Coronary Artery Disease and Inflammatory Activation Interfere with Peripheral Tissue Electrical Impedance Spectroscopy Characteristics—Initial Report
Abstract
:1. Introduction
2. Materials and Methods
2.1. Whole Blood Count Analysis
2.2. Electrical Impedance Spectroscopy (EIS)
2.3. Exclusion Criteria
2.4. Statistical Analysis
3. Results
3.1. Laboratory Results
3.2. EIS Parameters
3.3. Correlations with Inflammatory Indexes
Comparison of the CAD and Control Groups
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stoneman, M.; Raicu, V. Dielectric Spectroscopy Based Detection of Specific and Nonspecific Cellular Mechanisms. Sensors 2021, 21, 3177. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiari, S.; Manshadi, M.K.D.; Mansoorifar, A.; Beskok, A. A Microfluidic Dielectric Spectroscopy System for Characterization of Biological Cells in Physiological Media. Sensors 2022, 22, 463. [Google Scholar] [CrossRef] [PubMed]
- Bedard, C.; Piette, C.; Venance, L.; Destexhe, A. Extracellular and intracellular components of the impedance of neural tissue. Biophys. J. 2022, 121, 869–885. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.R.; Powell, T.; Sturridge, M.F.; Terrar, D.A.; Twist, V.W. Electrical properties and response to noradrenaline of individual heart cells isolated from human ventricular tissue. Cardiovasc. Res. 1986, 20, 869–876. [Google Scholar] [CrossRef]
- Schaefer, M.; Gross, W.; Ackemann, J.; Gebhard, M. The complex dielectric spectrum of heart tissue during ischemia. Bioelectrochemistry 2002, 58, 171–180. [Google Scholar] [CrossRef]
- van der Sande, F.M.; van de Wal-Visscher, E.R.; Stuard, S.; Moissl, U.; Kooman, J.P. Using Bioimpedance Spectroscopy to Assess Volume Status in Dialysis Patients. Blood Purif. 2020, 49, 178–184. [Google Scholar] [CrossRef]
- Aspromonte, N.; Cruz, D.N.; Ronco, C.; Valle, R. Role of Bioimpedance Vectorial Analysis in Cardio-Renal Syndromes. Semin. Nephrol. 2012, 32, 93–99. [Google Scholar] [CrossRef]
- Wang, H.; He, Y.; Yang, M.; Yan, Q.; You, F.; Fu, F.; Wang, T.; Huo, X.; Dong, X.; Shi, X. Dielectric properties of human liver from 10 Hz to 100 MHz: Normal liver, hepatocellular carcinoma, hepatic fibrosis and liver hemangioma. Biomed. Mater. Eng. 2014, 24, 2725–2732. [Google Scholar]
- Heileman, K.; Daoud, J.; Tabrizian, M. Dielectric spectroscopy as a viable biosensing tool for cell and tissue characterization and analysis. Biosens. Bioelectron. 2013, 49, 348–359. [Google Scholar] [CrossRef]
- Sekine, K.; Torii, N.; Kuroda, C.; Asami, K. Calculation of dielectric spectra of suspensions of rod-shaped cells using boundary element method. Bioelectrochemistry 2002, 57, 83–87. [Google Scholar] [CrossRef]
- Hernández-Bule, M.L.; Medel, E.; Colastra, C.; Roldán, R.; Úbeda, A. Response of neuroblastoma cells to RF currents as a function of the signal frequency. BMC Cancer 2019, 19, 889–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sieg, A.; Guy, R.; Delgado-Charro, M.B. Noninvasive and Minimally Invasive Methods for Transdermal Glucose Monitoring. Diabetes Technol. Ther. 2005, 7, 174–197. [Google Scholar] [CrossRef]
- Das, N.; Alexandrov, S.; Gilligan, K.E.; Dwyer, R.M.; Saager, R.B.; Ghosh, N.; Leahy, M. Characterization of nanosensitive multifractality in submicron scale tissue morphology and its alteration in tumor progression. J. Biomed. Opt. 2021, 26, 016003–016014. [Google Scholar] [PubMed]
- Markx, G.H.; Davey, C.L. The dielectric properties of biological cells at radiofrequencies: Applications in biotechnology. Enzym. Microb. Technol. 1999, 25, 161–171. [Google Scholar] [CrossRef]
- Hagiwara, M.; Maruyama, H.; Akiyama, M.; Koh, I.; Arai, F. Weakening of resistance force by cell–ECM interactions regulate cell migration directionality and pattern formation. Commun. Biol. 2021, 4, 808–821. [Google Scholar] [CrossRef] [PubMed]
- Severino, P.; D’Amato, A.; Pucci, M.; Infusino, F.; Birtolo, L.I.; Mariani, M.V.; LaValle, C.; Maestrini, V.; Mancone, M.; Fedele, F. Ischemic Heart Disease and Heart Failure: Role of Coronary Ion Channels. Int. J. Mol. Sci. 2020, 21, 3167. [Google Scholar] [CrossRef] [PubMed]
- Christodoulidis, G.; Vittorio, T.J.; Fudim, M.; Lerakis, S.; Kosmas, C.E. Inflammation in Coronary Artery Disease. Cardiol. Rev. 2014, 22, 279–288. [Google Scholar] [CrossRef]
- Freitas, I.A.; Lima, N.A.; Silva, G.B.D., Jr.; Castro, R.L., Jr.; Patel, P.; Lima, C.C.V.; Lino, D.O.D.C. Novel biomarkers in the prognosis of patients with atherosclerotic coronary artery disease. Rev. Port. Cardiol. (Engl. Ed.) 2020, 39, 667–672. [Google Scholar] [CrossRef]
- Ikonomidis, I.; Michalakeas, C.; Parissis, J.; Paraskevaidis, I.; Ntai, K.; Papadakis, I.; Anastasiou-Nana, M.; Lekakis, J. Inflammatory markers in coronary artery disease. Biofactors 2012, 38, 320–328. [Google Scholar] [CrossRef]
- Urbanowicz, T.; Michalak, M.; Olasińska-Wiśniewska, A.; Rodzki, M.; Witkowska, A.; Gąsecka, A.; Buczkowski, P.; Perek, B.; Jemielity, M. Neutrophil Counts, Neutrophil-to-Lymphocyte Ratio, and Systemic Inflammatory Response Index (SIRI) Predict Mortality after Off-Pump Coronary Artery Bypass Surgery. Cells 2022, 11, 1124. [Google Scholar] [CrossRef]
- Brzeziński, J.; Oszkinis, G.; Marzec, E. Dielectric relaxation of a protein–water system in atherosclerotic artery wall. Med. Biol. Eng. Comput. 2007, 45, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Magar, H.S.; Hassan, R.Y.A.; Mulchandani, A. Electrochemical Impedance Spectroscopy (EIS): Principles, Construction, and Biosensing Applications. Sensors 2021, 21, 6578. [Google Scholar] [CrossRef] [PubMed]
- Brunauer, G.C.; Meindl, A.; Rotter, B.; Gruber, A.; Slouka, C.; Schnabel, T.; Petutschnigg, A. Electrochemical Impedance Spectroscopy for Microbiological Pro-Cesses: On the Way to a Monitoring Tool for the Determination of Biomass. Biomed. J. Sci. Tech. Res. 2021, 35, 27548–27557. [Google Scholar] [CrossRef]
- Slouka, C.; Wurm, D.J.; Brunauer, G.; Welzl-Wachter, A.; Spadiut, O.; Fleig, J.; Christoph Herwig, C. A Novel Application for Low Frequency Electrochemical Impedance Spectroscopy as an Online Process Monitoring Tool for Viable Cell Concentrations. Sensors 2016, 16, 1900. [Google Scholar] [CrossRef]
- Raicu, V.Y.; Feldman, Y. Dielectric Relaxation in Biological Systems, Physical Principles, Methods, and Applications; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Dean, D.; Ramanathan, T.; Machado-Aranda, D.; Sundararajan, R. Electrical impedance spectroscopy study of biological tissues. J. Electrost. 2008, 66, 165–177. [Google Scholar] [CrossRef]
- Noitz, M.; Steinkellner, C.; Willingshofer, M.P.; Szasz, J.; Dünser, M. Die Rolle der Mikrozirkulation in der Entstehung des Or-ganversagens [The role of the microcirculation in the pathogenesis of organ dysfunction]. Dtsch. Med. Wochenschr. 2022, 147, 17–25. [Google Scholar]
- Ince, C.; De Backer, D.; Mayeux, P.R. Microvascular Dysfunction in the Critically III. Crit. Care Clin. 2020, 36, 323–331. [Google Scholar] [CrossRef]
- Ansari, J.; Gavins, F.N.E. The impact of thrombo-inflammation on the cerebral microcirculation. Microcirculation 2021, 28, e12689. [Google Scholar] [CrossRef]
- Marzec, E.; Olszewski, J. Dielectric response to thermal denaturation of lenses in healthy and diabetic rabbits. Bioelectrochemistry 2021, 142, 107923–107930. [Google Scholar] [CrossRef]
- Yamada, Y.; Hirata, K.; Iida, N.; Kanda, A.; Shoji, M.; Yoshida, T.; Myachi, M.; Akagi, R. Membrane capacitance and characteristic frequency are associated with contractile properties of skeletal muscle. Med. Eng. Phys. 2022, 106, 103832–103838. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhang, X.; Beckel, J.; de Groat, W.C.; Tai, C. Intracellular sodium concentration and membrane potential oscillation in axonal conduction block induced by high-frequency biphasic stimulation. J. Neural Eng. 2022, 19, 046024. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Liu, X.; Yuan, Y.; Zhang, T.; Kiya, R.; Yang, Y.; Suzuki, K.; Tanaka, Y.; Li, M.; Hosokawa, Y.; et al. Assessment of the electrical penetration of cell membranes using four-frequency impedance cytometry. Microsyst. Nanoeng. 2022, 8, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Zhang, Y.; Niu, Z.; Chi, Y.; Huang, Q. Transcriptomic responses of peripheral blood cells to coronary artery disease. Biosci. Trends 2018, 12, 354–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Parameter | CAD Patients n = 29 | Control Group n = 10 | p |
|---|---|---|---|
| Age (years) (median (Q1–Q3)) | 69 (65–72) | 66 (62–69) | 0.786 |
| Sex (male/female) (n,%) | 29/0 | 10/0 | -- |
| NYHA class (median (Q1–Q3) | 2 (1–2) | 2 (1–2) | -- |
| Height (cm) (median (Q1–Q3)) | 175 (170–175) | 176 (168–180) | 0.698 |
| Weight (kg) (median (Q1–Q3)) | 86 (79–83) | 98 (90–112) | 0.134 |
| Co-morbidities: | |||
| Arterial Hypertension (n,%) | 26 (89%) | 10 (100%) | 0.567 |
| Diabetes Mellitus (n,%) | 8 (28%) | 5 (50%) | 0.356 |
| COPD (n,%) | 3 (10%) | 1 (10%) | 0.976 |
| PAD (n,%) | 0 (0%) | 0 (0%) | -- |
| Hypercholesterolemia (n,%) | 19 (66%) | 9 (90%) | 0.456 |
| Kidney Failure (n,%) | 2 (7%) | 0 | 0.187 |
| Atrial Fibrillation (n,%) | 4 (13%) | 0 | 0.061 |
| Stroke in Medical History (n,%) | 1 (3%) | 0 | 0.094 |
| Echocardiography | |||
| LV (mm) (median (Q1–Q3)) | 45 (43–49) | 50 (47–52) | 0.126 |
| RV (mm) (median (Q1–Q3)) | 30 (26–33) | 32 (31–33) | 0.436 |
| LA (mm) (median (Q1–Q3)) | 37 (33–40) | 40 (38–45) | 0.438 |
| LVEF (%) (median (Q1–Q3)) | 60 (60–60) | 60 (60–60) | 0.976 |
| Parameter | CAD before Surgery (A1) | CAD after Surgery (A2) | Control Group (B) | p A1 vs. A2 | p A1 vs. B | p A2 vs. B |
|---|---|---|---|---|---|---|
| Laboratory | ||||||
| WBC (K/uL) (median (Q1–Q3)) | 7.7 (6.3–9.1) | 7.4 (6.1–8.6) | 7.7 (6.3–1.1) | 0.229 | 0.035 * | 0.098 |
| N (K/uL) (median (Q1–Q3)) | 5.1 (4.1–6.7) | 4.5 (3.9–5.4) | 4.3 (3.1–5.2) | 0.025 * | 0.022 * | 0.403 |
| L (K/uL) (median (Q1–Q3)) | 1.6 (1.2–1.7) | 1.7 (1.3–2.0) | 1.4 (1.1–1.8) | 0.166 | 0.412 | 0.139 |
| NLR (median (Q1–Q3)) | 3.6 (3–4.7) | 2.8 (2.4–3.1) | 2.9 (2.5–3.5) | 0.002 * | 0.079 | 0.541 |
| M (K/uL) (median (Q1–Q3)) | 0.5 (0.4–0.6) | 0.6 (0.5–0.8) | 0.4 (0.4–0.5) | 0.020 * | 0.152 | 0.004 * |
| MLR (median (Q1–Q3)) | 0.4 (0.3–0.4) | 0.4 (0.3–0.4) | 0.3 (0.3–0.4) | <0.001 * | 0.327 | 0.062 |
| SIRI (median (Q1–Q3)) | 1.9 (1.4–2.4) | 1.8 (1.4–2.1) | 1.4 (1.0–1.7) | 0.179 | 0.056 | 0.094 |
| Eo (K/uL) (median (Q1–Q3)) | 0.17 (0.08–0.16) | 0.35 (0.19–0.35) | 0.3 (0.1–0.4) | <0.001 * | 0.017 * | 0.618 |
| LUC (K/uL) (median (Q1–Q3)) | 0.12 (0.1–0.18) | 0.18 (0.13–0.24) | 0.13 (0.12–0.15) | 0.003 * | 0.923 | 0.059 |
| Hb (mmol/L) (median (Q1–Q3)) | 9.2 (8–9.5) | 6.9 (6.4–7.2) | 9.1 (8.3–9.6) | <0.001 * | 0.664 | <0.001 * |
| MCV (fl) (median (Q1–Q3)) | 92 (90–97) | 94 (91–97) | 93 (91–96) | 0.281 | 0.746 | <0.001 * |
| RBc (M/uL) (median (Q1–Q3)) | 4.5 (4.3–4.8) | 3.52 (3.31–3.86) | 4.7 (4.6–4.9) | <0.001 * | 0.234 | <0.001 * |
| Hct (%) (median (Q1–Q3)) | 43 (40–44) | 33 (30–35) | 45 (41–47) | <0.001 * | 0.061 | <0.001 * |
| MCHC (mmol/L) (median (Q1–Q3)) | 21.1 (20.4–21.6) | 21 (30–35) | 20.3 (19.8–20.9) | 0.006 * | 0.009 * | 0.131 |
| RDW (%) (median (Q1–Q3)) | 13.7 (13.1–14.1) | 14 (13.5–19.9) | 14 (13.6–14.5) | 0.001 * | 0.129 | 0.910 |
| Plt (K/uL) (median (Q1–Q3)) | 231 (192–279) | 221 (214–306) | 197 (186–261) | 0.006 * | 0.499 | 0.130 |
| SII (median (Q1–Q3)) | 800 (621–1098) | 653 (608–803) | 645 (505–849) | 0.018 * | 0.119 | 0.585 |
| AISI (median (Q1–Q3)) | 465 (258–571) | 433 (311–544) | 287 (261–328) | 0.503 | 0.104 | 0.031 * |
| MPV (fl) (median (Q1–Q3)) | 9 (8.4–9.7) | 8.5 (8.1–8.9) | 9.1 (8.8–9.9) | 0.039 * | 0.310 | 0.016 * |
| Lipid profile | ||||||
| Total cholesterol (mmol/L) (median (Q1–Q3)) | 3.45 (314–3.95) | - | 5.1 (4.1–5.7) | - | 0.049 * | - |
| LDL fraction (mmol/L) (median (Q1–Q3)) | 1.85 (1.49–2.35) | - | 2.95 (1.6–3.41) | - | 0.274 | - |
| HDL fraction (mmol/L) (median (Q1–Q3)) | 0.88 (0.84–1.28) | - | 1.1 (1.0–1.3) | - | 0.378 | - |
| Triglycerides (mmol/L) (median (Q1–Q3)) | 1.2 (0.82–1.69) | - | 1.4 (1.1–1.6) | - | 0.421 | - |
| Liver function: | ||||||
| ALT (U/L) (median (Q1–Q3)) | 29 (23–35) | 31 (20–44) | 31 (21–44) | 0.768 | 0.835 | 0.913 |
| Kidney function: | ||||||
| Creatinine (umol/L) (median (Q1–Q3)) | 88 (72–103) | 76 (71–82) | 55 (79–87) | 0.028 * | 0.949 | 0.111 |
| Myocardial markers: | MAX | |||||
| Troponin -I (ng/mL) (median (Q1–Q3)) | 0.012 (0.005–0.02) | 1.99 (1.389–2.378) | 0.014 (0.006–0.03) | <0.001 | 0.867 | <0.001 |
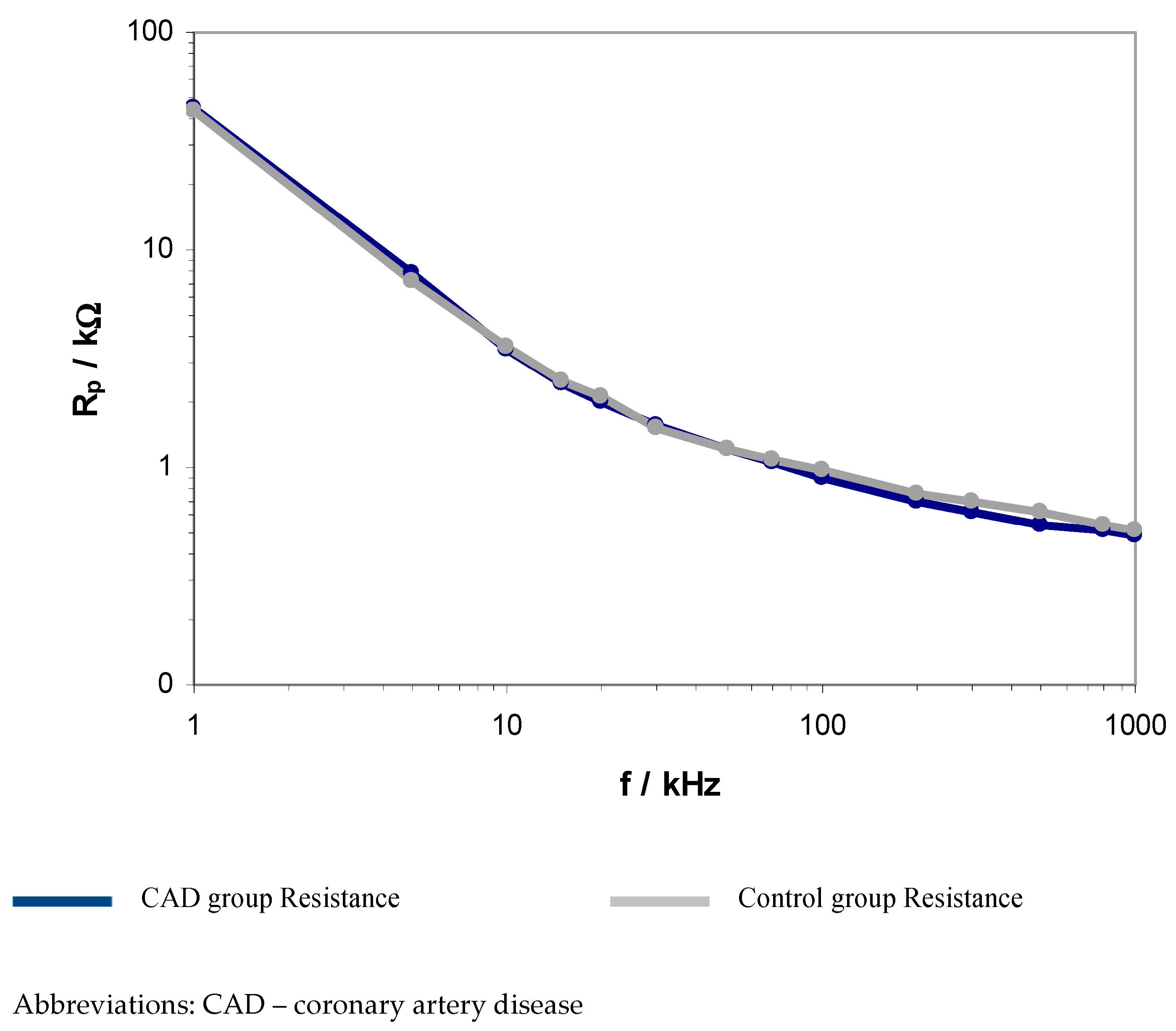
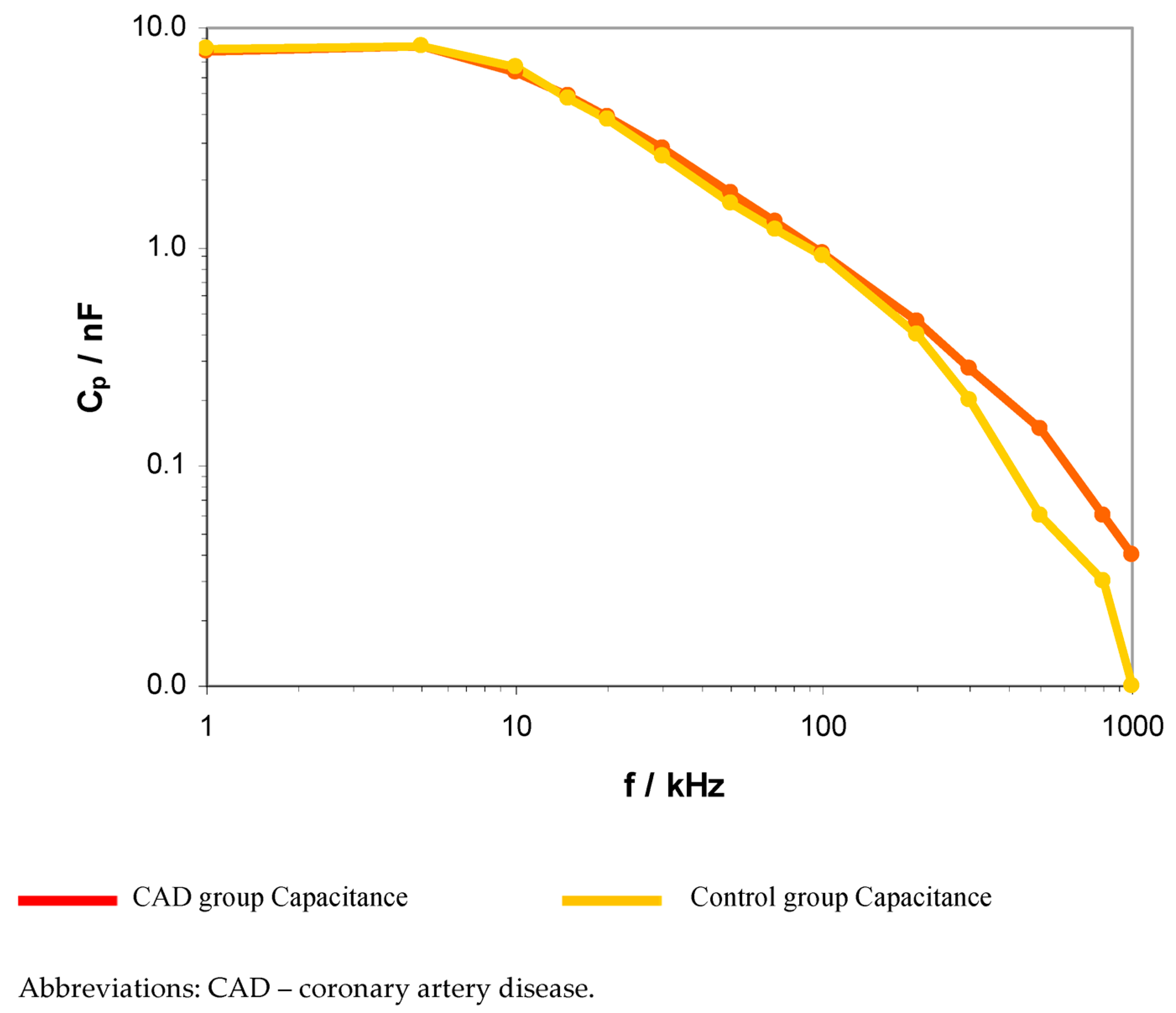
| Frequency f | CAD Group (A1) | Control Group (B) | p | |||
|---|---|---|---|---|---|---|
| Rp (kΩ) | Cp (nF) | Rp (kΩ) | Cp (nF) | p Rp | p Cp | |
| 1 kHz | 45.3 (35.7–52.7) | 7.7 (5.9–8.5) | 43.5 (41–49) | 8 (6.0–9.1) | 0.772 | 0.515 |
| 5 kHz | 7.8 (6.13–10.45) | 8.3 (7.1–9.9) | 7.22 (5.7–8.4) | 8.2 (7.6–8.2) | 0.106 | 0.502 |
| 10 kHz | 3.5 (2.9–3.8) | 6.3 (5.8–7) | 3.6 (2.7–4.1) | 6.6 (6–6.9) | 0.306 | 0.193 |
| 15 kHz | 2.5 (2.1–2.8) | 4.9 (4.5–5.3) | 2.5 (1.9–2.7) | 4.8 (4.4–4.8) | 0.032 * | 0.974 |
| 20 kHz | 2 (1.7–2.4) | 3.9 (3.7–4.1) | 2.1 (1.6–2.2) | 3.8 (3.5–4.1) | 0.079 | 0.987 |
| 30 kHz | 1.5 (1.3–1.6) | 2.8 (2.6–3.0) | 1.4 (1.3–1.6) | 2.6 (2.4–2.7) | 0.880 | 0.129 |
| 50 kHz | 1.2 (1–1.3) | 1.7 (1.6–1.9) | 1.2 (1–1.2) | 1.6 (1.3–1.7) | 0.065 | 0.879 |
| 70 kHz | 1 (0.9–1.1) | 1.3 (1.2–1.4) | 1.1 (1–1.2) | 1 (0.1–1.1) | 0.943 | 0.678 |
| 100 kHz | 0.9 (0.8–1.0) | 0.9 (0.9–1.0) | 0.9 (0.9–1.0) | 0.9 (0.9–1.1) | 0.066 | 0.508 |
| 200 kHz | 0.7 (0.6–0.7) | 0.5 (0.4–0.5) | 0.8 (0.8–0.9) | 0.4 (0.2–0.7) | 0.035 * | 0.001 * |
| 300 kHz | 0.6 (0.6–0.7) | 0.3 (0.3–0.3) | 0.7 (0.7–0.7) | 0.2 (0.2–0.2) | 0.045 * | 0.008 * |
| 500 kHz | 0.5 (0.4–0.6) | 0.1 (0.05–0.2) | 0.6 (0.5–0.6) | 0.06 (0.06–0.07) | 0.047 * | 0.003 * |
| 800 kHZ | 0.51 (0.48–0.53) | 0.06 (0.05–0.08) | 0.55 (0.41–0.63) | 0.03 (0.02–0.04) | 0.224 | 0.004 * |
| 1 MHZ | 0.49 (0.45–0.52) | 0.04 (0.03–0.05) | 0.51 (0.48–0.52) | 0.01 (0.01–0.01) | 0.628 | 0.001 * |
| Frequency f | Inflammatory Index | Rp/Cp | CAD Group (A1) | Control Group (B) | ||
|---|---|---|---|---|---|---|
| Spearman’s Rho | p | Spearman’s Rho | p | |||
| 1 kHz | NLR | Rp | 0.045 | 0.815 | −0.571 | 0.084 |
| 5 kHz | NLR | Rp | −0.254 | 0.749 | −0.628 | 0.052 |
| 10 kHz | NLR | Rp | −0.405 | 0.101 | −0.246 | 0.493 |
| 15 kHz | NLR | Rp | −0.329 | 0.059 | −0.238 | 0.724 |
| 20 kHz | NLR | Rp | 0.049 | 0.587 | −0.337 | 0.136 |
| 30 kHz | NLR | Rp | −0.317 | 0.038 * | −0.628 | 0.052 |
| 50 kHz | NLR | Rp | −0.252 | 0.069 | −0.261 | 0.466 |
| 70 kHz | NLR | Rp | −0.235 | 0.529 | −0.409 | 0.241 |
| 100 kHz | NLR | Rp | −0.368 | 0.060 | −0.523 | 0.121 |
| 200 kHz | NLR | Rp | 0.007 | 0.465 | −0.415 | 0.233 |
| 300 kHz | NLR | Rp | 0.009 | 0.538 | −0.175 | 0.629 |
| 500 kHz | NLR | Rp | −0.161 | 0.047 * | −0.525 | 0.119 |
| 800 kHz | NLR | Rp | 0.270 | 0.611 | −0.174 | 0.631 |
| 1 MHz | NLR | Rp | 0.028 | 0.067 | −0.174 | 0.630 |
| 1 kHz | NLR | Cp | −0.126 | 0.514 | −0.281 | 0.433 |
| 5 kHz | NLR | Cp | 0.311 | 0.343 | −0.246 | 0.493 |
| 10 kHz | NLR | Cp | 0.354 | 0.029 * | −0.534 | 0.802 |
| 15 kHz | NLR | Cp | 0.147 | 0.082 | −0.238 | 0.724 |
| 20 kHz | NLR | Cp | 0.388 | 0.856 | −0.506 | 0.136 |
| 30 kHz | NLR | Cp | −0.317 | 0.094 | −0.628 | 0.052 |
| 50 kHz | NLR | Cp | 0.167 | 0.187 | −0.261 | 0.466 |
| 70 kHz | NLR | Cp | 0.381 | 0.354 | −0.409 | 0.241 |
| 100 kHz | NLR | Cp | 0.197 | 0.049 * | −0.523 | 0.121 |
| 200 kHz | NLR | Cp | 0.166 | 0.978 | 0.233 | 0.031 |
| 300 kHz | NLR | Cp | 0.09 | 0.972 | −0.175 | 0.629 |
| 500 kHz | NLR | Cp | −0.162 | 0.402 | −0.525 | 0.119 |
| 800 kHz | NLR | Cp | 0.345 | 0.312 | 0.323 | 0.631 |
| 1 MHz | NLR | Cp | −0.166 | 0.883 | −0.640 | 0.631 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbanowicz, T.; Michalak, M.; Marzec, E.; Komosa, A.; Filipiak, K.J.; Olasińska-Wiśniewska, A.; Witkowska, A.; Rodzki, M.; Tykarski, A.; Jemielity, M. Coronary Artery Disease and Inflammatory Activation Interfere with Peripheral Tissue Electrical Impedance Spectroscopy Characteristics—Initial Report. Int. J. Environ. Res. Public Health 2023, 20, 2745. https://doi.org/10.3390/ijerph20032745
Urbanowicz T, Michalak M, Marzec E, Komosa A, Filipiak KJ, Olasińska-Wiśniewska A, Witkowska A, Rodzki M, Tykarski A, Jemielity M. Coronary Artery Disease and Inflammatory Activation Interfere with Peripheral Tissue Electrical Impedance Spectroscopy Characteristics—Initial Report. International Journal of Environmental Research and Public Health. 2023; 20(3):2745. https://doi.org/10.3390/ijerph20032745
Chicago/Turabian StyleUrbanowicz, Tomasz, Michał Michalak, Ewa Marzec, Anna Komosa, Krzysztof J. Filipiak, Anna Olasińska-Wiśniewska, Anna Witkowska, Michał Rodzki, Andrzej Tykarski, and Marek Jemielity. 2023. "Coronary Artery Disease and Inflammatory Activation Interfere with Peripheral Tissue Electrical Impedance Spectroscopy Characteristics—Initial Report" International Journal of Environmental Research and Public Health 20, no. 3: 2745. https://doi.org/10.3390/ijerph20032745
APA StyleUrbanowicz, T., Michalak, M., Marzec, E., Komosa, A., Filipiak, K. J., Olasińska-Wiśniewska, A., Witkowska, A., Rodzki, M., Tykarski, A., & Jemielity, M. (2023). Coronary Artery Disease and Inflammatory Activation Interfere with Peripheral Tissue Electrical Impedance Spectroscopy Characteristics—Initial Report. International Journal of Environmental Research and Public Health, 20(3), 2745. https://doi.org/10.3390/ijerph20032745









