | 40.56611 |
| AIRE | 4 | CBX6 | 949.5727 | TUBA4A | 0.01979 | PCGF3 | 0.011594 | SCN1B | 1.2 | AIRE | 1.933961 | PLAT | 39.07808 |
| AUC | Sensitivity (%) | Specificity (%) | ||
|---|---|---|---|---|
| LUAD | ARHGAP5-AS1 | 0.595 | 30.65 | 100.00 |
| LINC00174 | 0.733 | 53.64 | 83.05 | |
| MEG3 | 0.620 | 40.75 | 84.75 | |
| PVT1 | 0.974 | 88.97 | 94.42 | |
| MEG3 + ARHGAP5-AS1 + LINC00174 + PVT1 | 0.976 | 93.83 | 91.53 | |
| MT1JP | 0.855 | 77.57 | 84.75 | |
| MT1L | 0.743 | 57.76 | 96.61 | |
| RPL23AP64 | 0.512 | 28.79 | 79.66 | |
| TMEM198B | 0.579 | 36.26 | 88.14 | |
| ZNF826P | 0.519 | 40.56 | 91.53 | |
| MT1L + MT1JP + RPL23AP64 + TMEM198B + ZNF826P | 0.834 | 89.35 | 66.10 | |
| All lncRNAs+all pseudogenes | 0.977 | 94.39 | 89.93 | |
| LUSC | ARHGAP5-AS1 | 0.768 | 54.18 | 93.88 |
| LINC00174 | 0.768 | 58.37 | 85.71 | |
| MEG3 | 0.525 | 46.61 | 71.43 | |
| PVT1 | 0.975 | 95.22 | 91.84 | |
| MEG3+ARHGAP5-AS1+LINC00174+PVT1 | 0.990 | 93.82 | 95.92 | |
| MT1JP | 0.939 | 84.06 | 93.88 | |
| MT1L | 0.795 | 65.94 | 95.92 | |
| RPL23AP64 | 0.721 | 66.53 | 69.39 | |
| TMEM198B | 0.541 | 25.50 | 95.92 | |
| ZNF826P | 0.765 | 66.33 | 91.84 | |
| MT1L + MT1JP + RPL23AP64 + TMEM198B + ZNF826P | 0.925 | 86.06 | 87.76 | |
| All lncRNAs+all pseudogenes | 0.994 | 95.42 | 97.96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, G.; Xie, D.; Hu, J.; Wu, T.; Cao, K.; Luo, X. Identification of Candidate lncRNA and Pseudogene Biomarkers Associated with Carbon-Nanotube-Induced Malignant Transformation of Lung Cells and Prediction of Potential Preventive Drugs. Int. J. Environ. Res. Public Health 2022, 19, 2936. https://doi.org/10.3390/ijerph19052936
Chang G, Xie D, Hu J, Wu T, Cao K, Luo X. Identification of Candidate lncRNA and Pseudogene Biomarkers Associated with Carbon-Nanotube-Induced Malignant Transformation of Lung Cells and Prediction of Potential Preventive Drugs. International Journal of Environmental Research and Public Health. 2022; 19(5):2936. https://doi.org/10.3390/ijerph19052936
Chicago/Turabian StyleChang, Guangtao, Dongli Xie, Jianchen Hu, Tong Wu, Kangli Cao, and Xiaogang Luo. 2022. "Identification of Candidate lncRNA and Pseudogene Biomarkers Associated with Carbon-Nanotube-Induced Malignant Transformation of Lung Cells and Prediction of Potential Preventive Drugs" International Journal of Environmental Research and Public Health 19, no. 5: 2936. https://doi.org/10.3390/ijerph19052936






