COVID-19 Mortality in the Colorado Center for Personalized Medicine Biobank
Abstract
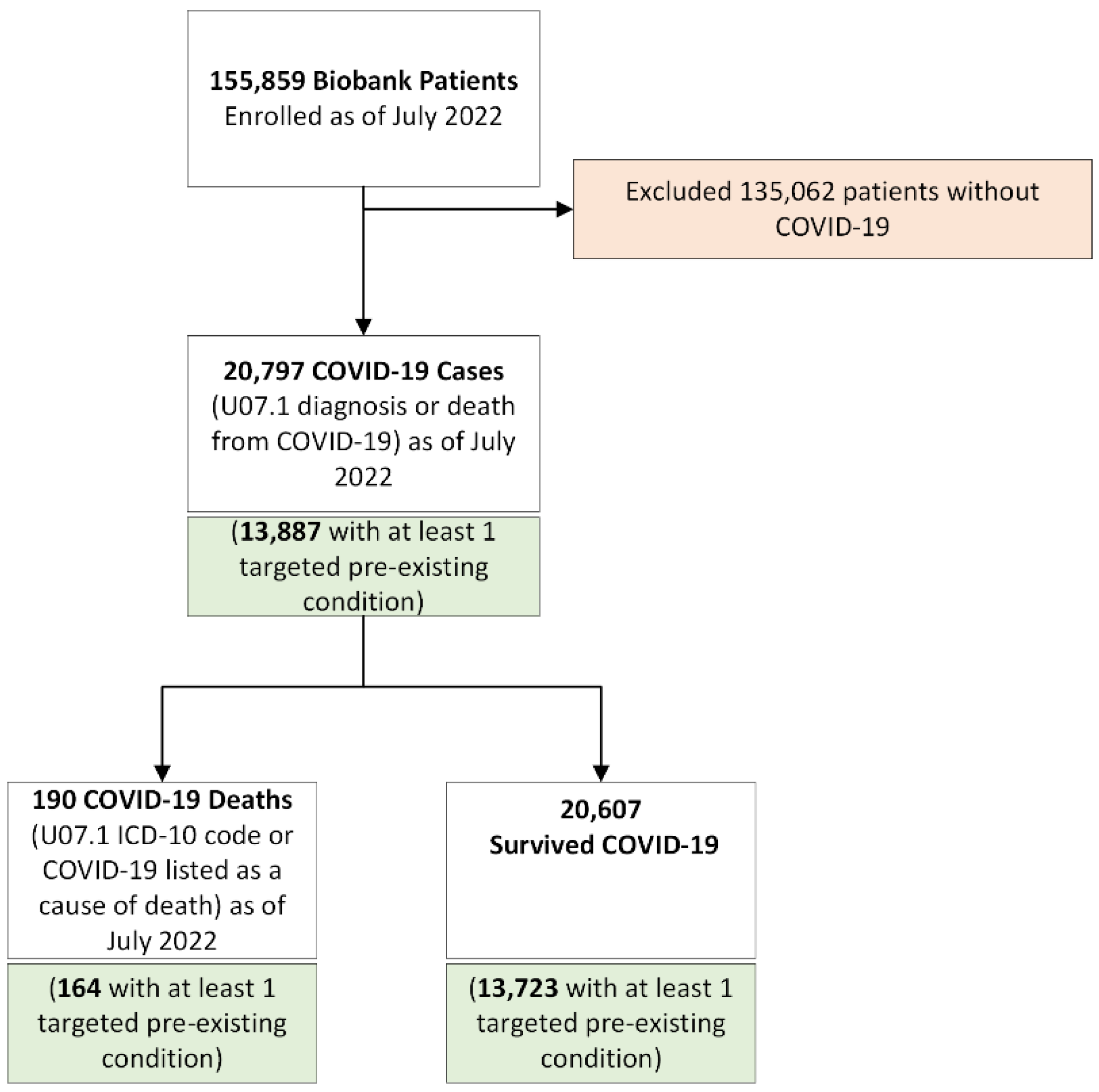
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Pre-Existing Conditions
2.3. Genotyping
2.4. Mortality and Other Covariates
2.5. Statistical Analyses
3. Results
3.1. Descriptive Statistics
3.2. Mortality and Case Fatality Rates
3.3. Multiple Logistic Regression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Johns Hopkins University & Medicine. Mortality Analyses. Available online: https://coronavirus.jhu.edu/data/mortality (accessed on 20 January 2023).
- The Sex, Gender and COVID-19 Project. Men, Sex, Gender, and COVID-19. Available online: https://globalhealth5050.org/the-sex-gender-and-covid-19-project/men-sex-gender-and-covid-19/ (accessed on 20 January 2023).
- Sisk, B.; Cull, W.; Harris, J.M.; Rothenburger, A.; Olson, L. National Trends of Cases of COVID-19 in Children Based on US State Health Department Data. Pediatrics 2020, 146, e2020027425. [Google Scholar] [CrossRef] [PubMed]
- CDC COVID-19 Response Team; Bialek, S.; Boundy, E.; Bowen, V.; Chow, N.; Chon, A.; Dowling, N.; Ellington, S.; Gierke, R.; Hall, A.; et al. Severe Outcomes Among Patients with Coronavirus Disease 2019 (COVID-19)—United States, February 12-March 16, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 343–346. [Google Scholar] [CrossRef]
- Renelus, B.D.; Khoury, N.C.; Chandrasekaran, K.; Bekele, E.; Briggs, W.M.; Ivanov, A.; Mohanty, S.R.; Jamorabo, D.S. Racial Disparities in COVID-19 Hospitalization and In-hospital Mortality at the Height of the New York City Pandemic. J. Racial Ethn. Health Disparities 2021, 8, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Zhonghua, L.; Xing, B.; Zhi, Z. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi 2020, 41, 145–151. [Google Scholar] [CrossRef]
- Kastora, S.; Patel, M.; Carter, B.; Delibegovic, M.; Myint, P.K. Impact of diabetes on COVID-19 mortality and hospital outcomes from a global perspective: An umbrella systematic review and meta-analysis. Endocrinol. Diabetes Metab. 2022, 5, e00338. [Google Scholar] [CrossRef]
- Başı, N.B.; Metin, S.; Sevinç, S.A.; Peker, N.; Çınar, A.S.; Salkaya, A.; Altuntaş, Y.; Özdemir, H.M. The Effect of Diabetes Mellitus on Mortality in Patients Hospitalized Intensive Care Unit in Covid-19 Pandemic. Acta Biomed. 2022, 93, e2022068. [Google Scholar] [CrossRef]
- Tian, W.; Jiang, W.; Yao, J.; Nicholson, C.J.; Li, R.H.; Sigurslid, H.H.; Wooster, L.; Rotter, J.I.; Guo, X.; Malhotra, R. Predictors of mortality in hospitalized COVID-19 patients: A systematic review and meta-analysis. J. Med. Virol. 2020, 92, 1875–1883. [Google Scholar] [CrossRef]
- Dai, L.S.; Zhu, M.P.; Li, Y.M.; Zhou, H.M.; Liao, H.L.; Cheng, P.P.; Xia, X.Y.; Yao, X.Y.; Zhang, H.J.; Liu, X.Q.; et al. Hypertension Exacerbates Severity and Outcomes of COVID-19 in Elderly Patients: A Retrospective Observational Study. Curr. Med. Sci. 2022, 42, 561–568. [Google Scholar] [CrossRef]
- Parohan, M.; Yaghoubi, S.; Seraji, A.; Javanbakht, M.H.; Sarraf, P.; Djalali, M. Risk factors for mortality in patients with Coronavirus disease 2019 (COVID-19) infection: A systematic review and meta-analysis of observational studies. Aging Male 2020, 23, 1416–1424. [Google Scholar] [CrossRef]
- Kaiser, J. Found: Genes that sway the course of the coronavirus. Science 2020, 370, 275–276. [Google Scholar] [CrossRef]
- Lehrer, S.; Rheinstein, P.H. ABO blood groups, COVID-19 infection and mortality. Blood Cells Mol. Dis. 2021, 89, 102571. [Google Scholar] [CrossRef] [PubMed]
- Wiley, L.K.; Shortt, J.A.; Roberts, E.R.; Lowery, J.; Kudron, E.; Lin, M.; Mayer, D.A.; Wilson, M.P.; Brunetti, T.M.; Chavan, S.; et al. Building a Vertically-Integrated Genomic Learning Health System: The Colorado Center for Personalized Medicine Biobank. medRxiv 2022. [Google Scholar] [CrossRef]
- Johnson, R.K.; Marker, K.M.; Mayer, D.; Shortt, J.; Kao, D.; Barnes, K.C.; Lowery, J.T.; Gignoux, C.R. COVID-19 Surveillance in the Biobank at the Colorado Center for Personalized Medicine: Observational Study. JMIR Public Health Surveill. 2022, 8, e37327. [Google Scholar] [CrossRef]
- Carroll, R.J.; Bastarache, L.; Denny, J.C. R PheWAS: Data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics 2014, 30, 2375–2376. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, M.; Gu, T.; Mack, J.A.; Sankar, S.P.; Patil, S.; Valley, T.S.; Singh, K.; Nallamothu, B.K.; Kheterpal, S.; Lisabeth, L.; et al. A phenome-wide association study (PheWAS) of COVID-19 outcomes by race using the electronic health records data in Michigan Medicine. J. Clin. Med. 2021, 10, 1351. [Google Scholar] [CrossRef]
- Taliun, D.; Harris, D.N.; Kessler, M.D.; Carlson, J.; Szpiech, Z.A.; Torres, R.; Taliun, S.A.G.; Corvelo, A.; Gogarten, S.M.; Kang, H.M.; et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 2021, 590, 290–299. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef]
- McLachlan, S.; Giambartolomei, C.; White, J.; Charoen, P.; Wong, A.; Finan, C.; Engmann, J.; Shah, T.; Hersch, M.; Podmore, C.; et al. Replication and Characterization of Association between ABO SNPs and Red Blood Cell Traits by Meta-Analysis in Europeans. PLoS ONE 2016, 11, e0156914. [Google Scholar] [CrossRef]
- Danielsen, A.C.; Lee, K.M.N.; Boulicault, M.; Rushovich, T.; Gompers, A.; Tarrant, A.; Reiches, M.; Shattuck-Heidorn, H.; Miratrix, L.W.; Richardson, S.S. Sex disparities in COVID-19 outcomes in the United States: Quantifying and contextualizing variation. Soc. Sci. Med. 2022, 294, 114716. [Google Scholar] [CrossRef]
- U.S. Bureau of Labor Statistics. Household Data Annual Averages: Employed Persons by Detailed Occupation, Sex, Race, and Hispanic or Latino Ethnicity; U.S. Bureau of Labor Statistics: Washington, DC, USA, 2020. [Google Scholar]
- Ahmed, F.; Cisewski, J. Quarterly Provisional Estimates for Selected Indicators of Mortality, 2019-Quarter 3, 2021; US Department of Health and Human Services: Atlanta, GA, USA, 2021. [Google Scholar]
- CDPHE. Vital Statistics Program. Colorado Births and Deaths 2020; Colorado Department of Public Health and Environment: Denver, CO, USA, 2020. [Google Scholar]
- Mohammed, I.; Nauman, A.; Paul, P.; Ganesan, S.; Chen, K.-H.; Jalil, S.M.S.; Jaouni, S.H.; Kawas, H.; Khan, W.A.; Vattoth, A.L.; et al. The efficacy and effectiveness of the COVID-19 vaccines in reducing infection, severity, hospitalization, and mortality: A systematic review. Hum. Vaccines Immunother. 2022, 18, 2027160. [Google Scholar] [CrossRef] [PubMed]
- Aljuaid, M.; Alotair, H.; Alnajjar, F.; Alonazi, W.; Sharaf, H.; Sheshah, E.; Alashgar, L.; Alshaikh, M. Risk factors associated with in-hospital mortality patients with COVID-19 in Saudi Arabia. PLoS ONE 2022, 17, e0270062. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, S.; Turgutalp, K.; Arici, M.; Odabas, A.R.; Altiparmak, M.R.; Aydin, Z.; Cebeci, E.; Basturk, T.; Soypacaci, Z.; Sahin, G.; et al. Mortality analysis of COVID-19 infection in chronic kidney disease, haemodialysis and renal transplant patients compared with patients without kidney disease: A nationwide analysis from Turkey. Nephrol. Dial. Transpl. 2020, 35, 2083–2095. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Ludwig, J.; Howell, T. A retrospective cohort study on COVID-19 at 2 Los Angeles hospitals: Older age, low triage oxygenation, and chronic kidney disease among the top risk factors associated with in-hospital mortality. PLoS ONE 2022, 17, e0268688. [Google Scholar] [CrossRef]
- Perico, L.; Benigni, A.; Casiraghi, F.; Ng, L.F.P.; Renia, L.; Remuzzi, G. Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat. Rev. Nephrol. 2021, 17, 46–64. [Google Scholar] [CrossRef]

| Death from COVID-19 (N = 190) | Survived COVID-19 (N = 20,607) | COVID-19 Cases (N = 20,797) | p-Value * | Biobank (N = 155,859) | |
|---|---|---|---|---|---|
| Age, mean (SD) | 69.6 (12.9) | 50.8 (16.0) | 51.0 | <0.001 | 52.6 (16.7) |
| Age (years), N (%) | <0.001 | ||||
| 18–49 | 16 (8.42) | 10,181 (49.4) | 10,197 | 70,564 (45.3) | |
| 50–64 | 42 (22.1) | 5562 (27.0) | 5604 | 40,275 (25.8) | |
| 65–79 | 83 (43.7) | 4167 (20.2) | 4250 | 37,710 (24.2) | |
| 80+ | 49 (25.8) | 697 (3.4) | 746 | 7310 (4.7) | |
| Race/ethnicity, N (%) | 0.07 | ||||
| Non-Hispanic White | 138 (72.6) | 16,025 (77.8) | 16,163 | 125,670 (80.6) | |
| Non-Hispanic Black | 14 (7.4) | 869 (4.2) | 883 | 6359 (4.1) | |
| Hispanic | 30 (15.8) | 2597 (12.6) | 2627 | 14,065 (9.0) | |
| Other | 8 (4.2) | 1116 (5.4) | 1124 | 9765 (6.3) | |
| Sex, N (%) | <0.001 | ||||
| Male | 113 (59.5) | 7282 (35.3) | 7395 | 61,348 (39.4) | |
| Female | 77 (40.5) | 13,325 (64.7) | 13,402 | 94,511 (60.6) | |
| Pre-existing conditions, N (%) | |||||
| Cardiovascular Disease | 152 (80.0) | 9799 (47.6) | 9951 | <0.001 | 63,518 (40.8) |
| Diabetes | 73 (38.4) | 3435 (16.7) | 3508 | <0.001 | 20,862 (13.4) |
| Respiratory disease | 118 (62.1) | 9414 (45.7) | 9532 | <0.001 | 50,230 (32.2) |
| Blood type, N (%) | <0.001 ** | ||||
| A | 32 (16.8) | 1933 (9.4) | 1965 | 13,378 (8.6) | |
| O | 26 (13.7) | 1838 (8.9) | 1864 | 13,262 (8.5) | |
| AB | <10 | -- | -- | 1363 (0.9) | |
| B | <10 | -- | -- | 3479 (2.2) | |
| Not genotyped | 125 (65.8) | 16,177 (78.5) | 16,302 | 124,377 (79.8) |
| Rate per 100,000, (N) | |
|---|---|
| Biobank | |
| All-cause mortality | 3442 (5334) |
| Cause-specific mortality | |
| COVID-19 | 122 (190) |
| Cardiovascular | 318 (495) |
| Diabetes | 68 (106) |
| Respiratory | 138 (215) |
| Other * | 2777 (4328) |
| Had COVID-19 | Died from COVID-19 | Case Fatality (%) | |
|---|---|---|---|
| Biobank population | 20,797 | 190 | 0.91 |
| Pre-existing condition | |||
| Cardiovascular | 9799 | 152 | 1.55 |
| Diabetes | 3435 | 73 | 2.13 |
| Respiratory | 9414 | 118 | 1.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brice, A.N.; Vanderlinden, L.A.; Marker, K.M.; Mayer, D.; Lin, M.; Rafaels, N.; Shortt, J.A.; Romero, A.; Lowery, J.T.; Gignoux, C.R.; et al. COVID-19 Mortality in the Colorado Center for Personalized Medicine Biobank. Int. J. Environ. Res. Public Health 2023, 20, 2368. https://doi.org/10.3390/ijerph20032368
Brice AN, Vanderlinden LA, Marker KM, Mayer D, Lin M, Rafaels N, Shortt JA, Romero A, Lowery JT, Gignoux CR, et al. COVID-19 Mortality in the Colorado Center for Personalized Medicine Biobank. International Journal of Environmental Research and Public Health. 2023; 20(3):2368. https://doi.org/10.3390/ijerph20032368
Chicago/Turabian StyleBrice, Amanda N., Lauren A. Vanderlinden, Katie M. Marker, David Mayer, Meng Lin, Nicholas Rafaels, Jonathan A. Shortt, Alex Romero, Jan T. Lowery, Christopher R. Gignoux, and et al. 2023. "COVID-19 Mortality in the Colorado Center for Personalized Medicine Biobank" International Journal of Environmental Research and Public Health 20, no. 3: 2368. https://doi.org/10.3390/ijerph20032368
APA StyleBrice, A. N., Vanderlinden, L. A., Marker, K. M., Mayer, D., Lin, M., Rafaels, N., Shortt, J. A., Romero, A., Lowery, J. T., Gignoux, C. R., & Johnson, R. K. (2023). COVID-19 Mortality in the Colorado Center for Personalized Medicine Biobank. International Journal of Environmental Research and Public Health, 20(3), 2368. https://doi.org/10.3390/ijerph20032368





