Vaccinomics to Design a Multi-Epitopes Vaccine for Acinetobacter baumannii
Abstract
1. Introduction
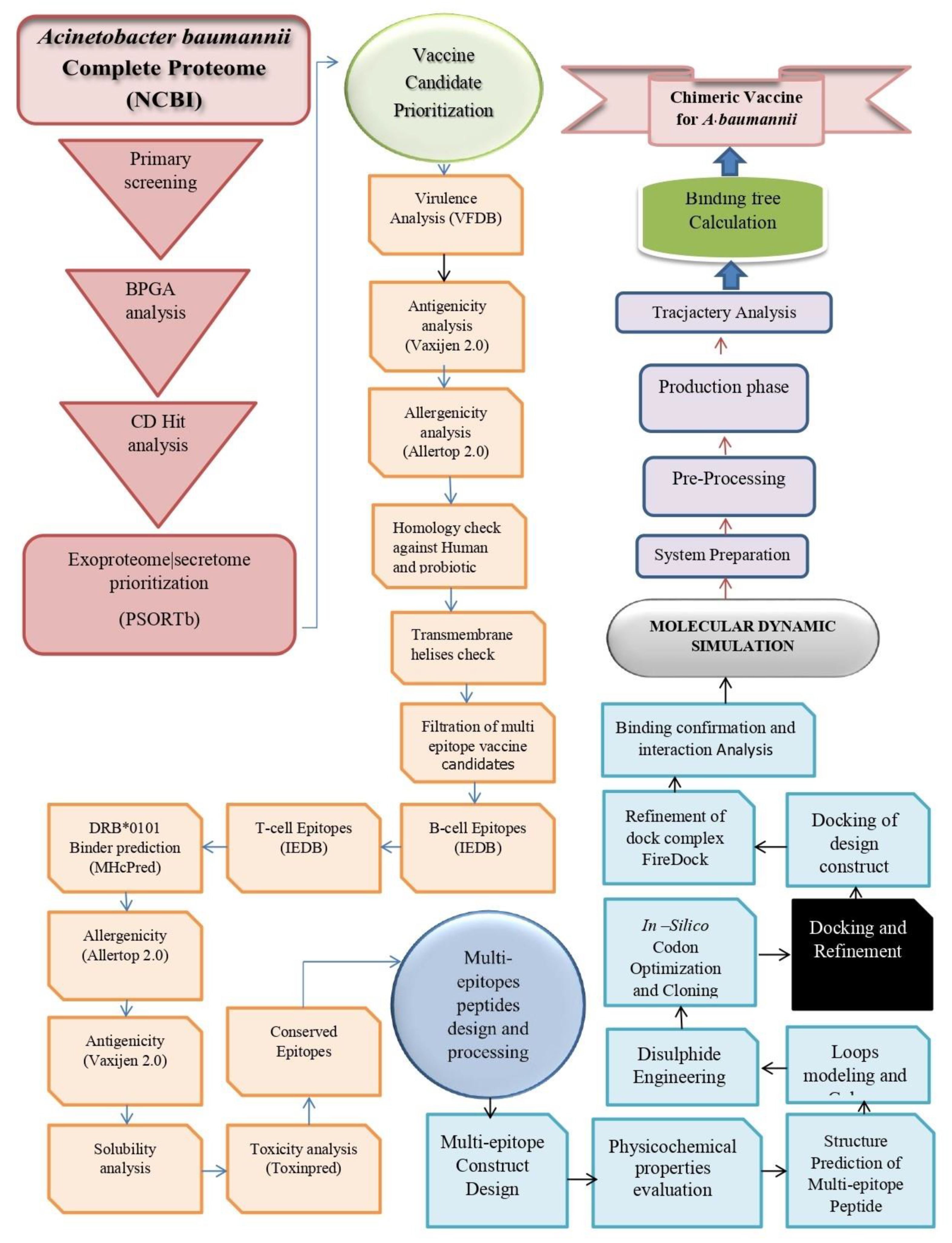
2. Research Methodology
2.1. Complete Genome Retrieval
2.2. Pre-Screening Phase
2.3. Vaccine Epitopes Prioritization Phase
2.4. Design of a Multi-Epitopes Vaccine
2.5. Simulating Host Immunity against Vaccine
2.6. Docking and Refinement
2.7. Molecular Dynamic Analysis
2.8. Calculation of TLR4-Vaccine Binding Energies
3. Results
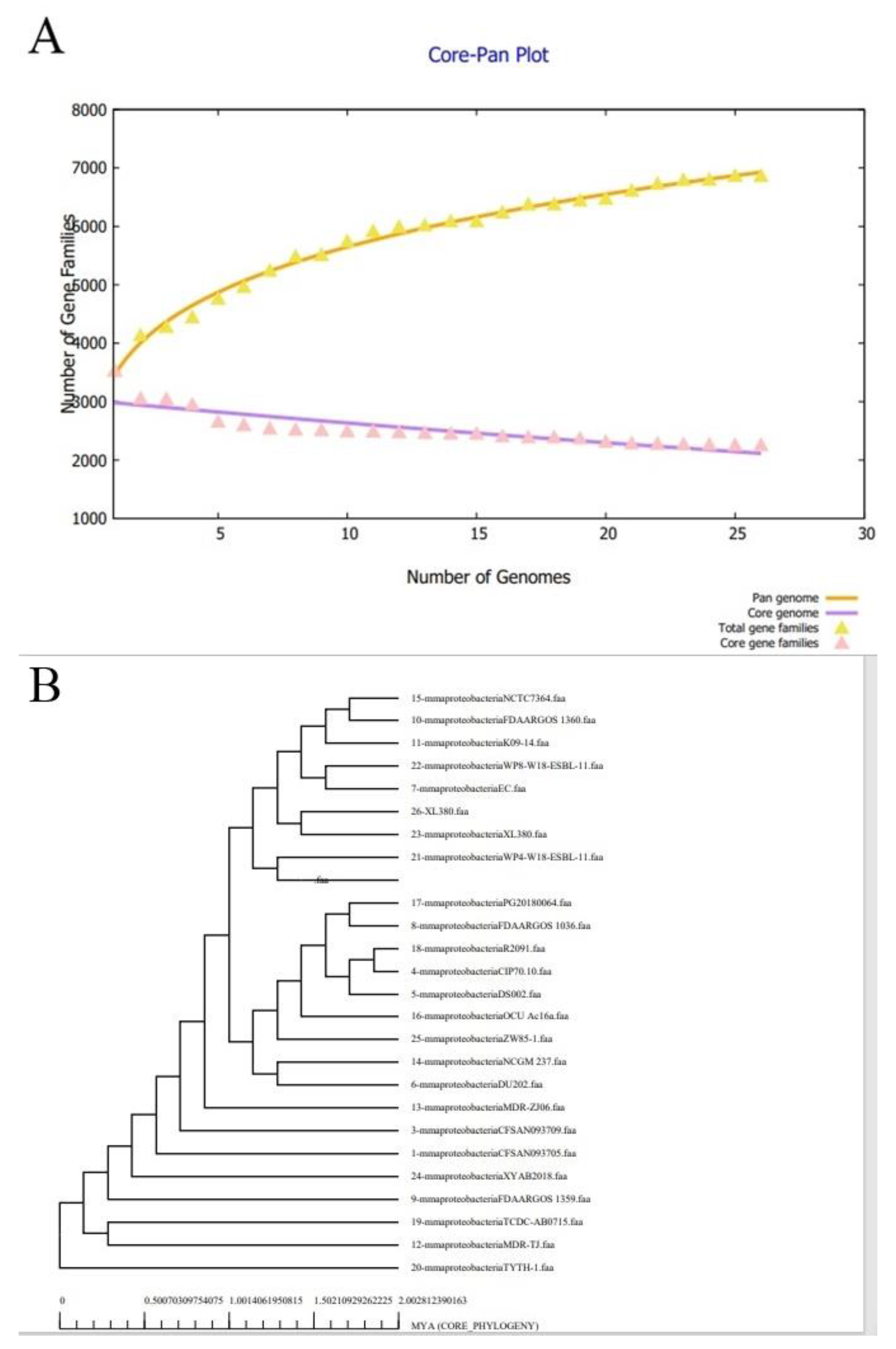
3.1. Bacterial Pan-Genome Analysis
3.2. CD HIT and PSORTB Analysis
3.3. Antigenicity, Allergenicity, Human and Normal Microbiota Similarity, and Transmembrane Helices and Stability Analysis
3.4. Physiochemical Properties
3.5. Prediction of B Cell Epitopes
3.6. Epitope Filtration Phase
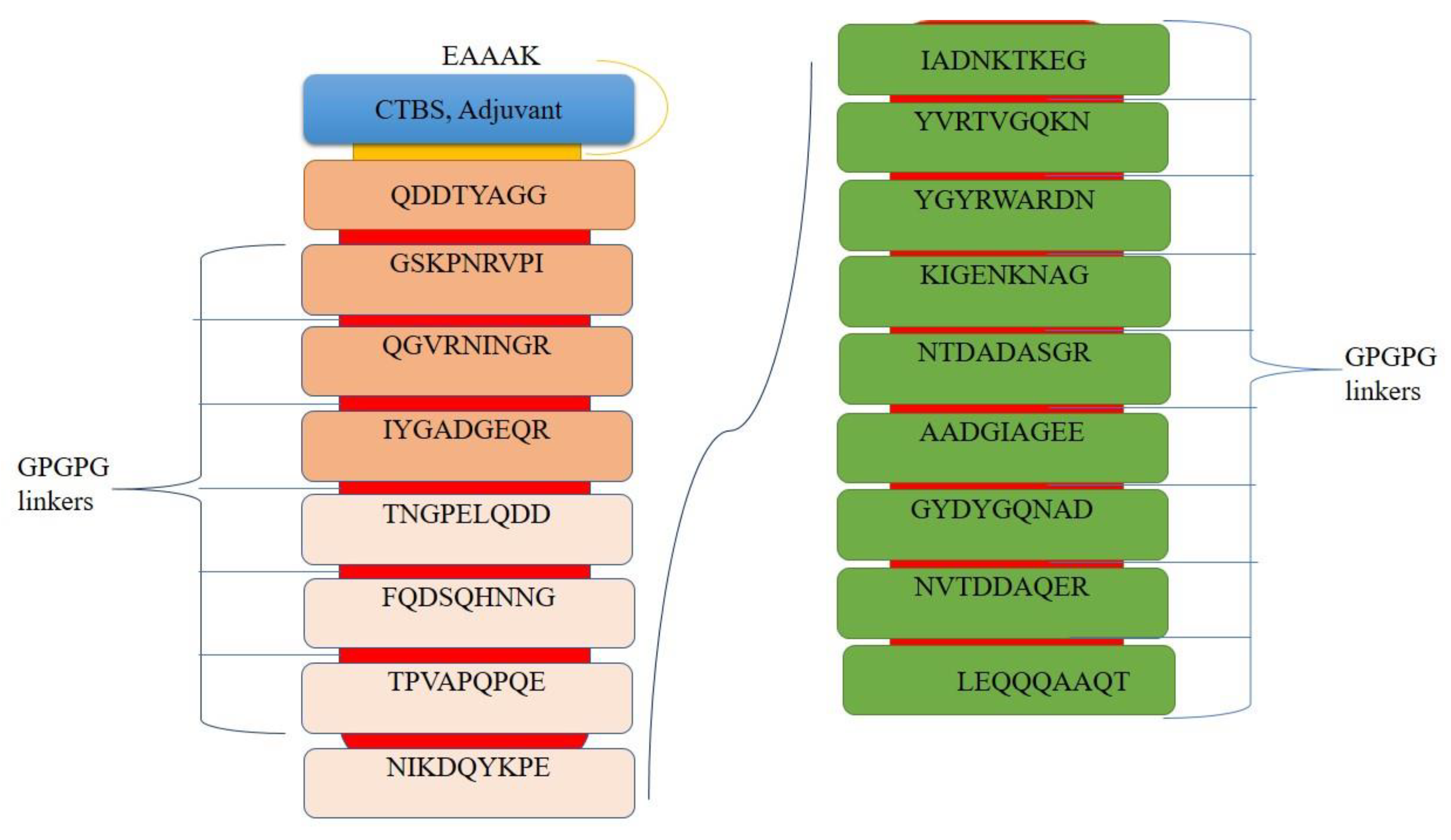
3.7. Multi-Epitopes Vaccine Construct Designing
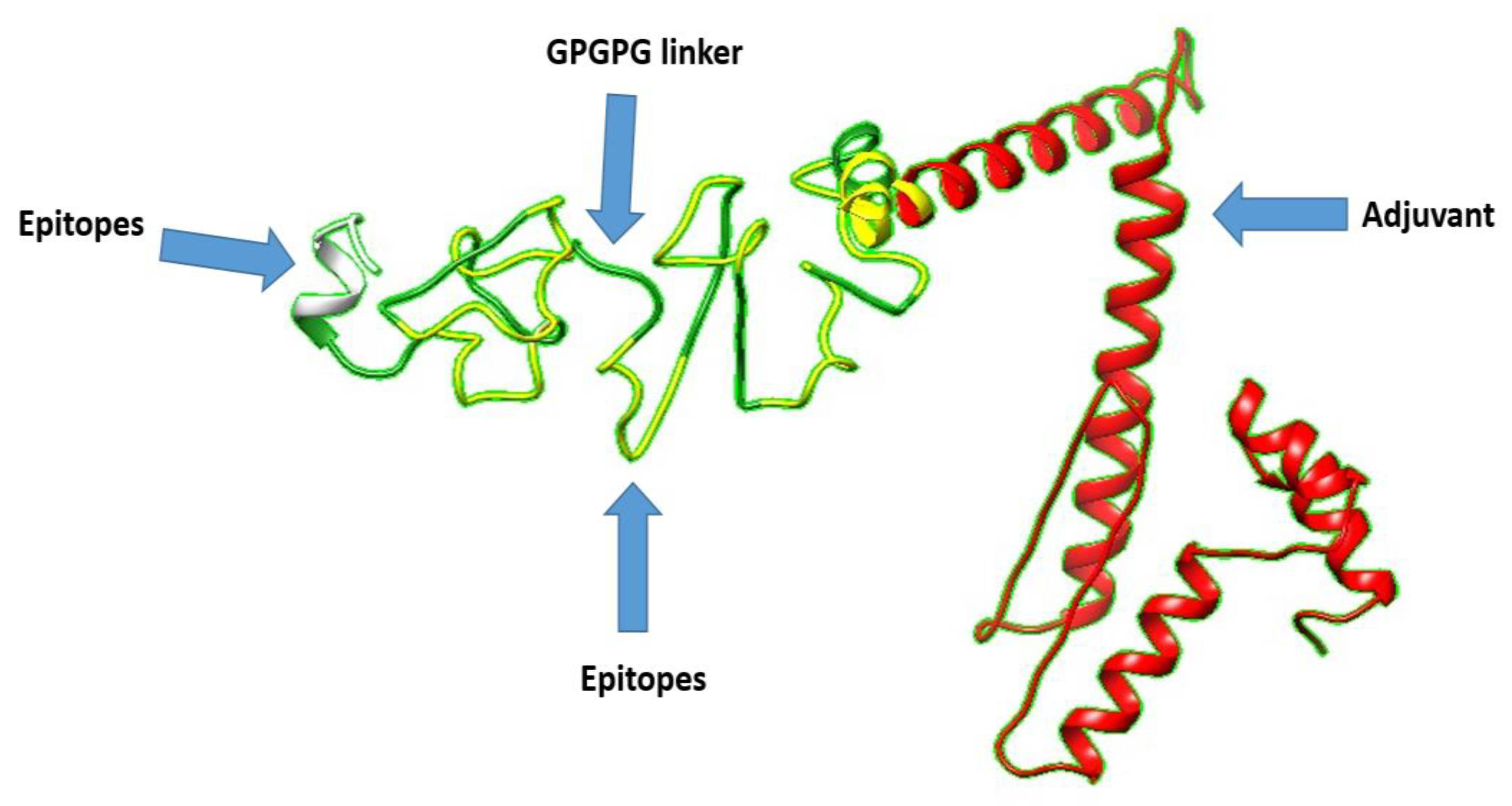
3.8. 3D Structure Prediction
3.9. Loops Modeling and Refinement
3.10. Disulfide Engineering
3.11. Codon Optimization
3.12. Molecular Docking
3.13. Refinement of Docked Complexes
3.14. Chemical Interactions of the Vaccine with MHC-I, MHC-II, and TLR-4
3.15. Molecular Dynamic Simulation
3.16. Calculation of Vaccine-Receptors Binding Energies
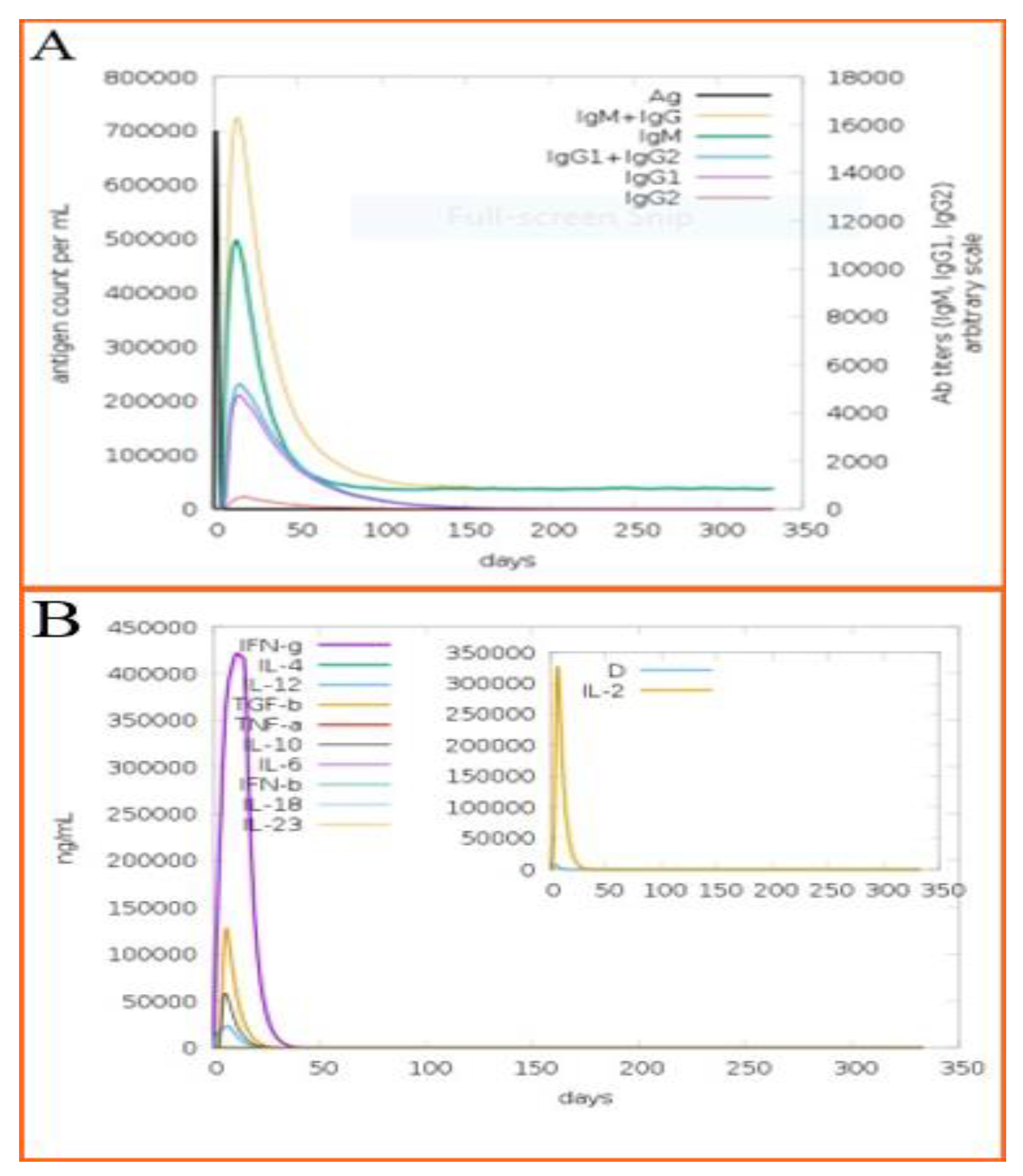
3.17. Immune Simulation of the Designed Vaccine
4. Discussion
5. Concluding Remarks and Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alanis, A.J. Resistance to antibiotics: Are we in the post-antibiotic era? Arch. Med. Res. 2005, 36, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global contributors to antibiotic resistance. J. Glob. Infect. Dis. 2019, 11, 36. [Google Scholar] [PubMed]
- Kulik, E.M.; Thurnheer, T.; Karygianni, L.; Walter, C.; Sculean, A.; Eick, S. Antibiotic susceptibility patterns of aggregatibacter actinomycetemcomitans and porphyromonas gingivalis strains from different decades. Antibiotics 2019, 8, 253. [Google Scholar] [CrossRef] [PubMed]
- PCAST. National Action Plan for Combatting Antibiotic-Resistant Bacteria; White House: Washington, DC, USA, 2015.
- Ventola, C.L. The antibiotic resistance crisis: Part 2: Management strategies and new agents. Pharm. Ther. 2015, 40, 344. [Google Scholar]
- Brooks, B.D.; Brooks, A.E. Therapeutic strategies to combat antibiotic resistance. Adv. Drug Deliv. Rev. 2014, 78, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, G. Strategies to Overcome Antimicrobial Resistance (AMR) Making Use of Non-Essential Target Inhibitors: A Review. Int. J. Mol. Sci. 2019, 20, 5844. [Google Scholar] [CrossRef]
- Yanfen, D.; Li, T.; Wan, Y.; Liao, P. Signal Molecule-Dependent Quorum-Sensing and Quorum-Quenching Enzymes in Bacteria. Crit. Rev. Eukaryot. Gene Expr. 2014, 24, 117–132. [Google Scholar]
- Hall, N. Advanced Sequencing Technologies and Their Wider Impact in Microbiology. J. Exp. Biol. 2007, 210, 1518–1525. [Google Scholar] [CrossRef]
- Afreenish, H.; Usman, J.; Kaleem, F.; Khan, A.; Hussain, Z. In Vitro Activity of Aminoglycosides, Lactam-Lactamases Inhibitor Combinations and Tetracyclines against Multi-Drug Resistant Acinetobacter Baumannii, Isolated from a Tertiary Care Hospital. J. Microbiol. Antimicrob. 2010, 2, 47–50. [Google Scholar]
- Kadri, S.S. Key Takeaways from the US CDC’s 2019 Antibiotic Resistance Threats Report for Frontline Providers. Crit. Care Med. 2020, 48, 939–945. [Google Scholar]
- Emmett, K.; Perkins, M.D.; Small, P.; Hanson, C.; Reed, S.; Cunningham, J.; Aledort, J.E.; Hillborne, L.; Rafael, M.E.; Girosi, F. Reducing the Global Burden of Tuberculosis: The Contribution of Improved Diagnostics. Nature 2006, 444, 49–57. [Google Scholar]
- Mobarki, N.; Almerabi, B.; Hattan, A. Antibiotic Resistance Crisis. Int. J. Med. Dev. Ctries 2019, 40, 561–564. [Google Scholar] [CrossRef]
- Oved, K. Fighting AMR with Host Immune Response Technology. Drug Discov. Today 2021, 26, 2081. [Google Scholar] [CrossRef] [PubMed]
- ProtParam. Available online: https://bio.tools/protparam (accessed on 21 April 2022).
- Mousumi, S.; Sarkar, A. Review on Multiple Facets of Drug Resistance: A Rising Challenge in the 21st Century. J. Xenobiotics 2021, 11, 197–214. [Google Scholar]
- Taylor, D. The Pharmaceutical Industry and the Future of Drug Development. 2015. Available online: https://pubs.rsc.org/en/content/chapterhtml/2015/bk9781782621898-00001?isbn=978-1-78262-189-8 (accessed on 22 April 2022).
- Lantian, Z.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory Mechanisms and Promising Applications of Quorum Sensing-Inhibiting Agents in Control of Bacterial Biofilm Formation. Front. Microbiol. 2020, 2558. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645. [Google Scholar] [CrossRef]
- Kennedy, D.A.; Read, A.F.; Read, A.F. Why does drug resistance readily evolve but vaccine resistance does not? Proc. R. Soc. B Biol. Sci. 2017, 284, 20162562. [Google Scholar] [CrossRef]
- Goldsby, R.A.; Kindt, T.J.; Osborne, B.A.; Kuby, J. Chapter 2: Cells and Organs of the Immune System. In Immunology, 5th ed.; W. H. Freeman and Company: New York, NY, USA, 2003; pp. 24–56. Available online: https://www.scirp.org/(S(351jmbntvnsjt1aadkposzje))/reference/ReferencesPapers.aspx?ReferenceID=1913594 (accessed on 22 April 2022).
- Plotkin, S. History of vaccination. Proc. Natl. Acad. Sci. USA 2014, 111, 12283–12287. [Google Scholar] [CrossRef]
- Moriel, D.G.; Scarselli, M.; Serino, L.; Mora, M.; Rappuoli, R.; Masignani, V. Genome-based vaccine development: A short cut for the future. Hum. Vaccines 2008, 4, 184–188. [Google Scholar] [CrossRef][Green Version]
- Baseer, S.; Ahmad, S.; Ranaghan, K.E.; Azam, S.S. Towards a peptide-based vaccine against Shigella sonnei: A subtractive reverse vaccinology based approach. Biologicals 2017, 50, 87–99. [Google Scholar] [CrossRef]
- Eliopoulos, G.M.; Maragakis, L.L.; Perl, T.M. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 2008, 46, 1254–1263. [Google Scholar]
- Gellings, P.S.; Wilkins, A.A.; Morici, L.A. Recent Advances in the Pursuit of an Effective Acinetobacter baumannii Vaccine. Pathogens 2020, 9, 1066. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [PubMed]
- Sanober, G.; Ahmad, S.; Azam, S.S. Identification of plausible drug targets by investigating the druggable genome of MDR Staphylococcus epidermidis. Gene Rep. 2017, 7, 147–153. [Google Scholar] [CrossRef]
- Naz, A.; Awan, F.M.; Obaid, A.; Muhammad, S.A.; Paracha, R.Z.; Ahmad, J.; Ali, A. Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: A reverse vaccinology based approach. Infect. Genet. Evol. 2015, 32, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Kumar Jaiswal, A.; Tiwari, S.; Jamal, S.B.; Barh, D.; Azevedo, V.; Soares, S.C. An in silico identification of common putative vaccine candidates against treponema pallidum: A reverse vaccinology and subtractive genomics based approach. Int. J. Mol. Sci. 2017, 18, 402. [Google Scholar] [CrossRef]
- Johri, S.; Solanki, J.; Cantu, V.A.; Fellows, S.R.; Edwards, R.A.; Moreno, I.; Vyas, A.; Dinsdale, E.A. ‘Genome skimming’ with the MinION hand-held sequencer identifies CITES-listed shark species in India’s exports market. Sci. Rep. 2019, 9, 4476. [Google Scholar] [CrossRef]
- Butt, A.M.; Tahir, S.; Nasrullah, I.; Idrees, M.; Lu, J.; Tong, Y. Mycoplasma genitalium: A comparative genomics study of metabolic pathways for the identification of drug and vaccine targets. Infect. Genet. Evol. 2012, 12, 53–62. [Google Scholar] [CrossRef]
- Sikic, K.; Carugo, O. Protein sequence redundancy reduction: Comparison of various method. Bioinformation 2010, 5, 234. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Rizwan, M.; Naz, A.; Ahmad, J.; Naz, K.; Obaid, A.; Parveen, T.; Ahsan, M.; Ali, A. VacSol: A high throughput in silico pipeline to predict potential therapeutic targets in prokaryotic pathogens using subtractive reverse vaccinology. BMC Bioinform. 2017, 18, 106. [Google Scholar] [CrossRef] [PubMed]
- Azam, S.S.; Shamim, A. An insight into the exploration of druggable genome of Streptococcus gordonii for the identification of novel therapeutic candidates. Genomics 2014, 104, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Ranaghan, K.E.; Azam, S.S. Combating tigecycline resistant Acinetobacter baumannii: A leap forward towards multi-epitope based vaccine discovery. Eur. J. Pharm. Sci. 2019, 132, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hara, Y.; Mohamed, R.; Nathan, S. Immunogenic Burkholderia pseudomallei outer membrane proteins as potential candidate vaccine targets. PLoS ONE 2009, 4, e6496. [Google Scholar] [CrossRef]
- Collins, B.S. Gram-negative outer membrane vesicles in vaccine development. Discov. Med. 2011, 12, 7–15. [Google Scholar]
- Rashid, M.I.; Naz, A.; Ali, A.; Andleeb, S. Prediction of vaccine candidates against Pseudomonas aeruginosa: An integrated genomics and proteomics approach. Genomics 2017, 109, 274–283. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Kaushik, D.K.; Sehgal, D. Developing antibacterial vaccines in genomics and proteomics era. Scand. J. Immunol. 2008, 67, 544–552. [Google Scholar] [CrossRef]
- Nain, Z.; Abdulla, F.; Rahman, M.M.; Karim, M.M.; Khan, M.S.A.; Sayed, S.B.; Mahmud, S.; Rahman, S.M.R.; Sheam, M.M.; Haque, Z. Proteome-wide screening for designing a multi-epitope vaccine against emerging pathogen Elizabethkingia anophelis using immunoinformatic approaches. J. Biomol. Struct. Dyn. 2020, 38, 4850–4867. [Google Scholar] [CrossRef]
- ExPASy. Available online: https://www.expasy.org/ (accessed on 22 April 2022).
- Guruprasad, K.; Reddy, B.V.B.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. Des. Sel. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Tusnady, G.E.; Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001, 17, 849–850. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, G.; Kumar, K.; Jain, P.; Ramachandran, S.; Ramachandran, S. SPAAN: A software for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics 2004, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Wadood, A.; Jamal, A.; Riaz, M.; Khan, A.; Uddin, R.; Jelani, M.; Azam, S.S. Subtractive genome analysis for in silico identification and characterization of novel drug targets in Streptococcus pneumonia strain JJA. Microb. Pathog. 2018, 115, 194–198. [Google Scholar] [CrossRef]
- He, Y.; Xiang, Z.; Mobley, H.L.T. Vaxign: The first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J. Biomed. Biotechnol. 2010, 2010, 297505. [Google Scholar] [CrossRef]
- Ali, A.; Naz, A.; Soares, S.C.; Bakhtiar, M.; Tiwari, S.; Hassan, S.S.; Hanan, F.; Ramos, R.; Pereira, U.; Barh, D. Pan-Genome analysis of human gastric pathogen H. pylori: Comparative genomics and pathogenomics approaches to identify regions associated with pathogenicity and prediction of potential core therapeutic targets. BioMed Res. Int. 2015, 2015, 139580. [Google Scholar] [CrossRef]
- Naz, K.; Naz, A.; Ashraf, S.T.; Rizwan, M.; Ahmad, J.; Baumbach, J.; Ali, A. PanRV: Pangenome-reverse vaccinology approach for identifications of potential vaccine candidates in microbial pangenome. BMC Bioinform. 2019, 20, 123. [Google Scholar] [CrossRef]
- Hassan, A.; Naz, A.; Obaid, A.; Paracha, R.Z.; Naz, K.; Awan, F.M.; Muhmmad, S.A.; Janjua, H.A.; Ahmad, J.; Ali, A. Pangenome and immuno-proteomics analysis of Acinetobacter baumannii strains revealed the core peptide vaccine targets. BMC Genom. 2016, 17, 732. [Google Scholar] [CrossRef]
- Vita, R.; Overton, J.A.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015, 43, D405–D412. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Travers, P.; Walport, M.; Shlomchik, M.J. The major histocompatibility complex and its functions. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Garg, A.; Gupta, D. VirulentPred: A SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinform. 2008, 9, 62. [Google Scholar] [CrossRef]
- Dimitrov, I.; Flower, D.R.; Doytchinova, I. AllerTOP-a server for in silico prediction of allergens. BMC Bioinform. 2013, 14, S4. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Joshi, M.D.; Singhania, S.; Ramsey, K.H.; Murthy, A.K. Peptide vaccine: Progress and challenges. Vaccines 2014, 2, 515–536. [Google Scholar] [CrossRef] [PubMed]
- Skwarczynski, M.; Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 2016, 7, 842–854. [Google Scholar] [CrossRef]
- Baldauf, K.J.; Royal, J.; Hamorsky, K.T.; Matoba, N. Cholera Toxin B: One Subunit with Many Pharmaceutical Applications. Toxins 2015, 7, 974–996. [Google Scholar] [CrossRef] [PubMed]
- ProtParam, K.E. ExPASy-ProtParam Tool. 2017. Available online: https://web.expasy.org/protparam/ (accessed on 1 April 2022).
- Cheng, J.; Randall, A.Z.; Sweredoski, M.J.; Baldi, P. SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Res. 2005, 33, W72–W76. [Google Scholar] [CrossRef] [PubMed]
- Giardine, B.; Riemer, C.; Hardison, R.C.; Burhans, R.; Elnitski, L.; Shah, P.; Zhang, Y.; Blankenberg, D.; Albert, I.; Taylor, J.; et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005, 15, 1451–1455. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Craig, D.B.; Dombkowski, A.A. Disulfide by Design 2.0: A web-based tool for disulfide engineering in proteins. BMC Bioinform. 2013, 14, 346. [Google Scholar] [CrossRef]
- Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D.C.; Jahn, D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005, 33, W526–W531. [Google Scholar] [CrossRef]
- Rapin, N.; Lund, O.; Castiglione, F. C-Immsim 10.1 Server. 2012. Available online: https://www.iac.cnr.it/~filippo/projects/c-immsim-online.html (accessed on 22 April 2022).
- Rapin, N.; Lund, O.; Bernaschi, M.; Castiglione, F. Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE 2010, 5, e9862. [Google Scholar] [CrossRef]
- Morris, G.M.; Lim-Wilby, M. Molecular docking. In Molecular Modeling of Proteins; Springer: Berlin/Heidelberg, Germany, 2008; pp. 365–382. [Google Scholar]
- Solanki, V.; Tiwari, M.; Tiwari, V. Prioritization of potential vaccine targets using comparative proteomics and designing of the chimeric multi-epitope vaccine against Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 5240. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Yamakawa, N.; Akashi-Takamura, S.; Miyake, K.; Shimizu, T. Structural analyses of human Toll-like receptor 4 polymorphisms D299G and T399I. J. Biol. Chem. 2012, 287, 40611–40617. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef]
- Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: Fast interaction refinement in molecular docking. Proteins Struct. Funct. Bioinforma. 2007, 69, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Andleeb, S.; Imtiaz-Ud-Din; Rauf, M.K.; Azam, S.S.; Badshah, A.; Sadaf, H.; Raheel, A.; Tahir, M.N.; Raza, S. A one-pot multicomponent facile synthesis of dihydropyrimidin-2(1: H)-thione derivatives using triphenylgermane as a catalyst and its binding pattern validation. RSC Adv. 2016, 6, 79651–79661. [Google Scholar] [CrossRef]
- Case, D.A.; Cerutti, D.S.; Cheateham, T.E.; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Greene, D.; Homeyer, N.; et al. AMBER16 Package, University of California, San Francisco. 2016. Available online: https://ambermd.org/doc12/Amber16.pdf (accessed on 22 April 2022).
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Antechamber: An accessory software package for molecular mechanical calculations. J. Am. Chem. Soc. 2001, 222, U403. [Google Scholar]
- Brice, A.R.; Dominy, B.N. Examining electrostatic influences on base-flipping: A comparison of TIP3P and GB solvent models. Commun. Comput. Phys. 2013, 13, 223–237. [Google Scholar] [CrossRef]
- Kerrigan, J.E. AMBER 10.0 Introductory Tutorial. 2009. Available online: https://ambermd.org/ (accessed on 1 April 2022).
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. The FF14SB force field. Amber 2014, 14, 29–31. [Google Scholar]
- Lavenda, B.H. Statistical Physics: A Probabilistic Approach; Courier Dover Publications: Mineola, NY, USA, 2016; ISBN 0486810313. [Google Scholar]
- Case, D.A.; Cheatham, T.E., III; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Lemak, A.S.; Balabaev, N.K. On the Berendsen thermostat. Mol. Simul. 1994, 13, 177–187. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Tahir ul Qamar, M.; Zhu, X.; Khan, M.S.; Xing, F.; Chen, L.L. Pan-genome: A promising resource for noncoding RNA discovery in plants. Plant Genome 2020, 13, e20046. [Google Scholar] [CrossRef]
- Ul Qamar, M.T.; Zhu, X.; Xing, F.; Chen, L.-L.L.; Tahir Ul Qamar, M.; Zhu, X.; Xing, F.; Chen, L.-L.L. ppsPCP: A plant presence/absence variants scanner and pan-genome construction pipeline. Bioinformatics 2019, 35, 4156–4158. [Google Scholar] [CrossRef]
- Araújo, C.L.; Alves, J.; Nogueira, W.; Pereira, L.C.; Gomide, A.C.; Ramos, R.; Azevedo, V.; Silva, A.; Folador, A. Prediction of new vaccine targets in the core genome of Corynebacterium pseudotuberculosis through omics approaches and reverse vaccinology. Gene 2019, 702, 36–45. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Kristensen, D.M.; Makarova, K.S.; Wolf, Y.I.; Koonin, E. V Microbial genome analysis: The COG approach. Brief. Bioinform. 2019, 20, 1063–1070. [Google Scholar] [CrossRef]
- Kardani, K.; Bolhassani, A.; Namvar, A. An overview of in silico vaccine design against different pathogens and cancer. Expert Rev. Vaccines 2020, 19, 699–726. [Google Scholar] [CrossRef]
- Dorosti, H.; Eslami, M.; Negahdaripour, M.; Ghoshoon, M.B.; Gholami, A.; Heidari, R.; Dehshahri, A.; Erfani, N.; Nezafat, N.; Ghasemi, Y. Vaccinomics approach for developing multi-epitope peptide pneumococcal vaccine. J. Biomol. Struct. Dyn. 2019, 37, 3524–3535. [Google Scholar] [CrossRef] [PubMed]
- Bragazzi, N.L.; Gianfredi, V.; Villarini, M.; Rosselli, R.; Nasr, A.; Hussein, A.; Martini, M.; Behzadifar, M. Vaccines meet big data: State-ofthe- Art and future prospects. From the classical 3is (“isolate-inactivate-inject”) vaccinology 1.0 to vaccinology 3.0, vaccinomics, and Beyond: A historical overview. Front. Public Health 2018, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.D.; Burton, D.R. Toward an AIDS vaccine. Science 2008, 320, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Longbrake, E.E.; Cantoni, C.; Chahin, S.; Cignarella, F.; Cross, A.H.; Piccio, L. Dimethyl fumarate induces changes in B-and T-lymphocyte function independent of the effects on absolute lymphocyte count. Mult. Scler. J. 2018, 24, 728–738. [Google Scholar] [CrossRef]
- Ahmad, B.; Ashfaq, U.A.; Rahman, M.; Masoud, M.S.; Yousaf, M.Z. Conserved B and T cell epitopes prediction of ebola virus glycoprotein for vaccine development: An immuno-informatics approach. Microb. Pathog. 2019, 132, 243–253. [Google Scholar] [CrossRef]
- Choga, W.-T.; Anderson, M.; Phinius, B.-B.; Mbangiwa, T.; Bell, T.-G.; Seatla, K.-K.; Musonda, R.-M.; Moyo, S.; Blackard, J.-T.; Gaseitsiwe, S. A25 Impact of polymorphism in the hepatitis B surface gene on human leukocyte antigen (HLA) class II. Virus Evol. 2019, 5, vez002–vez024. [Google Scholar] [CrossRef]
- Tahir ul Qamar, M.; Ahmad, S.; Fatima, I.; Ahmad, F.; Shahid, F.; Naz, A.; Abbasi, S.W.; Khan, A.; Mirza, M.U.; Ashfaq, U.A.; et al. Designing multi-epitope vaccine against Staphylococcus aureus by employing subtractive proteomics, reverse vaccinology and immuno-informatics approaches. Comput. Biol. Med. 2021, 132, 104389. [Google Scholar] [CrossRef]
- Ul Qamar, M.T.; Bari, A.; Adeel, M.M.; Maryam, A.; Ashfaq, U.A.; Du, X.; Muneer, I.; Ahmad, H.I.; Wang, J.; Tahir Ul Qamar, M.; et al. Peptide vaccine against chikungunya virus: Immuno-informatics combined with molecular docking approach. J. Transl. Med. 2018, 16, 298. [Google Scholar] [CrossRef]
- Ullah, A.; Ahmad, S.; Ismail, S.; Afsheen, Z.; Khurram, M.; Tahir ul Qamar, M.; AlSuhaymi, N.; Alsugoor, M.H.; Allemailem, K.S. Towards A Novel Multi-Epitopes Chimeric Vaccine for Simulating Strong Immune Responses and Protection against Morganella morganii. Int. J. Environ. Res. Public Health 2021, 18, 10961. [Google Scholar] [CrossRef]
- Naz, S.; Ahmad, S.; Abbasi, S.W.; Ismail, S.; Waseem, S.; Ul Qamar, M.T.; Almatroudi, A.; Ali, Z. Identification of immunodominant epitopes in allelic variants VK210 and VK247 of Plasmodium Vivax Circumsporozoite immunogen. Infect. Genet. Evol. 2021, 96, 105120. [Google Scholar] [CrossRef]
- Fatima, I.; Ahmad, S.; Abbasi, S.W.; Ashfaq, U.A.; Shahid, F.; ul Qamar, M.T.; Rehman, A.; Allemailem, K.S. Designing of a multi-epitopes-based peptide vaccine against rift valley fever virus and its validation through integrated computational approaches. Comput. Biol. Med. 2021, 141, 105151. [Google Scholar] [CrossRef] [PubMed]
- Jaydari, A.; Nazifi, N.; Forouharmehr, A. Computational design of a novel multi-epitope vaccine against Coxiella burnetii. Hum. Immunol. 2020, 81, 596–605. [Google Scholar] [CrossRef] [PubMed]
- MacLean, R.C.; San Millan, A. The evolution of antibiotic resistance. Science 2019, 365, 1082–1083. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-Resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Wareth, G.; Brandt, C.; Sprague, L.D.; Neubauer, H.; Pletz, M.W. Spatio-temporal distribution of Acinetobacter baumannii in Germany—A comprehensive systematic review of studies on resistance development in humans (2000–2018). Microorganisms 2020, 8, 375. [Google Scholar] [CrossRef]
- Bambini, S.; Rappuoli, R. The use of genomics in microbial vaccine development. Drug Discov. Today 2009, 14, 252–260. [Google Scholar] [CrossRef]
- Bidmos, F.A.; Siris, S.; Gladstone, C.A.; Langford, P.R. Bacterial Vaccine Antigen Discovery in the Reverse Vaccinology 2.0 Era: Progress and Challenges. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Mora, M.; Veggi, D.; Santini, L.; Pizza, M.; Rappuoli, R. Reverse vaccinology. Drug Discov. Today 2003, 8, 459–464. [Google Scholar] [CrossRef]
- Adu-Bobie, J.; Capecchi, B.; Serruto, D.; Rappuoli, R.; Pizza, M. Two years into reverse vaccinology. Vaccine 2003, 21, 605–610. [Google Scholar] [CrossRef]
- Vaishnav, N.; Gupta, A.; Paul, S.; John, G.J. Overview of computational vaccinology: Vaccine development through information technology. J. Appl. Genet. 2015, 56, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Albutti, A. An integrated computational framework to design a multi-epitopes vaccine against Mycobacterium tuberculosis. Sci. Rep. 2021, 11, 21929. [Google Scholar] [CrossRef] [PubMed]
- Maione, D.; Margarit, I.; Rinaudo, C.D.; Masignani, V.; Mora, M.; Scarselli, M.; Tettelin, H.; Brettoni, C.; Iacobini, E.T.; Rosini, R. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science 2005, 309, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz Çolak, Ç. Computational Design of a Multi-epitope Vaccine Against Clostridium chauvoei: An Immunoinformatics Approach. Int. J. Pept. Res. Ther. 2021, 27, 2639–2649. [Google Scholar] [CrossRef]
- Ul Qamar, M.T.; Shahid, F.; Aslam, S.; Ashfaq, U.A.; Aslam, S.; Fatima, I.; Fareed, M.M.; Zohaib, A.; Chen, L.-L. Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2. Infect. Dis. Poverty 2020, 9, 1–14. [Google Scholar] [CrossRef]
- Chiang, M.-H.; Sung, W.-C.; Lien, S.-P.; Chen, Y.-Z.; Lo, A.F.; Huang, J.-H.; Kuo, S.-C.; Chong, P. Identification of novel vaccine candidates against Acinetobacter baumannii using reverse vaccinology. Hum. Vaccin. Immunother. 2015, 11, 1065–1073. [Google Scholar] [CrossRef]
- Wintachai, P.; Phaonakrop, N.; Roytrakul, S.; Naknaen, A.; Pomwised, R.; Voravuthikunchai, S.P.; Surachat, K.; Smith, D.R. Enhanced antibacterial effect of a novel Friunavirus phage vWU2001 in combination with colistin against carbapenem-resistant Acinetobacter baumannii. Sci. Rep. 2022, 12, 2633. [Google Scholar] [CrossRef]
- Ayoub Moubareck, C.; Hammoudi Halat, D. Insights into Acinetobacter baumannii: A review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef]


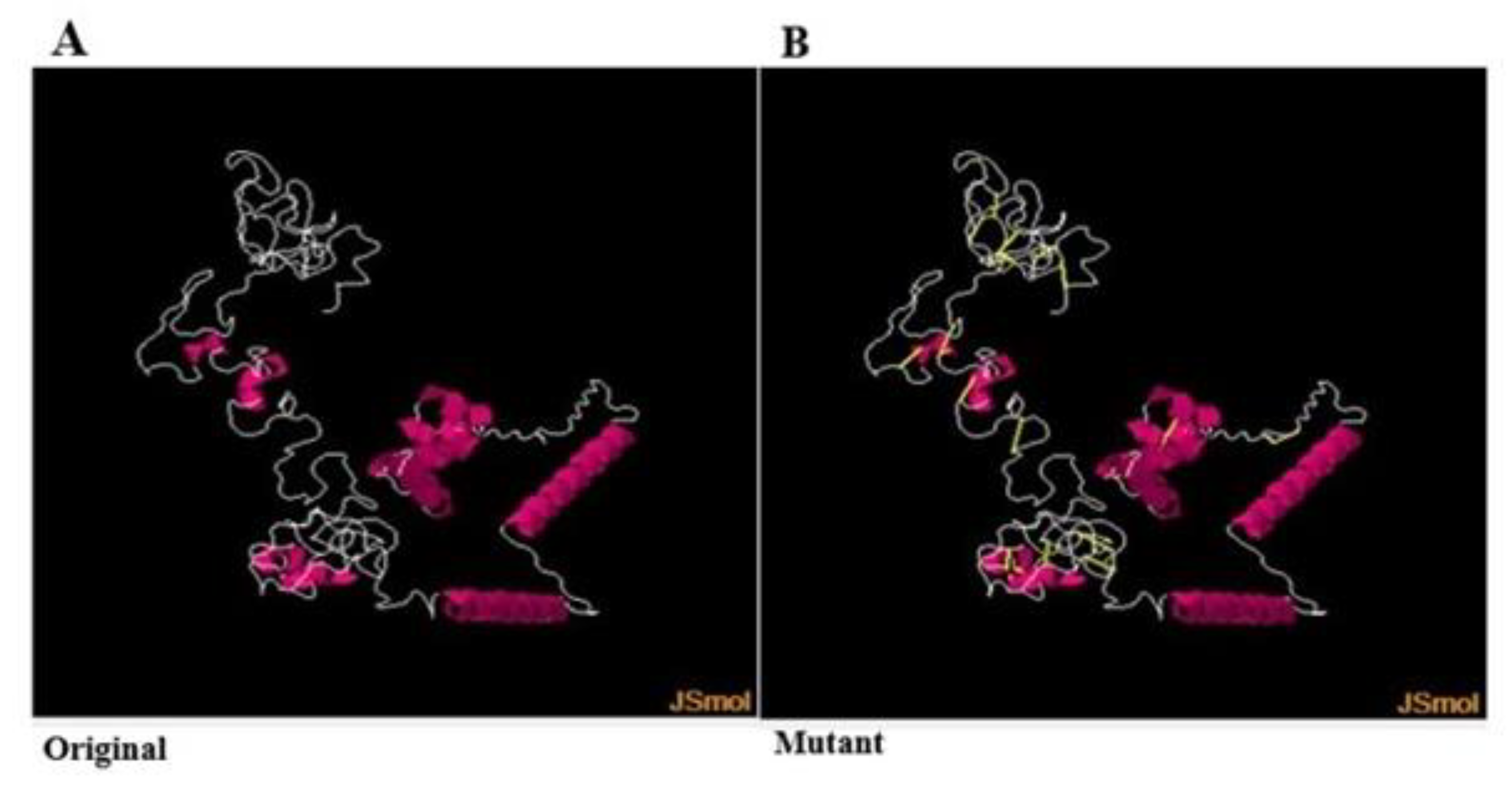


| Outer Membrane Proteins | Encoded Proteins | Amino Acid | Molecular Weight | Gravy | Aliphatic Index | Instability Index | Theoretical (PI) |
|---|---|---|---|---|---|---|---|
| >core/109/1/Org1_Gene719 | Type II toxin-antitoxin system antitoxin maze3 | 1071 | 115.65 | −0.336 | 87.06 | 30.04 | 9.35 |
| >core/298/1/Org1_Gene3311 | Methionine synthase [Mycobacterium tuberculosis | 775 | 84.63 | −0.391 | 76.68 | 28.67 | 8.58 |
| >core/315/1/Org1_Gene2327 | Hypothetical protein | 758 | 80.95 | −0.222 | 96.37 | 30.48 | 8.86 |
| >core/982/1/Org1_Gene2259 | MULTISPECIES: 1-acyl-sn-glycerol-3-phosphate acyltransferase | 484 | 52.26 | −0.311 | 92.02 | 39.59 | 9.02 |
| >core/1058/1/Org1_Gene66 | Nuclear transport factor 2 family protein [Mycobacterium tuberculosis | 471 | 48.39 | −0.015 | 100.81 | 32.8 | 5.59 |
| >core/2148/1/Org1_Gene1404 | Hypothetical protein K60_024290 [Mycobacterium tuberculosis variant bovis BCG str. Korea 1168P | 356 | 38.97 | −0.396 | 80.93 | 32 | 5.32 |
| >core/3446/1/Org1_Gene2677 | Chromosome segregation protein SMC | 266 | 30.44 | −0.415 | 85.23 | 41.44 | 9.01 |
| >core/4300/1/Org1_Gene2340 | NlpC/P60 family peptidoglycan endopeptidase RipB | 217 | 22.13 | −0.288 | 84.61 | 30.23 | 9.3 |
| >core/1212/7/Org7_Gene3542 | MULTISPECIES: zf-HC2 domain-containing protein [Mycobacterium tuberculosis complex | 451 | 49.38 | −0.42 | 70.35 | 21 | 5.66 |
| >core/3840/27/Org27_Gene62 | MULTISPECIES: hypothetical protein | 244 | 26.05 | −0.347 | 80.82 | 42.43 | 6.44 |
| >core/4074/16/Org16_Gene452 | Ribosomal protein S7 from Mycobacterium tuberculosis | 211 | 25.4 | 0.08 | 112.81 | 31.57 | 5.74 |
| Extracellular Proteins | |||||||
| >core/331/1/Org1_Gene3212 | DUF427 domain-containing protein | 742 | 83.32 | −0.556 | 69.46 | 35.45 | 6.15 |
| >core/5806/1/Org1_Gene1744 | D-alanyl-D-alanine carboxypeptidase | 143 | 15.5 | -0.09 | 84.06 | 25.08 | 5.52 |
| Periplasmic Proteins | |||||||
| >core/3114/1/Org1_Gene3080 | D-alanyl-D-alanine carboxypeptidase dacb2 | 288 | 31.25 | −0.318 | 95.14 | 19.93 | 5.93 |
| >core/466/2/Org2_Gene2561 | Glycine cleavage system aminomethyltransferase gcvt | 661 | 69.36 | −0.241 | 78.11 | 35.08 | 5.46 |
| >core/1121/3/Org3_Gene1840 | MULTISPECIES: multidrug efflux SMR transporter Mmr | 463 | 49.7 | −0.114 | 96.2 | 35.66 | 9.34 |
| >core/2579/16/Org16_Gene524 | Conserved protein of uncharacterised function | 324 | 36.57 | −0.223 | 92.96 | 30.15 | 5.81 |
| >core/4275/27/Org27_Gene2284 | Site-2 protease family protein [Mycobacterium tuberculosis | 219 | 24.1 | −0.401 | 76.8 | 21.59 | 5.88 |
| Epitopes | DRB*0101 Binder Score | Antigenicity | Allergenicity | Water Solubility | Toxicity |
|---|---|---|---|---|---|
| LEQQQAAQT | 7.635 | Probable antigenic | Non-Allergic | Good water soluble | Non-toxic |
| QDDTYAGGQ | 12.74 | ||||
| GSKPNRVPI | 13.55 | ||||
| QGVRNINGR | 17.02 | ||||
| IYGADGEQR | 63.53 | ||||
| TNGPELQDD | 41.78 | ||||
| FQDSQHNNG | 5.85 | ||||
| TPVAPQPQE | 52.97 | ||||
| NIKDQYKPE | 22.86 | ||||
| IADNKTKEG | 77.62 | ||||
| YVRTVGQKN | 47.32 | ||||
| YGYRWARDN | 6.41 | ||||
| KIGENKNAG | 83.18 | ||||
| NTDADASGR | 3.29 | ||||
| AADGIAGEE | 36.73 | ||||
| GYDYGQNAD | 27.42 | ||||
| NVTDDAQER | 1.17 |
| Rank | Solution Number | Global Energy | Attractive VdW | Repulsive VdW | ACE | HB |
|---|---|---|---|---|---|---|
| 1 | 7 | −17.35 | −6.20 | −1.65 | −4.99 | −1.25 |
| 2 | 5 | 25.32 | −33.83 | 17.69 | 13.88 | −0.26 |
| 3 | 9 | 436.06 | −52.43 | 675.26 | −3.37 | −6.49 |
| 4 | 1 | 1559.57 | −83.37 | 2066.11 | 15.99 | −8.89 |
| 5 | 6 | 2178.22 | −50.99 | 2740.59 | 29.10 | −11.89 |
| 6 | 10 | 4719.15 | −72.39 | 6014.70 | 16.31 | −8.98 |
| 7 | 8 | 6838.22 | −94.58 | 8700.09 | 6.24 | −14.44 |
| 8 | 3 | 13,952.77 | −95.41 | 17,578.41 | 16.24 | −17.85 |
| 9 | 4 | 13,994.61 | −146.04 | 17,861.02 | −19.01 | −27.64 |
| 10 | 2 | 22,692.17 | −198.31 | 28,778.51 | 3.39 | −32.98 |
| Rank | Solution Number | Global Energy | Attractive VdW | Repulsive VdW | ACE | HB |
|---|---|---|---|---|---|---|
| 1 | 8 | −2.99 | −4.29 | 1.49 | 4.17 | −1.27 |
| 2 | 7 | 7.20 | −1.75 | 0.00 | 1.92 | 0.00 |
| 3 | 3 | 20.83 | −4.65 | 5.59 | 1.20 | −0.27 |
| 4 | 9 | 23.60 | −5.65 | 0.78 | 4.63 | −0.30 |
| 5 | 5 | 35.96 | −25.25 | 36.41 | 14.29 | −3.90 |
| 6 | 10 | 164.06 | −15.38 | 199.72 | 13.40 | −1.43 |
| 7 | 4 | 1426.58 | −34.55 | 1827.26 | 1.63 | −2.65 |
| 8 | 6 | 2708.43 | −65.54 | 3541.68 | −9.12 | −2.25 |
| 9 | 1 | 7877.71 | −87.43 | 9990.68 | 12.16 | −9.37 |
| 10 | 2 | 10,558.78 | −89.75 | 13,391.88 | −3.44 | −16.32 |
| Rank | Solution Number | Global Energy | Attractive VdW | Repulsive VdW | ACE | HB |
|---|---|---|---|---|---|---|
| 1 | 2 | −3.22 | −26.77 | 7.43 | 21.39 | −2.28 |
| 2 | 7 | 7.57 | −2.84 | 0.00 | 1.28 | 0.00 |
| 3 | 9 | 10.66 | −15.81 | 32.02 | 2.23 | −1.76 |
| 4 | 10 | 28.75 | −11.25 | 3.62 | 13.23 | −0.50 |
| 5 | 6 | 29.86 | −11.03 | 1.79 | 10.45 | −1.72 |
| 6 | 1 | 915.94 | −48.21 | 1190.12 | 17.81 | −4.33 |
| 7 | 5 | 3386.99 | −52.35 | 4311.65 | 8.99 | −5.38 |
| 8 | 4 | 4737.61 | −121.11 | 6190.84 | −3.73 | −16.92 |
| 9 | 3 | 7186.19 | −93.88 | 9131.91 | 16.17 | −11.81 |
| 10 | 8 | 7763.02 | −85.23 | 9873.19 | 2.20 | −7.86 |
| Vaccine Complex | Interactive Residues |
|---|---|
| MHC-I | Asn346, Asn347, Arg131, Glu58, Glu60, Glu161, Glu53, Ile 46, Lys127, Leu 126, Met138, Pro57, Ser132, Try135, Thr124, Leu64, Phe8, Try50, Try133 |
| MHC-II | Ile187, Ser218, Ser240, Val203 |
| TLR-4 | Asn58, Asp84, Asn176, Asn173, Asn44, Cys 29, Glu42, Arg69, Asn112, Asp84, Arg67, Cys40, Cys148, Cys29, Tyr70, Leu17, Asp35, Thr92, Gln91, Glu136, Gln91, Glu31, Glu179, Glu143, Glu144, Glu111, Glu178, Glu31, His179, His8, Ile138, Tyr46, Glu89, Ile93, Lys108, Pro28, Pro113, Phe64, Phe147, Pro145, Pro142, Phe77, Pro78, Pro78, Leu74, Met40, Ser172, Ser141, Ser141, Ser126, Thr110, Leu85, Lys47, Lys30, Lys130, Tyr46, Leu66, Thr15, Leu152, Lys153, Ser105, Thr106, Lys57, Thr56, Thr37, Tyr38, Thr37, Val133, Thr37, Val35, Val30, Pro28, Val35, Val30. |
| Energy Parameter | TLR-4-Vaccine Complex | MHC-I-Vaccine Complex | MHC-II-Vaccine Complex |
|---|---|---|---|
| MM-GBSA | |||
| VDWAALS | −97.12 | −106.08 | −107 |
| EEL | −105.68 | −85.68 | −88.07 |
| EGB | 48.01 | 32.39 | 28.24 |
| ESURF | −15.04 | −19.25 | −15.11 |
| Delta G gas | −202.8 | −191.76 | −195.07 |
| Delta G solv | 32.97 | 13.14 | 13.13 |
| Delta Total | −169.83 | −178.62 | −181.94 |
| MM-PBSA | |||
| VDWAALS | −97.12 | −98.36 | −117.66 |
| EEL | −105.68 | −88.07 | −54.05 |
| EPB | 43.25 | 23 | 28.28 |
| ENPOLAR | −9 | −14.19 | −17.1 |
| Delta G gas | −202.8 | −191.76 | −195.07 |
| Delta G solv | 34.25 | 8.81 | 11.18 |
| Delta Total | −168.55 | −182.95 | −183.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
ud-din, M.; Albutti, A.; Ullah, A.; Ismail, S.; Ahmad, S.; Naz, A.; Khurram, M.; Haq, M.u.; Afsheen, Z.; Bakri, Y.E.; et al. Vaccinomics to Design a Multi-Epitopes Vaccine for Acinetobacter baumannii. Int. J. Environ. Res. Public Health 2022, 19, 5568. https://doi.org/10.3390/ijerph19095568
ud-din M, Albutti A, Ullah A, Ismail S, Ahmad S, Naz A, Khurram M, Haq Mu, Afsheen Z, Bakri YE, et al. Vaccinomics to Design a Multi-Epitopes Vaccine for Acinetobacter baumannii. International Journal of Environmental Research and Public Health. 2022; 19(9):5568. https://doi.org/10.3390/ijerph19095568
Chicago/Turabian Styleud-din, Miraj, Aqel Albutti, Asad Ullah, Saba Ismail, Sajjad Ahmad, Anam Naz, Muhammad Khurram, Mahboob ul Haq, Zobia Afsheen, Youness El Bakri, and et al. 2022. "Vaccinomics to Design a Multi-Epitopes Vaccine for Acinetobacter baumannii" International Journal of Environmental Research and Public Health 19, no. 9: 5568. https://doi.org/10.3390/ijerph19095568
APA Styleud-din, M., Albutti, A., Ullah, A., Ismail, S., Ahmad, S., Naz, A., Khurram, M., Haq, M. u., Afsheen, Z., Bakri, Y. E., Salman, M., Shaker, B., & Tahir ul Qamar, M. (2022). Vaccinomics to Design a Multi-Epitopes Vaccine for Acinetobacter baumannii. International Journal of Environmental Research and Public Health, 19(9), 5568. https://doi.org/10.3390/ijerph19095568










