A Novel Ex Vivo Approach Based on Proteomics and Biomarkers to Evaluate the Effects of Chrysene, MEHP, and PBDE-47 on Loggerhead Sea Turtles (Caretta caretta)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Treatment Reagents
2.3. Tissue Exposure
2.4. Sample Preparation for 2D-PAGE
2.5. 2-DE and Image Analysis
2.6. Biomarker Analysis in Blood
2.6.1. TAS
2.6.2. Respiratory Burst
2.6.3. Lysozyme Activity
2.6.4. Serum Hemolytic Complement Assay
2.6.5. ENA Assay
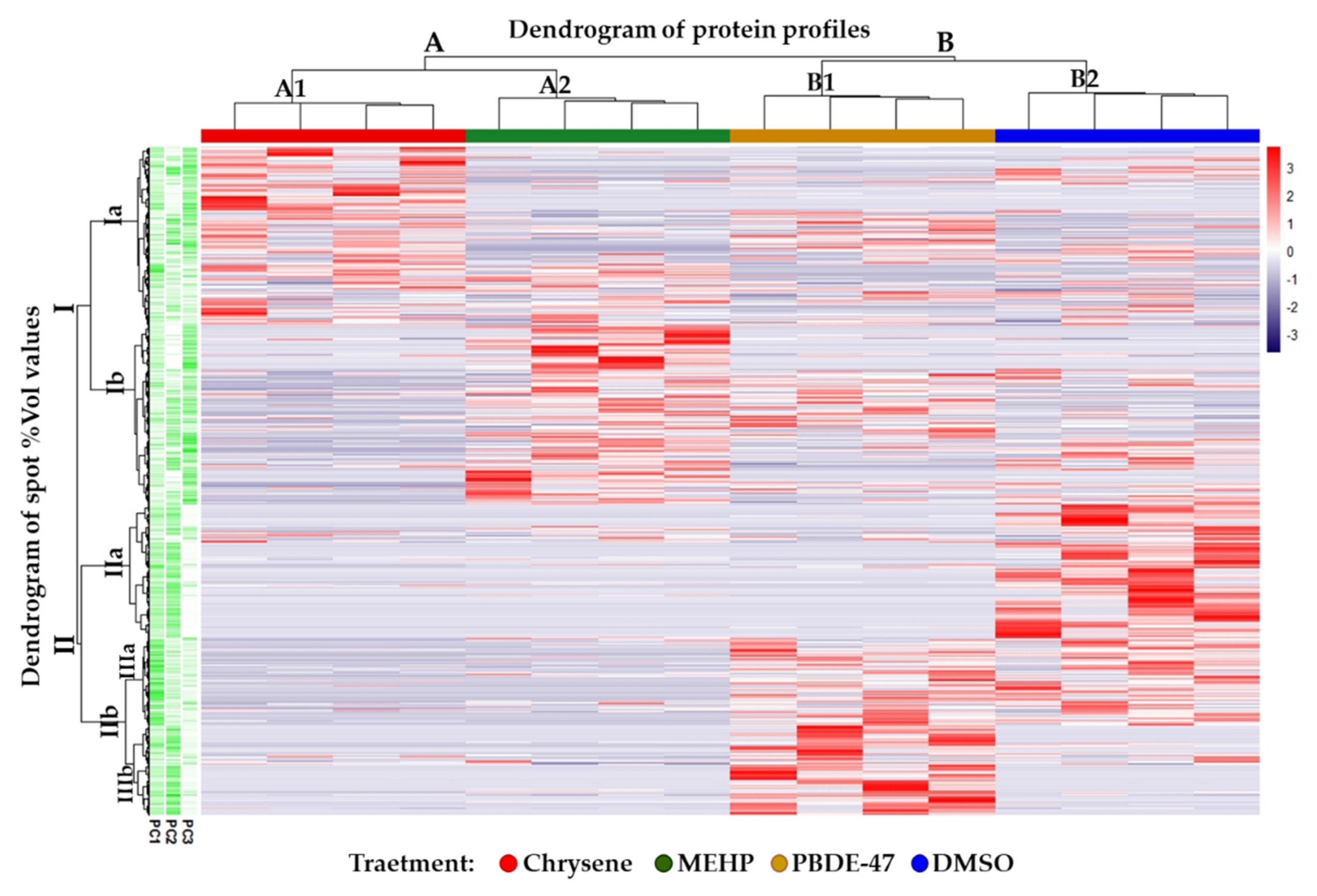
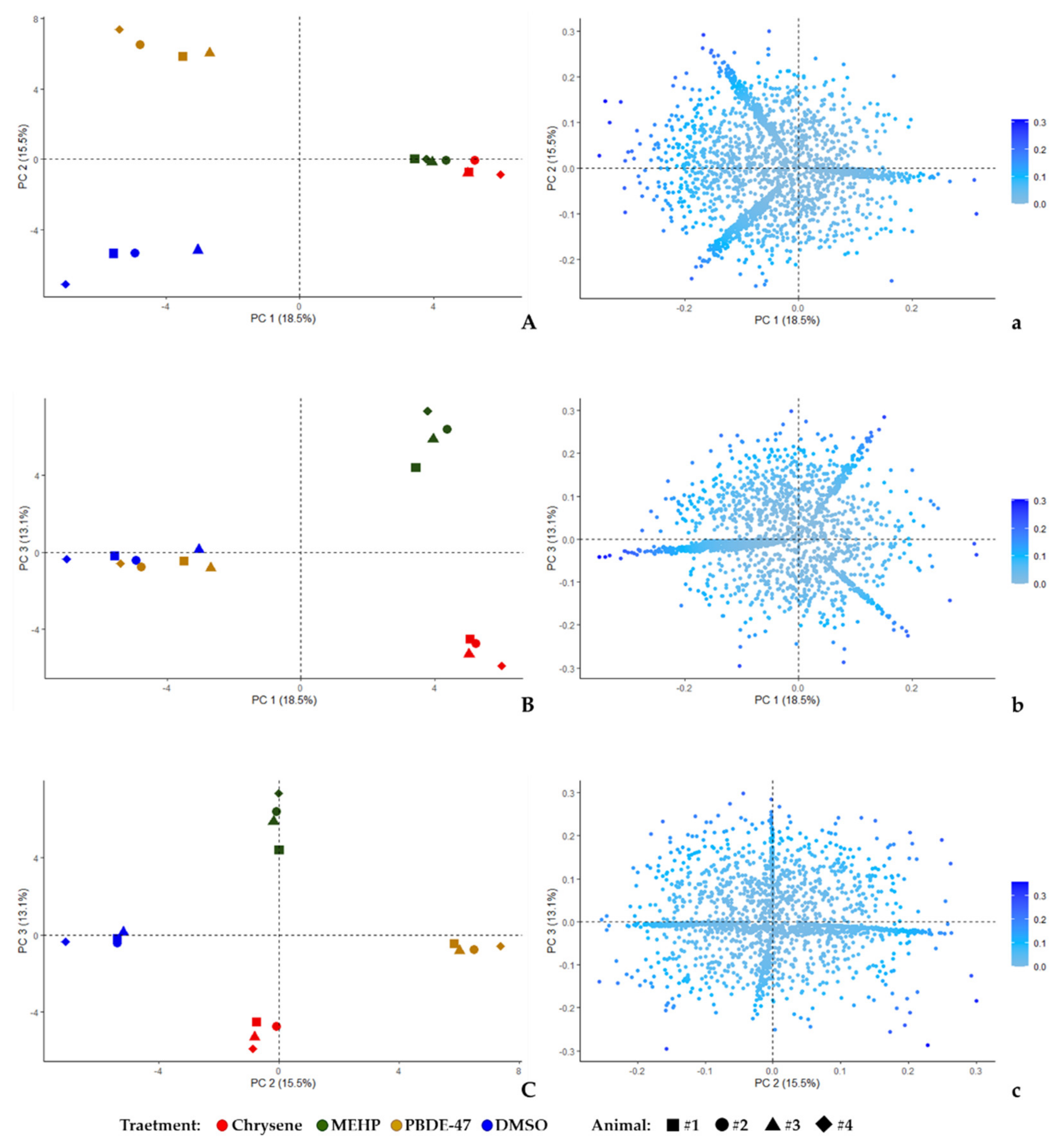
2.7. Statistical Analysis
2.8. Protein Identification by MALDI TOF/TOF
3. Results and Discussion
3.1. Proteomic Characterization of Chrysene, MEHP, and PBDE-47 Effects on Ex Vivo Skin-Samples
3.1.1. Glutathione S Transferase P
3.1.2. Mimecan
3.1.3. Peptidyl-Prolyl Cis-Trans Isomerase
3.1.4. Protein S100-A6
3.2. Biomarkers in Whole Blood Treated with Chrysene, MEHP, and PBDE-47
3.2.1. Total Antioxidant Status
3.2.2. Respiratory Burst
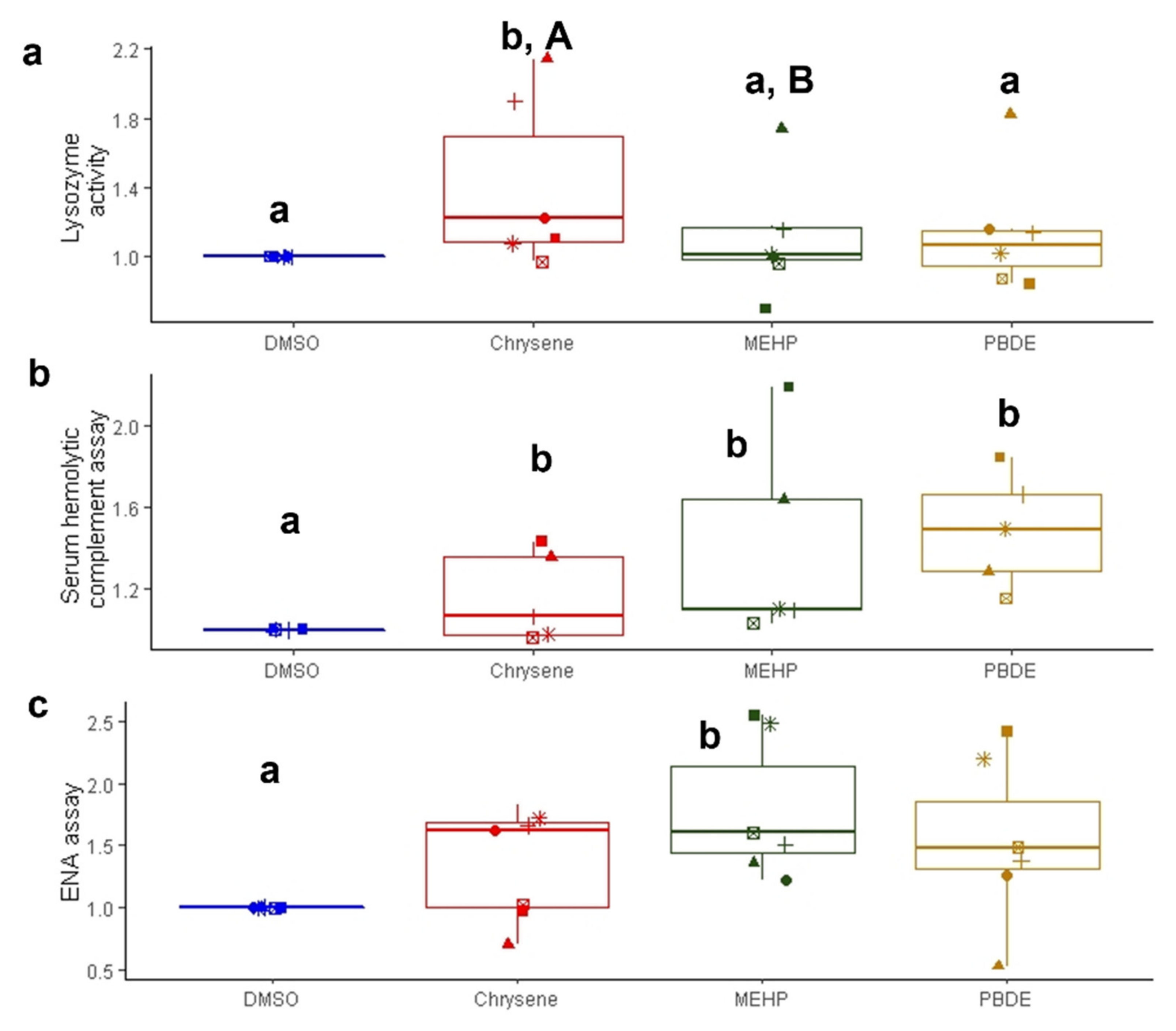
3.2.3. Lysozyme Activity
3.2.4. Serum Hemolytic Complement Assay
3.2.5. ENA Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Camedda, A.; Matiddi, M.; Vianello, A.; Coppa, S.; Bianchi, J.; Silvestri, C.; Palazzo, L.; Massaro, G.; Atzori, F.; Ruiu, A.; et al. Polymer Composition Assessment Suggests Prevalence of Single-Use Plastics among Items Ingested by Loggerhead Sea Turtles in the Western Mediterranean Sub-Region. Environ. Pollut. 2022, 292, 118274. [Google Scholar] [CrossRef] [PubMed]
- Sala, B.; Balasch, A.; Eljarrat, E.; Cardona, L. First Study on the Presence of Plastic Additives in Loggerhead Sea Turtles (Caretta Caretta) from the Mediterranean Sea. Environ. Pollut. 2021, 283, 117108. [Google Scholar] [CrossRef] [PubMed]
- Campani, T.; Baini, M.; Giannetti, M.; Cancelli, F.; Mancusi, C.; Serena, F.; Marsili, L.; Casini, S.; Fossi, M.C. Presence of Plastic Debris in Loggerhead Turtle Stranded along the Tuscany Coasts of the Pelagos Sanctuary for Mediterranean Marine Mammals (Italy). Mar. Pollut. Bull. 2013, 74, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Squadrone, S.; Pederiva, S.; Bezzo, T.; Sartor, R.M.; Battuello, M.; Nurra, N.; Griglione, A.; Brizio, P.; Abete, M.C. Microplastics as Vectors of Metals Contamination in Mediterranean Sea. Environ. Sci. Pollut. Res. 2021. [Google Scholar] [CrossRef]
- Karapanagioti, H.K.; Endo, S.; Ogata, Y.; Takada, H. Diffuse Pollution by Persistent Organic Pollutants as Measured in Plastic Pellets Sampled from Various Beaches in Greece. Mar. Pollut. Bull. 2011, 62, 312–317. [Google Scholar] [CrossRef]
- Dasgupta, S.; Peng, X.; Xu, H.; Ta, K.; Chen, S.; Li, J.; Du, M. Deep Seafloor Plastics as the Source and Sink of Organic Pollutants in the Northern South China Sea. Sci. Total Environ. 2021, 765, 144228. [Google Scholar] [CrossRef]
- Camacho, M.; Herrera, A.; Gómez, M.; Acosta-Dacal, A.; Martínez, I.; Henríquez-Hernández, L.A.; Luzardo, O.P. Organic Pollutants in Marine Plastic Debris from Canary Islands Beaches. Sci. Total Environ. 2019, 662, 22–31. [Google Scholar] [CrossRef]
- Lambiase, S.; Serpe, F.P.; Pilia, M.; Fiorito, F.; Iaccarino, D.; Gallo, P.; Esposito, M. Polychlorinated Organic Pollutants (PCDD/Fs and DL-PCBs) in Loggerhead (Caretta Caretta) and Green (Chelonia Mydas) Turtles from Central-Southern Tyrrhenian Sea. Chemosphere 2021, 263, 128226. [Google Scholar] [CrossRef]
- Renaguli, A.; Fernando, S.; Holsen, T.M.; Hopke, P.K.; Adams, D.H.; Balazs, G.H.; Jones, T.T.; Work, T.M.; Lynch, J.M.; Crimmins, B.S. Characterization of Halogenated Organic Compounds in Pelagic Sharks and Sea Turtles Using a Nontargeted Approach. Environ. Sci. Technol. 2021, 55, 16390–16401. [Google Scholar] [CrossRef]
- Guerranti, C.; Baini, M.; Casini, S.; Focardi, S.E.; Giannetti, M.; Mancusi, C.; Marsili, L.; Perra, G.; Fossi, M.C. Pilot Study on Levels of Chemical Contaminants and Porphyrins in Caretta Caretta from the Mediterranean Sea. Mar. Environ. Res. 2014, 100, 33–37. [Google Scholar] [CrossRef]
- Savoca, D.; Arculeo, M.; Barreca, S.; Buscemi, S.; Caracappa, S.; Gentile, A.; Persichetti, M.F.; Pace, A. Chasing Phthalates in Tissues of Marine Turtles from the Mediterranean Sea. Mar. Pollut. Bull. 2018, 127, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Casini, S.; Caliani, I.; Giannetti, M.; Marsili, L.; Maltese, S.; Coppola, D.; Bianchi, N.; Campani, T.; Ancora, S.; Caruso, C.; et al. First Ecotoxicological Assessment of Caretta Caretta (Linnaeus, 1758) in the Mediterranean Sea Using an Integrated Nondestructive Protocol. Sci. Total Environ. 2018, 631, 1221–1233. [Google Scholar] [CrossRef] [PubMed]
- Caliani, I.; Campani, T.; Giannetti, M.; Marsili, L.; Casini, S.; Fossi, M.C. First Application of Comet Assay in Blood Cells of Mediterranean Loggerhead Sea Turtle (Caretta Caretta). Mar. Environ. Res. 2014, 96, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Caliani, I.; Poggioni, L.; D’Agostino, A.; Fossi, M.C.; Casini, S. An Immune Response-Based Approach to Evaluate Physiological Stress in Rehabilitating Loggerhead Sea Turtle. Vet. Immunol. Immunopathol. 2019, 207, 18–24. [Google Scholar] [CrossRef]
- Lillicrap, A.; Belanger, S.; Burden, N.; Pasquier, D.D.; Embry, M.R.; Halder, M.; Lampi, M.A.; Lee, L.; Norberg-King, T.; Rattner, B.A.; et al. Alternative Approaches to Vertebrate Ecotoxicity Tests in the 21st Century: A Review of Developments over the Last 2 Decades and Current Status. Environ. Toxicol. Chem. 2016, 35, 2637–2646. [Google Scholar] [CrossRef]
- Frouin, H.; Lebeuf, M.; Hammill, M.; Masson, S.; Fournier, M. Effects of Individual Polybrominated Diphenyl Ether (PBDE) Congeners on Harbour Seal Immune Cells in Vitro. Mar. Pollut. Bull. 2010, 60, 291–298. [Google Scholar] [CrossRef]
- Lv, Q.-Y.; Wan, B.; Guo, L.-H.; Zhao, L.; Yang, Y. In Vitro Immune Toxicity of Polybrominated Diphenyl Ethers on Murine Peritoneal Macrophages: Apoptosis and Immune Cell Dysfunction. Chemosphere 2015, 120, 621–630. [Google Scholar] [CrossRef]
- Narayanan, S. Lysis of Whole Blood in Vitro Causes DNA Strand Breaks in Human Lymphocytes. Mutagenesis 2001, 16, 455–459. [Google Scholar] [CrossRef][Green Version]
- Ikonomopoulou, M.P.; Olszowy, H.; Hodge, M.; Bradley, A.J. The Effect of Organochlorines and Heavy Metals on Sex Steroid-Binding Proteins in Vitro in the Plasma of Nesting Green Turtles, Chelonia Mydas. J. Comp. Physiol. B 2009, 179, 653–662. [Google Scholar] [CrossRef]
- Cocci, P.; Capriotti, M.; Mosconi, G.; Palermo, F.A. Effects of Endocrine Disrupting Chemicals on Estrogen Receptor Alpha and Heat Shock Protein 60 Gene Expression in Primary Cultures of Loggerhead Sea Turtle (Caretta Caretta) Erythrocytes. Environ. Res. 2017, 158, 616–624. [Google Scholar] [CrossRef]
- Rousselet, E.; Levin, M.; Gebhard, E.; Higgins, B.M.; DeGuise, S.; Godard-Codding, C.A.J. Polychlorinated Biphenyls (PCBs) Modulate Both Phagocytosis and NK Cell Activity in Vitro in Juvenile Loggerhead Sea Turtles (Caretta Caretta). J. Toxicol. Environ. Health Part A 2017, 80, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Finlayson, K.A.; van de Merwe, J.P. Differences in Marine Megafauna in Vitro Sensitivity Highlights the Need for Species-Specific Chemical Risk Assessments. Aquat. Toxicol. 2021, 239, 105939. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.; Wang, M.; Wang, W.; Alonso Aguirre, A.; Lu, Y. Validation of an in Vitro Cytotoxicity Test for Four Heavy Metals Using Cell Lines Derived from a Green Sea Turtle (Chelonia Mydas). Cell Biol. Toxicol. 2010, 26, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Pascual-Anaya, J.; Zadissa, A.; Li, W.; Niimura, Y.; Huang, Z.; Li, C.; White, S.; Xiong, Z.; Fang, D.; et al. The Draft Genomes of Soft-Shell Turtle and Green Sea Turtle Yield Insights into the Development and Evolution of the Turtle-Specific Body Plan. Nat. Genet. 2013, 45, 701–706. [Google Scholar] [CrossRef]
- Webb, S.J.; Zychowski, G.V.; Bauman, S.W.; Higgins, B.M.; Raudsepp, T.; Gollahon, L.S.; Wooten, K.J.; Cole, J.M.; Godard-Codding, C. Establishment, Characterization, and Toxicological Application of Loggerhead Sea Turtle (Caretta Caretta) Primary Skin Fibroblast Cell Cultures. Environ. Sci. Technol. 2014, 48, 14728–14737. [Google Scholar] [CrossRef]
- Speer, R.M.; Wise, C.F.; Young, J.L.; Aboueissa, A.-M.; Martin Bras, M.; Barandiaran, M.; Bermúdez, E.; Márquez-D’Acunti, L.; Wise, J.P. The Cytotoxicity and Genotoxicity of Particulate and Soluble Hexavalent Chromium in Leatherback Sea Turtle Lung Cells. Aquat. Toxicol. 2018, 198, 149–157. [Google Scholar] [CrossRef]
- Finlayson, K.A.; Leusch, F.D.L.; Limpus, C.J.; van de Merwe, J.P. Towards the Development of Standardised Sea Turtle Primary Cell Cultures for Toxicity Testing. Ecotoxicol. Environ. Saf. 2019, 173, 63–70. [Google Scholar] [CrossRef]
- Finlayson, K.A.; Leusch, F.D.L.; van de Merwe, J.P. Cytotoxicity of Organic and Inorganic Compounds to Primary Cell Cultures Established from Internal Tissues of Chelonia Mydas. Sci. Total Environ. 2019, 664, 958–967. [Google Scholar] [CrossRef]
- Finlayson, K.A.; Madden Hof, C.A.; van de Merwe, J.P. Development and Application of Species-Specific Cell-Based Bioassays to Assess Toxicity in Green Sea Turtles. Sci. Total Environ. 2020, 747, 142095. [Google Scholar] [CrossRef]
- Alam, S.K.; Brim, M.S. Organochlorine, PCB, PAH, and Metal Concentrations in Eggs of Loggerhead Sea Turtles (Caretta Caretta) from Northwest Florida, USA. J. Environ. Sci. Health Part B 2000, 35, 705–724. [Google Scholar] [CrossRef]
- Bucchia, M.; Camacho, M.; Santos, M.R.D.; Boada, L.D.; Roncada, P.; Mateo, R.; Ortiz-Santaliestra, M.E.; Rodríguez-Estival, J.; Zumbado, M.; Orós, J.; et al. Plasma Levels of Pollutants Are Much Higher in Loggerhead Turtle Populations from the Adriatic Sea than in Those from Open Waters (Eastern Atlantic Ocean). Sci. Total Environ. 2015, 523, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Camacho, M.; Boada, L.D.; Orós, J.; Calabuig, P.; Zumbado, M.; Luzardo, O.P. Comparative Study of Polycyclic Aromatic Hydrocarbons (PAHs) in Plasma of Eastern Atlantic Juvenile and Adult Nesting Loggerhead Sea Turtles (Caretta Caretta). Mar. Pollut. Bull. 2012, 64, 1974–1980. [Google Scholar] [CrossRef] [PubMed]
- Camacho, M.; Luzardo, O.P.; Boada, L.D.; López Jurado, L.F.; Medina, M.; Zumbado, M.; Orós, J. Potential Adverse Health Effects of Persistent Organic Pollutants on Sea Turtles: Evidences from a Cross-Sectional Study on Cape Verde Loggerhead Sea Turtles. Sci. Total Environ. 2013, 458, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Camacho, M.; Boada, L.D.; Orós, J.; López, P.; Zumbado, M.; Almeida-González, M.; Luzardo, O.P. Monitoring Organic and Inorganic Pollutants in Juvenile Live Sea Turtles: Results from a Study of Chelonia Mydas and Eretmochelys Imbricata in Cape Verde. Sci. Total Environ. 2014, 481, 303–310. [Google Scholar] [CrossRef]
- Cocci, P.; Mosconi, G.; Palermo, F.A. Gene Expression Profiles of Putative Biomarkers in Juvenile Loggerhead Sea Turtles (Caretta Caretta) Exposed to Polycyclic Aromatic Hydrocarbons. Environ. Pollut. 2019, 246, 99–106. [Google Scholar] [CrossRef]
- Sinaei, M.; Zare, R. Polycyclic Aromatic Hydrocarbons (PAHs) and Some Biomarkers in the Green Sea Turtles (Chelonia Mydas). Mar. Pollut. Bull. 2019, 146, 336–342. [Google Scholar] [CrossRef]
- Keller, J.M.; Alava, J.J.; Aleksa, K.; Young, B.; Kucklick, J.R. Spatial Trends of Polybrominated Diphenyl Ethers (PBDEs) in Loggerhead Sea Turtle Eggs and Plasma. Organohalogen Compd. 2005, 67, 2. [Google Scholar]
- Van de Merwe, J.P.; Hodge, M.; Olszowy, H.A.; Whittier, J.M.; Lee, S.Y. Using Blood Samples to Estimate Persistent Organic Pollutants and Metals in Green Sea Turtles (Chelonia Mydas). Mar. Pollut. Bull. 2010, 60, 579. [Google Scholar] [CrossRef]
- Alava, J.J.; Keller, J.M.; Wyneken, J.; Crowder, L.; Scott, G.; Kucklick, J.R. Geographical Variation of Persistent Organic Pollutants in Eggs of Threatened Loggerhead Sea Turtles (Caretta Caretta) from Southeastern United States. Environ. Toxicol. Chem. 2011, 30, 1677–1688. [Google Scholar] [CrossRef]
- Savoca, D.; Arculeo, M.; Vecchioni, L.; Cambera, I.; Visconti, G.; Melfi, R.; Arizza, V.; Palumbo Piccionello, A.; Buscemi, S.; Pace, A. Can Phthalates Move into the Eggs of the Loggerhead Sea Turtle Caretta Caretta? The Case of the Nests on the Linosa Island in the Mediterranean Sea. Mar. Pollut. Bull. 2021, 168, 112395. [Google Scholar] [CrossRef]
- Bols, N.C.; Schirmer, K.; Joyce, E.M.; Dixon, D.G.; Greenberg, B.M.; Whyte, J.J. Ability of Polycyclic Aromatic Hydrocarbons to Induce 7-Ethoxyresorufin-o-Deethylase Activity in a Trout Liver Cell Line. Ecotoxicol. Environ. Saf. 1999, 44, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Clemons, J.H.; Allan, L.M.; Marvin, C.H.; Wu, Z.; McCarry, B.E.; Bryant, D.W.; Zacharewski, T.R. Evidence of Estrogen- and TCDD-Like Activities in Crude and Fractionated Extracts of PM 10 Air Particulate Material Using in Vitro Gene Expression Assays. Environ. Sci. Technol. 1998, 32, 1853–1860. [Google Scholar] [CrossRef]
- Jin, S.; Yang, F.; Hui, Y.; Xu, Y.; Lu, Y.; Liu, J. Cytotoxicity and Apoptosis Induction on RTG-2 Cells of 2,2′,4,4′-Tetrabromodiphenyl Ether (BDE-47) and Decabrominated Diphenyl Ether (BDE-209). Toxicol. Vitr. 2010, 24, 1190–1196. [Google Scholar] [CrossRef] [PubMed]
- Hennigar, S.R.; Myers, J.L.; Tagliaferro, A.R. Exposure of Alveolar Macrophages to Polybrominated Diphenyl Ethers Suppresses the Release of Pro-Inflammatory Products in Vitro. Exp. Biol. Med. 2012, 237, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Kocbach Bølling, A.; Ovrevik, J.; Samuelsen, J.T.; Holme, J.A.; Rakkestad, K.E.; Mathisen, G.H.; Paulsen, R.E.; Suárez Korsnes, M.; Becher, R. Mono-2-Ethylhexylphthalate (MEHP) Induces TNF-α Release and Macrophage Differentiation through Different Signalling Pathways in RAW264.7 Cells. Toxicol. Lett. 2012, 209, 43–50. [Google Scholar] [CrossRef]
- Guibert, E.; Prieur, B.; Cariou, R.; Courant, F.; Antignac, J.P.; Pain, B.; Brillard, J.P.; Froment, P. Effects of Mono-(2-Ethylhexyl) Phthalate (MEHP) on Chicken Germ Cells Cultured in Vitro. Environ. Sci. Pollut. Res. 2013, 20, 2771–2783. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Oakley, B.R.; Kirsch, D.R.; Morris, N.R. A Simplified Ultrasensitive Silver Stain for Detecting Proteins in Polyacrylamide Gels. Anal. Biochem. 1980, 105, 361–363. [Google Scholar] [CrossRef]
- Sinha, P.; Poland, J.; Schnölzer, M.; Rabilloud, T. A New Silver Staining Apparatus and Procedure for Matrix-Assisted Laser Desorption/Ionization-Time of Flight Analysis of Proteins after Two-Dimensional Electrophoresis. Proteomics 2001, 1, 835–840. [Google Scholar] [CrossRef]
- Miller, N.J.; Rice-Evans, C.; Davies, M.J.; Gopinathan, V.; Milner, A. A Novel Method for Measuring Antioxidant Capacity and Its Application to Monitoring the Antioxidant Status in Premature Neonates. Clin. Sci. 1993, 84, 407–412. [Google Scholar] [CrossRef]
- Secombes, C. Isolation of Salmonid Macrophages and Analysis of Their Killing Activity. Tech. Fish Immunol. 1990, 1, 137–154. [Google Scholar]
- Keller, J.M.; McClellan-Green, P.D.; Kucklick, J.R.; Keil, D.E.; Peden-Adams, M.M. Effects of Organochlorine Contaminants on Loggerhead Sea Turtle Immunity: Comparison of a Correlative Field Study and In Vitro Exposure Experiments. Environ. Health Perspect. 2006, 114, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.M. Complement and Complement Fixation. In Experimental Immunochemistry; Kabat, E.A., Mayer, M.M., Eds.; Charles C. Thomas Publisher: Springfield, IL, USA, 1961; pp. 133–241. [Google Scholar]
- Merchant, M.; Britton, A. Characterization of Serum Complement Activity of Saltwater (Crocodylus Porosus) and Freshwater (Crocodylus Johnstoni) Crocodiles. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2006, 143, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. Null 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Chaousis, S.; Leusch, F.D.L.; Nouwens, A.; Melvin, S.D.; van de Merwe, J.P. Changes in Global Protein Expression in Sea Turtle Cells Exposed to Common Contaminants Indicates New Biomarkers of Chemical Exposure. Sci. Total Environ. 2021, 751, 141680. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, H.M.; Sant, K.E.; Basnet, A.; Williams, L.M.; Moss, J.B.; Timme-Laragy, A.R. Embryonic Exposure to Mono(2-Ethylhexyl) Phthalate (MEHP) Disrupts Pancreatic Organogenesis in Zebrafish (Danio Rerio). Chemosphere 2018, 195, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Law, M.Y.L.; Moody, D.E. In Vitro Inhibition of Mouse and Rat Glutathione S-Transferases by Di(2-Ethylhexyl) Phthalate, Mono(2-Ethylhexyl) Phthalate, 2-Ethylhexanol, 2-Ethylhexanoic Acid and Clofibric Acid. Toxicol. Vitr. 1991, 5, 207–210. [Google Scholar] [CrossRef]
- Yan, C.; Huang, D.; Zhang, Y. The Involvement of ROS Overproduction and Mitochondrial Dysfunction in PBDE-47-Induced Apoptosis on Jurkat Cells. Exp. Toxicol. Pathol. 2011, 63, 413–417. [Google Scholar] [CrossRef]
- Montalbano, A.M.; Albano, G.D.; Anzalone, G.; Moscato, M.; Gagliardo, R.; Di Sano, C.; Bonanno, A.; Ruggieri, S.; Cibella, F.; Profita, M. Cytotoxic and Genotoxic Effects of the Flame Retardants (PBDE-47, PBDE-99 and PBDE-209) in Human Bronchial Epithelial Cells. Chemosphere 2020, 245, 125600. [Google Scholar] [CrossRef]
- Liu, C.; Wang, B.; Zhou, B.; Jian, X.; Zhang, X.; Wang, Y. The Responses of Oncorhynchus Mykiss Coping with BDE-47 Stress via PXR-Mediated Detoxification and Nrf2-Mediated Antioxidation System. Aquat. Toxicol. 2019, 207, 63–71. [Google Scholar] [CrossRef]
- Bentz, H.; Nathan, R.M.; Rosen, D.M.; Armstrong, R.M.; Thompson, A.Y.; Segarini, P.R.; Mathews, M.C.; Dasch, J.R.; Piez, K.A.; Seyedin, S.M. Purification and Characterization of a Unique Osteoinductive Factor from Bovine Bone. J. Biol. Chem. 1989, 264, 20805–20810. [Google Scholar] [CrossRef]
- Deckx, S.; Heymans, S.; Papageorgiou, A. The Diverse Functions of Osteoglycin: A Deceitful Dwarf, or a Master Regulator of Disease? FASEB J. 2016, 30, 2651–2661. [Google Scholar] [CrossRef] [PubMed]
- Tengryd, C.; Nielsen, S.H.; Cavalera, M.; Bengtsson, E.; Genovese, F.; Karsdal, M.; Dunér, P.; Orho-Melander, M.; Nilsson, J.; Edsfeldt, A.; et al. The Proteoglycan Mimecan Is Associated with Carotid Plaque Vulnerability and Increased Risk of Future Cardiovascular Death. Atherosclerosis 2020, 313, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Zhang, Q.-Y.; Li, X.-S.; Yu, H.-M.; Li, P.; Ma, J.-H.; Cao, H.-M.; Sun, F.; Zhao, S.-X.; Zheng, C.-X.; et al. The Expression of Mimecan in Adrenal Tissue Plays a Role in an Organism’s Responses to Stress. Aging 2021, 13, 13087–13107. [Google Scholar] [CrossRef]
- Camino, T.; Lago-Baameiro, N.; Bravo, S.B.; Molares-Vila, A.; Sueiro, A.; Couto, I.; Baltar, J.; Casanueva, E.F.; Pardo, M. Human Obese White Adipose Tissue Sheds Depot-Specific Extracellular Vesicles and Reveals Candidate Biomarkers for Monitoring Obesity and Its Comorbidities. Transl. Res. 2022, 239, 85–102. [Google Scholar] [CrossRef]
- Lee, N.J.; Ali, N.; Zhang, L.; Qi, Y.; Clarke, I.; Enriquez, R.F.; Brzozowska, M.; Lee, I.C.; Rogers, M.J.; Laybutt, D.R.; et al. Osteoglycin, a Novel Coordinator of Bone and Glucose Homeostasis. Mol. Metab. 2018, 13, 30–44. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, Y.; Lü, B.; Xu, E.; Huang, Q.; Lai, M. Differential Expression of Mimecan and Thioredoxin Domain–Containing Protein 5 in Colorectal Adenoma and Cancer: A Proteomic Study. Exp. Biol. Med. 2007, 232, 1152–1159. [Google Scholar] [CrossRef]
- Hu, X.; Li, Y.-Q.; Li, Q.-G.; Ma, Y.-L.; Peng, J.-J.; Cai, S.-J. Osteoglycin-Induced VEGF Inhibition Enhances T Lymphocytes Infiltrating in Colorectal Cancer. EBioMedicine 2018, 34, 35–45. [Google Scholar] [CrossRef]
- Hu, X.; Li, Y.-Q.; Li, Q.-G.; Ma, Y.-L.; Peng, J.-J.; Cai, S.-J. Osteoglycin (OGN) Reverses Epithelial to Mesenchymal Transition and Invasiveness in Colorectal Cancer via EGFR/Akt Pathway. J. Exp. Clin. Cancer Res. 2018, 37, 41. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, R.; Dong, M.; Zhang, Z.; Li, H.; Zhan, C.; Li, X. Osteoglycin (OGN) Inhibits Cell Proliferation and Invasiveness in Breast Cancer via PI3K/Akt/MTOR Signaling Pathway. OncoTargets Ther. 2019, 12, 10639–10650. [Google Scholar] [CrossRef]
- Starup-Linde, J.; Viggers, R.; Handberg, A. Osteoglycin and Bone—A Systematic Review. Curr. Osteoporos. Rep. 2019, 17, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Tasheva, E.S.; Ke, A.; Deng, Y.; Jun, C.; Takemoto, L.J.; Koester, A.; Conrad, G.W. Differentially Expressed Genes in the Lens of Mimecan-Null Mice. Mol. Vis. 2004, 10, 403–416. [Google Scholar] [PubMed]
- Moriggi, M.; Giussani, M.; Torretta, E.; Capitanio, D.; Sandri, M.; Leone, R.; De Palma, S.; Vasso, M.; Vozzi, G.; Tagliabue, E.; et al. ECM Remodeling in Breast Cancer with Different Grade: Contribution of 2D-DIGE Proteomics. Proteomics 2018, 18, 1800278. [Google Scholar] [CrossRef] [PubMed]
- Alfaro-Núñez, A.; Bertelsen, M.F.; Bojesen, A.M.; Rasmussen, I.; Zepeda-Mendoza, L.; Olsen, M.T.; Gilbert, M.T.P. Global Distribution of Chelonid Fibropapilloma-Associated Herpesvirus among Clinically Healthy Sea Turtles. BMC Evol. Biol. 2014, 14, 206. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.; Ariel, E.; Burgess, G.; Read, M. A Review of Fibropapillomatosis in Green Turtles (Chelonia Mydas). Vet. J. 2016, 212, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; Kim, J.K.; Noh, J.H.; Eun, J.W.; Bae, H.J.; Kim, M.G.; Chang, Y.G.; Shen, Q.; Kim, S.-J.; Kwon, S.H.; et al. Characteristic Molecular Signature for the Early Detection and Prediction of Polycyclic Aromatic Hydrocarbons in Rat Liver. Toxicol. Lett. 2013, 216, 1–8. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, International Agency for Research on Cancer (Ed.) Some Non-Heterocyclic Polycyclic Aromatic Hydrocarbons and Some Related Occupational Exposures. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; IARC Press: Lyon, France, 1987; Distributed by World Health Organization: Geneva, Switzerland, 2010; ISBN 978-92-832-1292-8. [Google Scholar]
- Alalaiwe, A.; Lin, Y.-K.; Lin, C.-H.; Wang, P.-W.; Lin, J.-Y.; Fang, J.-Y. The Absorption of Polycyclic Aromatic Hydrocarbons into the Skin to Elicit Cutaneous Inflammation: The Establishment of Structure–Permeation and in Silico–in Vitro–in Vivo Relationships. Chemosphere 2020, 255, 126955. [Google Scholar] [CrossRef]
- MacDonald, J.A.; Takai, Y.; Ishihara, O.; Seki, H.; Woods, D.C.; Tilly, J.L. Extracellular Matrix Signaling Activates Differentiation of Adult Ovary-Derived Oogonial Stem Cells in a Species-Specific Manner. Fertil. Steril. 2019, 111, 794–805. [Google Scholar] [CrossRef]
- Chen, A.Q.; Wang, Z.G.; Xu, Z.R.; Yu, S.D.; Yang, Z.G. Analysis of Gene Expression in Granulosa Cells of Ovine Antral Growing Follicles Using Suppressive Subtractive Hybridization. Anim. Reprod. Sci. 2009, 115, 39–48. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, W.; Li, Q.; Liu, J.; Zhang, T.; Zhou, T.; Li, L.; Wang, J.; Xu, H.; He, H. The Comprehensive Mechanisms Underlying Nonhierarchical Follicular Development in Geese (Anser Cygnoides). Anim. Reprod. Sci. 2015, 159, 131–140. [Google Scholar] [CrossRef]
- Neal, M.S.; Zhu, J.; Foster, W.G. Quantification of Benzo[a]Pyrene and Other PAHs in the Serum and Follicular Fluid of Smokers versus Non-Smokers. Reprod. Toxicol. 2008, 25, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Outerbridge, M.E.; O’Riordan, R.; Fort, D.J.; Davenport, J. Ecotoxicological Assessment of Diamondback Terrapin (Malaclemys Terrapin) Pond Habitat, Prey and Eggs in Bermuda. Mar. Pollut. Bull. 2016, 102, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.; Kang, M.; Huang, Q.; Fang, C.; Chen, Y.; Shen, H.; Dong, S. Exposure to DEHP and MEHP from Hatching to Adulthood Causes Reproductive Dysfunction and Endocrine Disruption in Marine Medaka (Oryzias Melastigma). Aquat. Toxicol. 2014, 146, 115–126. [Google Scholar] [CrossRef]
- Latini, G.; Verrotti, A.; Felice, C.D. DI-2-Ethylhexyl Phthalate and Endocrine Disruption: A Review. Curr. Drug Targets-Immune Endocr. Metab. Disord. 2004, 4, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, C.; Zhao, S.; Wang, B.; Wang, H.; Zhang, J.; Wang, Y.; Cheng, H.; Zhu, L.; Shen, R.; et al. Exposure to DEHP or Its Metabolite MEHP Promotes Progesterone Secretion and Inhibits Proliferation in Mouse Placenta or JEG-3 Cells. Environ. Pollut. 2020, 257, 113593. [Google Scholar] [CrossRef]
- Caporossi, L.; Viganò, P.; Paci, E.; Capanna, S.; Alteri, A.; Campo, G.; Pigini, D.; De Rosa, M.; Tranfo, G.; Papaleo, B. Female Reproductive Health and Exposure to Phthalates and Bisphenol A: A Cross Sectional Study. Toxics 2021, 9, 299. [Google Scholar] [CrossRef]
- Rowdhwal, S.S.S.; Chen, J. Toxic Effects of Di-2-Ethylhexyl Phthalate: An Overview. BioMed Res. Int. 2018, 2018, 1750368. [Google Scholar] [CrossRef]
- Dutta, S.; Haggerty, D.K.; Rappolee, D.A.; Ruden, D.M. Phthalate Exposure and Long-Term Epigenomic Consequences: A Review. Front. Genet. 2020, 11, 405. [Google Scholar] [CrossRef]
- Barron, M.G.; Schultz, I.R.; Hayton, W.L. Presystemic Branchial Metabolism Limits Di-2-Ethylhexyl Phthalate Accumulation in Fish. Toxicol. Appl. Pharmacol. 1989, 98, 49–57. [Google Scholar] [CrossRef]
- Wang, P.; Heitman, J. The cyclophilins. Genome Biol. 2005, 6, 226. [Google Scholar] [CrossRef][Green Version]
- Nath, P.R.; Isakov, N. Insights into Peptidyl-Prolyl Cis–Trans Isomerase Structure and Function in Immunocytes. Immunol. Lett. 2015, 163, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.P.; Hwang, Y.S.; Kim, Y.J.; Kwon, K.-S.; Kim, H.J.; Kim, K.; Chae, H.Z. Cyclophilin A Binds to Peroxiredoxins and Activates Its Peroxidase Activity. J. Biol. Chem. 2001, 276, 29826–29832. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.J.; Piao, Y.J.; Lim, M.J.; Kim, J.H.; Ha, J.; Choe, W.; Kim, S.S. Overexpressed Cyclophilin A in Cancer Cells Renders Resistance to Hypoxia- and Cisplatin-Induced Cell Death. Cancer Res. 2007, 67, 3654–3662. [Google Scholar] [CrossRef]
- Kim, H.; Oh, Y.; Kim, K.; Jeong, S.; Chon, S.; Kim, D.; Jung, M.H.; Pak, Y.K.; Ha, J.; Kang, I.; et al. Cyclophilin A Regulates JNK/P38-MAPK Signaling through Its Physical Interaction with ASK1. Biochem. Biophys. Res. Commun. 2015, 464, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Lauranzano, E.; Pozzi, S.; Pasetto, L.; Stucchi, R.; Massignan, T.; Paolella, K.; Mombrini, M.; Nardo, G.; Lunetta, C.; Corbo, M.; et al. Peptidylprolyl Isomerase A Governs TARDBP Function and Assembly in Heterogeneous Nuclear Ribonucleoprotein Complexes. Brain 2015, 138, 974–991. [Google Scholar] [CrossRef]
- Izumikawa, K.; Nobe, Y.; Yoshikawa, H.; Ishikawa, H.; Miura, Y.; Nakayama, H.; Nonaka, T.; Hasegawa, M.; Egawa, N.; Inoue, H.; et al. TDP-43 Stabilises the Processing Intermediates of Mitochondrial Transcripts. Sci. Rep. 2017, 7, 7709. [Google Scholar] [CrossRef]
- Song, F.; Zhang, X.; Ren, X.-B.; Zhu, P.; Xu, J.; Wang, L.; Li, Y.-F.; Zhong, N.; Ru, Q.; Zhang, D.-W.; et al. Cyclophilin A (CyPA) Induces Chemotaxis Independent of Its Peptidylprolyl Cis-Trans Isomerase Activity. J. Biol. Chem. 2011, 286, 8197–8203. [Google Scholar] [CrossRef]
- Brazin, K.N.; Mallis, R.J.; Fulton, D.B.; Andreotti, A.H. Regulation of the Tyrosine Kinase Itk by the Peptidyl-Prolyl Isomerase Cyclophilin A. Proc. Natl. Acad. Sci. USA 2002, 99, 1899–1904. [Google Scholar] [CrossRef]
- Dawar, F.U.; Xiong, Y.; Khattak, M.N.K.; Li, J.; Lin, L.; Mei, J. Potential Role of Cyclophilin A in Regulating Cytokine Secretion. J. Leukoc. Biol. 2017, 102, 989–992. [Google Scholar] [CrossRef]
- Bannon, J.H.; O’Donovan, D.S.; Kennelly, S.M.E.; Mc Gee, M.M. The Peptidyl Prolyl Isomerase Cyclophilin A Localizes at the Centrosome and the Midbody and Is Required for Cytokinesis. Cell Cycle 2012, 11, 1340–1353. [Google Scholar] [CrossRef]
- Volker, S.E.; Hedrick, S.E.; Feeney, Y.B.; Clevenger, C.V. Cyclophilin A Function in Mammary Epithelium Impacts Jak2/Stat5 Signaling, Morphogenesis, Differentiation, and Tumorigenesis in the Mammary Gland. Cancer Res. 2018, 78, 3877–3887. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, R.; Satoh, K.; Nakata, T.; Shindo, T.; Kikuchi, N.; Satoh, T.; Siddique, M.A.H.; Omura, J.; Sunamura, S.; Nogi, M.; et al. Identification of Celastrol as a Novel Therapeutic Agent for Pulmonary Arterial Hypertension and Right Ventricular Failure Through Suppression of Bsg (Basigin)/CyPA (Cyclophilin A). ATVB 2021, 41, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Takakura, H.; Mitamura, K.; Taga, A. Cyclophilin a Knokdown Inhibits Cell Migration and Invasion through the Suppression of Epithelial–Mesenchymal Transition in Colorectal Cancer Cells. Biochem. Biophys. Res. Commun. 2020, 526, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Nigro, P.; Pompilio, G.; Capogrossi, M.C. Cyclophilin A: A Key Player for Human Disease. Cell Death Dis. 2013, 4, e888. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.A.; Yetsko, K.; Whitmore, L.; Whilde, J.; Eastman, C.B.; Ramia, D.R.; Thomas, R.; Linser, P.; Creer, S.; Burkhalter, B.; et al. Environmental DNA Monitoring of Oncogenic Viral Shedding and Genomic Profiling of Sea Turtle Fibropapillomatosis Reveals Unusual Viral Dynamics. Commun. Biol. 2021, 4, 565. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Luo, D.; Peng, K.; Zeng, Y. Cyclophilin A: A Key Player for Etiological Agent Infection. Appl. Microbiol. Biotechnol. 2021, 105, 1365–1377. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Sowden, M.P.; Berk, B.C. Extracellular and Intracellular Cyclophilin A, Native and Post-Translationally Modified, Show Diverse and Specific Pathological Roles in Diseases. ATVB 2018, 38, 986–993. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, P.; Li, C.; Su, X.; Jin, C.; Li, Y.; Xu, Y.; Li, T. Characterisation of Immune-Related Gene Expression in Clam (Venerupis Philippinarum) under Exposure to Di(2-Ethylhexyl) Phthalate. Fish Shellfish. Immunol. 2013, 34, 142–146. [Google Scholar] [CrossRef]
- Marenholz, I.; Heizmann, C.W.; Fritz, G. S100 Proteins in Mouse and Man: From Evolution to Function and Pathology (Including an Update of the Nomenclature). Biochem. Biophys. Res. Commun. 2004, 322, 1111–1122. [Google Scholar] [CrossRef]
- Li, P.; Lv, X.; Zhang, Z.; Xie, S. S100A6/MiR193a Regulates the Proliferation, Invasion, Migration and Angiogenesis of Lung Cancer Cells through the P53 Acetylation. Am. J. Transl. Res. 2019, 11, 4634. [Google Scholar]
- Li, A.; Gu, Y.; Li, X.; Sun, H.; Zha, H.; Xie, J.; Zhao, J.; Huang, M.; Chen, L.; Peng, Q.; et al. S100A6 Promotes the Proliferation and Migration of Cervical Cancer Cells via the PI3K/Akt Signaling Pathway. Oncol. Lett. 2018, 15, 5685–5693. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Wu, R.; Zou, Z.; Wang, H.; Ye, L.; Li, H.; Yuan, S.; Li, X.; Zha, H.; Sun, H.; et al. S100A6 Stimulates Proliferation and Migration of Colorectal Carcinoma Cells through Activation of the MAPK Pathways. Int. J. Oncol. 2014, 44, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tang, M.; Ling, B.; Liu, S.; Zheng, Y.; Nie, C.; Yuan, Z.; Zhou, L.; Guo, G.; Tong, A.; et al. Increased Expression of S100A6 Promotes Cell Proliferation and Migration in Human Hepatocellular Carcinoma. J. Mol. Med. 2014, 92, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Sorci, G.; Giambanco, I. S100A6 Protein: Functional Roles. Cell. Mol. Life Sci. 2017, 74, 2749–2760. [Google Scholar] [CrossRef] [PubMed]
- Leśniak, W.; Szczepańska, A.; Kuźnicki, J. Calcyclin (S100A6) Expression Is Stimulated by Agents Evoking Oxidative Stress via the Antioxidant Response Element. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2005, 1744, 29–37. [Google Scholar] [CrossRef][Green Version]
- Zhu, L.; Kohda, F.; Nakahara, T.; Chiba, T.; Tsuji, G.; Hachisuka, J.; Ito, T.; Tu, Y.; Moroi, Y.; Uchi, H.; et al. Aberrant Expression of S100A6 and Matrix Metalloproteinase 9, but Not S100A2, S100A4, and S100A7, Is Associated with Epidermal Carcinogenesis. J. Dermatol. Sci. 2013, 72, 311–319. [Google Scholar] [CrossRef]
- Ivanov, B.; Leonard, A.; Deknudt, G. Blood Storage and the Rate of Chromosome Aberrations after in Vitro Exposure to Ionizing Radiations. Radiat. Res. 1973, 55, 469. [Google Scholar] [CrossRef]
- Fischer, J.R.; Schindel, M.; Lahm, H.; Drings, P. Tumour-Derived, Endocrine, Exogenous and Therapeutic Factors Differentially Modulate Cytokine Secretion in Whole Blood Cell Culture. Eur. J. Cancer 1997, 33, 1661–1667. [Google Scholar] [CrossRef]
- Krakauer, T.; Stephens, J.; Buckley, M.; Tate, M. Superantigen-Induced Cytokine Release from Whole-Blood Cell Culture as a Functional Measure of Drug Efficacy after Oral Dosing in Nonhuman Primates. Res. Vet. Sci. 2007, 83, 182–187. [Google Scholar] [CrossRef]
- Rodríguez, A.B.; Terrón, M.P.; Durán, J.; Ortega, E.; Barriga, C. Physiological Concentrations of Melatonin and Corticosterone Affect Phagocytosis and Oxidative Metabolism of Ring Dove Heterophils: Physiological Effect of Melatonin and Corticosterone in Ring Dove Heterophils. J. Pineal Res. 2001, 31, 31–38. [Google Scholar] [CrossRef]
- Terrón, M.P.; Paredes, S.D.; Barriga, C.; Ortega, E.; Rodríguez, A.B. Comparative Study of the Heterophil Phagocytic Function in Young and Old Ring Doves (Streptopelia Risoria) and Its Relationship with Melatonin Levels. J. Comp. Physiol. B 2004, 174, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, A.; Kiron, V.; Kobayashi, T.; Puangkaew, J.; Satoh, S.; Sugita, H. Immune Responses in Rainbow Trout Oncorhynchus Mykiss Induced by a Potential Probiotic Bacteria Lactobacillus Rhamnosus JCM 1136. Vet. Immunol. Immunopathol. 2004, 102, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, V.; Sampath, K.; Sekar, V. Adjuvant and Immunostimulatory Effects of β-Glucan Administration in Combination with Lipopolysaccharide Enhances Survival and Some Immune Parameters in Carp Challenged with Aeromonas Hydrophila. Vet. Immunol. Immunopathol. 2006, 114, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Mccoll, S.; Daniels, C. A Comparison Between the Inflammatory Mediators Produced by the Blue-Tongue Lizard (Tiliqua-Scincoides) and Human White Blood-Cells. Aust. J. Zool. 1988, 36, 209. [Google Scholar] [CrossRef]
- Rousselet, E.; Levin, M.; Gebhard, E.; Higgins, B.M.; DeGuise, S.; Godard-Codding, C.A.J. Evaluation of Immune Functions in Captive Immature Loggerhead Sea Turtles (Caretta Caretta). Vet. Immunol. Immunopathol. 2013, 156, 43–53. [Google Scholar] [CrossRef]
- Balfry, S.K.; Iwama, G.K. Observations on the Inherent Variability of Measuring Lysozyme Activity in Coho Salmon (Oncorhynchus Kisutch). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 138, 207–211. [Google Scholar] [CrossRef]
- Weeks, B.A.; Anderson, D.P.; DuFour, A.P.; Ann, F.; Goven, A.J.; Lahvls, G.P.; Gabriele, P. Immunological Biomarkers to Assess Environmental Stress. In Biomarkers; CRC Press: Boca Raton, FL, USA, 2018; pp. 211–234. ISBN 1-351-07027-4. [Google Scholar]
- Burton, J.E.; Dorociak, I.R.; Schwedler, T.E.; Rice, C.D. Circulating Lysozyme AND Hepatic Cyp1a Activities During a Chronic Dietary Exposure to Tributyltin (Tbt) and 3,3’,4,4’,5-Pentachlorobiphenyl (Pcb-126) Mixtures in Channel Catfish, Ictalurus Punctatus. J. Toxicol. Environ. Health Part A 2002, 65, 589–602. [Google Scholar] [CrossRef]
- Day, R.D.; Segars, A.L.; Arendt, M.D.; Lee, A.M.; Peden-Adams, M.M. Relationship of Blood Mercury Levels to Health Parameters in the Loggerhead Sea Turtle (Caretta Caretta). Environ. Health Perspect. 2007, 115, 1421–1428. [Google Scholar] [CrossRef]
- Walsh, C.J.; Leggett, S.R.; Carter, B.J.; Colle, C. Effects of Brevetoxin Exposure on the Immune System of Loggerhead Sea Turtles. Aquat. Toxicol. 2010, 97, 293–303. [Google Scholar] [CrossRef]
- Rodgers, M.L.; Toline, C.A.; Rice, C.D. Humoral Immune Responses to Select Marine Bacteria in Loggerhead Sea Turtles Caretta Caretta and Kemp’s Ridley Sea Turtles Lepidochelys Kempii from the Southeastern United States. J. Aquat. Anim. Health 2018, 30, 20–30. [Google Scholar] [CrossRef]
- Neuman-Lee, L.A.; Carr, J.; Vaughn, K.; French, S.S. Physiological Effects of Polybrominated Diphenyl Ether (PBDE-47) on Pregnant Gartersnakes and Resulting Offspring. Gen. Comp. Endocrinol. 2015, 219, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Monzón-Argüello, C.; Cardona, L.; Calabuig, P.; Camacho, M.; Crespo-Picazo, J.L.; García-Párraga, D.; Mayans, S.; Luzardo, O.P.; Orós, J.; Varo-Cruz, N. Supplemental Feeding and Other Anthropogenic Threats to Green Turtles (Chelonia Mydas) in the Canary Islands. Sci. Total Environ. 2018, 621, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-C.; Rosa Sarrias, M.; Lambris, J.D. Complement and Innate Immunity. Immunopharmacology 2000, 49, 187–198. [Google Scholar] [CrossRef]
- Zhong, R.; Wang, H.; Wu, X.; Cao, Y.; He, Z.; He, Y.; Liu, J. In Vitro Investigation of the Effect of Plasticizers on the Blood Compatibility of Medical Grade Plasticized Poly (Vinyl Chloride). J. Mater. Sci. Mater. Med. 2013, 24, 1985–1992. [Google Scholar] [CrossRef] [PubMed]
- Watanuki, H.; Gushiken, Y.; Sakai, M. In Vitro Modulation of Common Carp (Cyprinus Carpio L.) Phagocytic Cells by Di-n-Butyl Phthalate and Di-2-Ethylhexyl Phthalate. Aquat. Toxicol. 2003, 63, 119–126. [Google Scholar] [CrossRef]
- Wang, S.; Cao, Y.; Wang, S.; Cai, J.; Zhang, Z. DEHP Induces Immunosuppression through Disturbing Inflammatory Factors and CYPs System Homeostasis in Common Carp Neutrophils. Fish Shellfish Immunol. 2020, 96, 26–31. [Google Scholar] [CrossRef]
- Zhong, Y.F.; Wang, L.L.; Yin, L.L.; An, J.; Hou, M.L.; Zheng, K.W.; Zhang, X.Y.; Wu, M.H.; Yu, Z.Q.; Sheng, G.Y.; et al. Cytotoxic Effects and Oxidative Stress Response of Six PBDE Metabolites on Human L02 Cells. J. Environ. Sci. Health Part A 2011, 46, 1320–1327. [Google Scholar] [CrossRef]
- Pellacani, C.; Buschini, A.; Galati, S.; Mussi, F.; Franzoni, S.; Costa, L.G. Evaluation of DNA Damage Induced by 2 Polybrominated Diphenyl Ether Flame Retardants (BDE-47 and BDE-209) in SK-N-MC Cells. Int. J. Toxicol. 2012, 31, 372–379. [Google Scholar] [CrossRef]
- Bondy, G.S.; Lefebvre, D.E.; Aziz, S.; Cherry, W.; Coady, L.; MacLellan, E.; Armstrong, C.; Barker, M.; Cooke, G.; Gaertner, D.; et al. Toxicologic and Immunologic Effects of Perinatal Exposure to the Brominated Diphenyl Ether (BDE) Mixture DE-71 in the Sprague-Dawley Rat. Environ. Toxicol. 2013, 28, 215–228. [Google Scholar] [CrossRef]
- Arkoosh, M.R.; Boylen, D.; Dietrich, J.; Anulacion, B.F.; Ylitalo, G.; Bravo, C.F.; Johnson, L.L.; Loge, F.J.; Collier, T.K. Disease Susceptibility of Salmon Exposed to Polybrominated Diphenyl Ethers (PBDEs). Aquat. Toxicol. 2010, 98, 51–59. [Google Scholar] [CrossRef]
- Hutchinson, J.; Simmonds, M. Escalation of Threats to Marine Turtles. Oryx 1992, 26, 95–102. [Google Scholar] [CrossRef]
- Cocci, P.; Mosconi, G.; Bracchetti, L.; Nalocca, J.M.; Frapiccini, E.; Marini, M.; Caprioli, G.; Sagratini, G.; Palermo, F.A. Investigating the Potential Impact of Polycyclic Aromatic Hydrocarbons (PAHs) and Polychlorinated Biphenyls (PCBs) on Gene Biomarker Expression and Global DNA Methylation in Loggerhead Sea Turtles (Caretta Caretta) from the Adriatic Sea. Sci. Total Environ. 2018, 619, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Gaskill, S.J.; Bruce, E.D. Binary Mixtures of Polycyclic Aromatic Hydrocarbons Display Nonadditive Mixture Interactions in an In Vitro Liver Cell Model: Mixtures of PAHs Display Nonadditive Mixture Interactions. Risk Anal. 2016, 36, 968–991. [Google Scholar] [CrossRef]
- Tao, L.-P.; Li, X.; Zhao, M.-Z.; Shi, J.-R.; Ji, S.-Q.; Jiang, W.-Y.; Liang, Q.-J.; Lei, Y.-H.; Zhou, Y.-Y.; Cheng, R.; et al. Chrysene, a Four-Ring Polycyclic Aromatic Hydrocarbon, Induces Hepatotoxicity in Mice by Activation of the Aryl Hydrocarbon Receptor (AhR). Chemosphere 2021, 276, 130108. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Santiago, O.; Castillo, C.G.; Espinosa-Reyes, G.; Pérez-Maldonado, I.N.; González-Mille, D.J.; Cuevas-Díaz, M.D.C.; Ilizaliturri-Hernández, C.A. Giant Toads (Rhinella Marina) From the Industrial Zones of Low Basin of the Coatzacoalcos River (Veracruz, MX) Presents Genotoxicity in Erythrocytes. Bull. Environ. Contam. Toxicol. 2021, 108, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Esmaeilbeigi, M.; Kalbassi, M.R.; Seyedi, J.; Tayemeh, M.B.; Moghaddam, J.A. Intra and Extracellular Effects of Benzo [α] Pyrene on Liver, Gill and Blood of Caspian White Fish (Rutilus Frissi Kutum): Cyto-Genotoxicity and Histopathology Approach. Mar. Pollut. Bull. 2021, 163, 111942. [Google Scholar] [CrossRef]
- Kleinsasser, N.H.; Harréus, U.A.; Kastenbauer, E.R.; Wallner, B.C.; Sassen, A.W.; Staudenmaier, R.; Rettenmeier, A.W. Mono(2-Ethylhexyl)Phthalate Exhibits Genotoxic Effects in Human Lymphocytes and Mucosal Cells of the Upper Aerodigestive Tract in the Comet Assay. Toxicol. Lett. 2004, 148, 83–90. [Google Scholar] [CrossRef]
- Çağlayan, M.; Wilson, S.H. Oxidant and Environmental Toxicant-Induced Effects Compromise DNA Ligation during Base Excision DNA Repair. DNA Repair 2015, 35, 85–89. [Google Scholar] [CrossRef]
- Chang, Y.-J.; Tseng, C.-Y.; Lin, P.-Y.; Chuang, Y.-C.; Chao, M.-W. Acute Exposure to DEHP Metabolite, MEHP Cause Genotoxicity, Mutagenesis and Carcinogenicity in Mammalian Chinese Hamster Ovary Cells. Carcinogenesis 2017, 38, 336–345. [Google Scholar] [CrossRef]
- Iyama, T.; Wilson, D.M. DNA Repair Mechanisms in Dividing and Non-Dividing Cells. DNA Repair 2013, 12, 620–636. [Google Scholar] [CrossRef]
- Malarvannan, G.; Takahashi, S.; Isobe, T.; Kunisue, T.; Sudaryanto, A.; Miyagi, T.; Nakamura, M.; Yasumura, S.; Tanabe, S. Levels and Distribution of Polybrominated Diphenyl Ethers and Organochlorine Compounds in Sea Turtles from Japan. Mar. Pollut. Bull. 2011, 63, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Vijayasarathy, S.; Baduel, C.; Hof, C.; Bell, I.; del Mar Gómez Ramos, M.; Ramos, M.J.G.; Kock, M.; Gaus, C. Multi-Residue Screening of Non-Polar Hazardous Chemicals in Green Turtle Blood from Different Foraging Regions of the Great Barrier Reef. Sci. Total Environ. 2019, 652, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Ragland, J.M.; Arendt, M.D.; Kucklick, J.R.; Keller, J.M. Persistent Organic Pollutants in Blood Plasma of Satellite-tracked Adult Male Loggerhead Sea Turtles (Caretta caretta). Environ. Toxicol. Chem. 2011, 30, 1549–1556. [Google Scholar] [CrossRef] [PubMed]
- He, P.; He, W.; Wang, A.; Xia, T.; Xu, B.; Zhang, M.; Chen, X. PBDE-47-Induced Oxidative Stress, DNA Damage and Apoptosis in Primary Cultured Rat Hippocampal Neurons. Neurotoxicology 2008, 29, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.; Longo, A.; Di Sano, C.; Cigna, D.; Cibella, F.; Di Felice, G.; Colombo, P. In Vitro Exposure to 2,2′,4,4′-Tetrabromodiphenyl Ether (PBDE-47) Impairs Innate Inflammatory Response. Chemosphere 2019, 219, 845–854. [Google Scholar] [CrossRef] [PubMed]
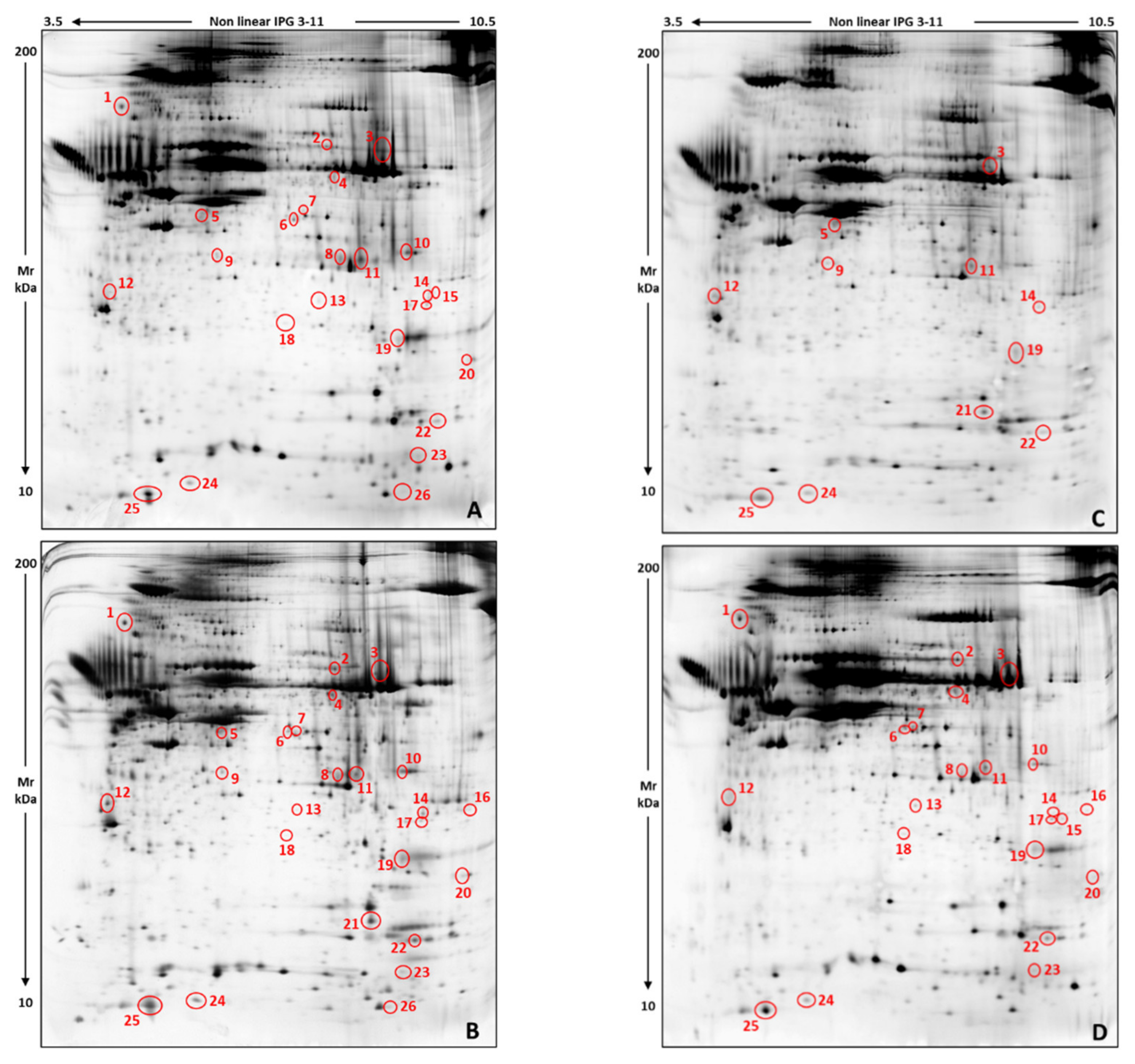

| Spot N. | Protein Identification | Chrysene %V Mean ± SD | MEHP %V Mean ± SD | PBDE-47 %V Mean ± SD | DMSO CTRL %V Mean ± SD | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Protein ID | NCBIprot, UniProtKB, or GenBank AN | Score | Expected | Detected Masses/Matched Masses | Seq. Coverage % | |||||
| 5 | Actin. cytoplasmic 2 isoform X3 (Cariama cristata) | XP_009695579.1 | 92 | 0.0094 | 7/15 | 28 | 0.4141 ± 0.1171 | 0.1477 ± 0.1181 | 0 ± 0 | 0.1343 ± 0.1045 |
| 6 | Alpha-enolase (Chelonia mydas) | XP_037737064.1 | 170 | 1.6 × 10−10 | 29/16 | 37 | 0 ± 0 | 0.3526 ± 0.09 | 0.1948 ± 0.0328 | 0.2042 ± 0.0774 |
| 8 | Mimecan (Chelonia mydas) | EMP39737.1 | 75 | 0.05 | 4/4 | 15 | 0 ± 0 | 0.0478 ± 0.0063 € | 0.3156 ± 0.2172 | 0.5189 ± 0.3459 € |
| 10 | Mimecan (Chelonia mydas) | XP_007055647.1 | 91 | 0.012 | 5/5 | 14 | 0 ± 0 | 0.7315 ± 0.1362 ∑ | 0.1901 ± 0.1724 ∑ | 0.3899 ± 0.1163 |
| 11 | Mimecan (Chelonia mydas) | XP_007055647.1 | 108 | 0.00026 | 6/6 | 17 | 0.2121 ± 0.1567 | 0.3246 ± 0.097 | 0.1487 ± 0.7414 Ω | 0.4872 ± 0.1932 Ω |
| 12 | Keratin. type I cytoskeletal 15 (Chelonia mydas) | XP_007062046.2 | 137 | 3.3 × 10−7 | 13/25 | 24 | 0.2075 ± 0.1123 * | 0.7221 ± 0.1899 * | 0.3926 ± 0.1333 | 0.5253 ± 0.2178 |
| 19 | Glutathione S-transferase P (Chelonia mydas) | XP_037755179.1 | 148 | 2.6 × 10−8 | 11/17 | 47 | 0.00962 ± 0.0572 # ¥ | 0.1725 ± 0.085 | 0.6684 ± 0.3708 ¥ | 0.2494 ± 0.1771 # |
| 22 | Peptidyl-prolyl cis-trans isomerase A (Chelonia mydas) | XP_007066086.1 | 90 | 0.017 | 6/14 | 29 | 0.1531 ± 0.4331 * | 0.8279 ± 0.1072 * | 0.3072 ± 0.1801 | 0.4633 ± 0.2387 |
| 23 | Hemoglobin subunit alpha-A (Caretta caretta) | Q10732 (Q10732.1) | 132 | 1.00 × 10−6 | 8/11 | 57 | 0 ± 0 | 0.2615 ± 0.0819 | 0.1461 ± 0.0151 | 0.5494 ± 0.271 |
| 25 | Protein S100-A6 (Chelonia mydas) | XP_037739342.1 | LNDAEIVGLMEDLDR Score: 136; E: 5 × 10−9 | 0.08945 ± 0.0079 * | 0.3056 ± 0.0909 * | 0.1904 ± 0.014 | 0.155 ± 0.0105 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianchi, L.; Casini, S.; Vantaggiato, L.; Di Noi, A.; Carleo, A.; Shaba, E.; Armini, A.; Bellucci, F.; Furii, G.; Bini, L.; et al. A Novel Ex Vivo Approach Based on Proteomics and Biomarkers to Evaluate the Effects of Chrysene, MEHP, and PBDE-47 on Loggerhead Sea Turtles (Caretta caretta). Int. J. Environ. Res. Public Health 2022, 19, 4369. https://doi.org/10.3390/ijerph19074369
Bianchi L, Casini S, Vantaggiato L, Di Noi A, Carleo A, Shaba E, Armini A, Bellucci F, Furii G, Bini L, et al. A Novel Ex Vivo Approach Based on Proteomics and Biomarkers to Evaluate the Effects of Chrysene, MEHP, and PBDE-47 on Loggerhead Sea Turtles (Caretta caretta). International Journal of Environmental Research and Public Health. 2022; 19(7):4369. https://doi.org/10.3390/ijerph19074369
Chicago/Turabian StyleBianchi, Laura, Silvia Casini, Lorenza Vantaggiato, Agata Di Noi, Alfonso Carleo, Enxhi Shaba, Alessandro Armini, Francesco Bellucci, Giovanni Furii, Luca Bini, and et al. 2022. "A Novel Ex Vivo Approach Based on Proteomics and Biomarkers to Evaluate the Effects of Chrysene, MEHP, and PBDE-47 on Loggerhead Sea Turtles (Caretta caretta)" International Journal of Environmental Research and Public Health 19, no. 7: 4369. https://doi.org/10.3390/ijerph19074369
APA StyleBianchi, L., Casini, S., Vantaggiato, L., Di Noi, A., Carleo, A., Shaba, E., Armini, A., Bellucci, F., Furii, G., Bini, L., & Caliani, I. (2022). A Novel Ex Vivo Approach Based on Proteomics and Biomarkers to Evaluate the Effects of Chrysene, MEHP, and PBDE-47 on Loggerhead Sea Turtles (Caretta caretta). International Journal of Environmental Research and Public Health, 19(7), 4369. https://doi.org/10.3390/ijerph19074369









