Predicting Fecal Indicator Bacteria Using Spatial Stream Network Models in A Mixed-Land-Use Suburban Watershed in New Jersey, USA
Abstract
1. Introduction
2. Materials and Methods
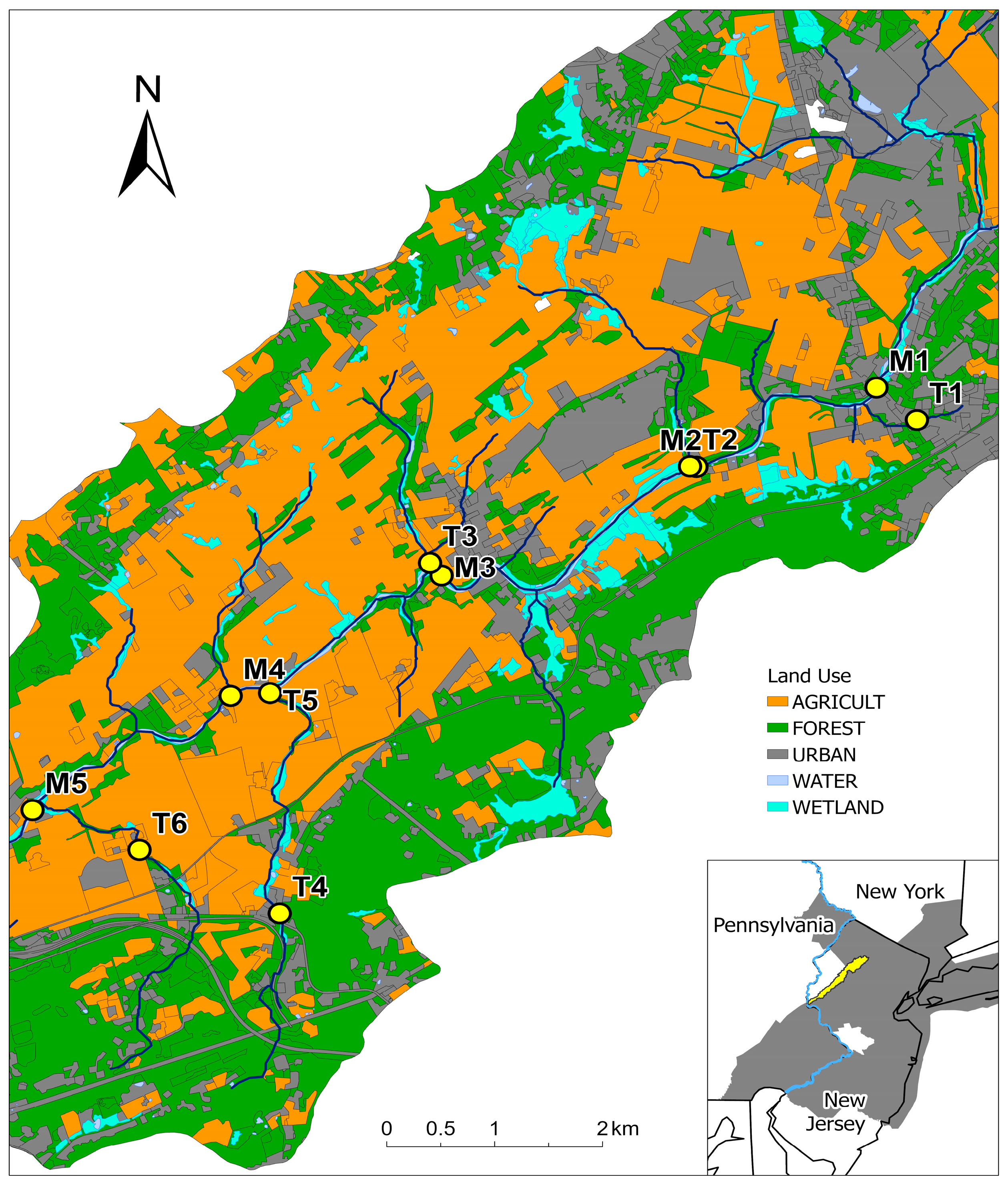
2.1. Water Quality, Land Use and Precipitation Data
2.2. Spatial Stream Network Modeling
2.3. Predicting E. coli Concentrations
3. Results
3.1. Microbial Water Quality and Land Use
3.2. Spatial Stream Network Modeling
3.3. Predicting E. coli Concentrations
4. Discussion
4.1. Spatial Stream Network Modeling Performance
4.2. Suburban Land Use and Microbial Water Quality
4.3. Extreme Weather Conditions and Microbial Water Quality
4.4. Predicting Fecal Contamination Hot Spots and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnone, R.D.; Walling, J.P. Waterborne pathogens in urban watersheds. J. Water Health 2007, 5, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Schneider, I.E. Urban Water Recreation: Experiences, Place Meanings, and Future Issues. In The Water Environment of Cities; Springer: Boston, MA, USA, 2009; pp. 125–140. [Google Scholar]
- USEPA. Health Effects Criteria for Fresh Recreational Waters; USEPA: Washington, DC, USA, 1984. [Google Scholar]
- USEPA. Recreational Water Quality Criteria; USEPA: Washington, DC, USA, 2012. [Google Scholar]
- Prüss, A. Review of epidemiological studies on health effects from exposure to recreational water. Int. J. Epidemiol. 1998, 27, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wymer, L.J.; Dufour, A.P. A model for estimating the incidence of swimming-related gastrointestinal illness as a function of water quality indicators. Environmetrics 2002, 13, 669–678. [Google Scholar] [CrossRef]
- Almeida, C.; González, S.O.; Mallea, M.; González, P. A recreational water quality index using chemical, physical and microbiological parameters. Environ. Sci. Pollut. Res. 2012, 19, 3400–3411. [Google Scholar] [CrossRef] [PubMed]
- Wymer, L.J.; Wade, T.J.; Dufour, A.P. Equivalency of risk for a modified health endpoint: A case from recreational water epidemiology studies. BMC Public Health 2013, 13, 459. [Google Scholar] [CrossRef]
- Keller, A.A.; Cavallaro, L. Assessing the US Clean Water Act 303(d) listing process for determining impairment of a waterbody. J. Environ. Manag. 2008, 86, 699–711. [Google Scholar] [CrossRef]
- Benham, B.L.; Baffaut, C.; Zeckoski, R.W.; Mankin, K.R.; Pachepsky, Y.A.; Sadeghi, A.M.; Brannan, K.M.; Soupir, M.L.; Habersack, M.J. Modeling bacteria fate and transport in watersheds to support TMDLs. Trans. ASABE 2006, 49, 987–1002. [Google Scholar] [CrossRef]
- National Research Council. Assessing the TMDL Approach to Water Quality Management; National Research Council: Washington, DC, USA, 2001. [Google Scholar]
- Cho, K.H.; Pachepsky, Y.A.; Oliver, D.M.; Muirhead, R.W.; Park, Y.; Quilliam, R.S.; Shelton, D.R. Modeling fate and transport of fecally-derived microorganisms at the watershed scale: State of the science and future opportunities. Water Res. 2016, 100, 38–56. [Google Scholar] [CrossRef]
- Jha, M.K.; Wolter, C.F.; Schilling, K.E.; Gassman, P.W. Assessment of Total Maximum Daily Load Implementation Strategies for Nitrate Impairment of the Raccoon River, Iowa. J. Environ. Qual. 2010, 39, 1317–1327. [Google Scholar] [CrossRef]
- Mas, D.M.L.; Ahlfeld, D.P. Comparing artificial neural networks and regression models for predicting faecal coliform concentrations. Hydrol. Sci. J. 2007, 52, 713–731. [Google Scholar] [CrossRef]
- Cha, Y.; Park, M.-H.; Lee, S.-H.; Kim, J.H.; Cho, K.H. Modeling spatiotemporal bacterial variability with meteorological and watershed land-use characteristics. Water Res. 2016, 100, 306–315. [Google Scholar] [CrossRef]
- Herrig, I.; Seis, W.; Fischer, H.; Regnery, J.; Manz, W.; Reifferscheid, G.; Böer, S. Prediction of fecal indicator organism concentrations in rivers: The shifting role of environmental factors under varying flow conditions. Environ. Sci. Eur. 2019, 31, 59. [Google Scholar] [CrossRef]
- Rossi, A.; Wolde, B.T.; Lee, L.H.; Wu, M. Prediction of recreational water safety using Escherichia coli as an indicator: Case study of the Passaic and Pompton rivers, New Jersey. Sci. Total. Environ. 2020, 714, 136814. [Google Scholar] [CrossRef] [PubMed]
- Herrig, I.M.; Böer, S.I.; Brennholt, N.; Manz, W. Development of multiple linear regression models as predictive tools for fecal indicator concentrations in a stretch of the lower Lahn River, Germany. Water Res. 2015, 85, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, M.F. Data Analysis and Statistics for Geography, Environmental Science, and Engineering; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Tobler, W.R. A Computer Movie Simulating Urban Growth in the Detroit Region. Econ. Geogr. 1970, 46, 234–240. [Google Scholar] [CrossRef]
- Peterson, E.E.; Hoef, J.V.; Isaak, D.J.; Falke, J.A.; Fortin, M.-J.; Jordan, C.E.; McNyset, K.; Monestiez, P.; Ruesch, A.S.; Sengupta, A.; et al. Modelling dendritic ecological networks in space: An integrated network perspective. Ecol. Lett. 2013, 16, 707–719. [Google Scholar] [CrossRef]
- Isaak, D.J.; Peterson, E.E.; Hoef, J.M.V.; Wenger, S.J.; Falke, J.A.; Torgersen, C.E.; Sowder, C.; Steel, E.A.; Fortin, M.-J.; Jordan, C.E.; et al. Applications of spatial statistical network models to stream data. WIREs Water 2014, 1, 277–294. [Google Scholar] [CrossRef]
- Money, E.S.; Carter, G.P.; Serre, M.L. Modern space/time geostatistics using river distances: Data integration of turbidity and E. coli measurements to assess fecal contamination along the Raritan River in New Jersey. Environ. Sci. Technol. 2009, 43, 3736–3742. [Google Scholar] [CrossRef] [PubMed]
- USEPA. Using Microbial Source Tracking to Support TMDL Development and Implementation; USEPA: Washington, DC, USA, 2011. [Google Scholar]
- Isaak, D.J.; Hoef, J.M.V.; Peterson, E.E.; Horan, D.L.; Nagel, D.E. Scalable population estimates using spatial-stream-network (SSN) models, fish density surveys, and national geospatial database frameworks for streams. Can. J. Fish. Aquat. Sci. 2017, 74, 147–156. [Google Scholar] [CrossRef]
- Scown, M.W.; McManus, M.G.; Carson, J.H., Jr.; Nietch, C.T. Improving Predictive Models of In-Stream Phosphorus Concentration Based on Nationally-Available Spatial Data Coverages. JAWRA J. Am. Water Resour. Assoc. 2017, 53, 944–960. [Google Scholar] [CrossRef]
- ElSaadani, M.; Krajewski, W.F.; Zimmerman, D.L. River network based characterization of errors in remotely sensed rainfall products in hydrological applications. Remote Sens. Lett. 2018, 9, 743–752. [Google Scholar] [CrossRef]
- Gendaszek, A.S.; Dunham, J.B.; Torgersen, C.E.; Hockman-Wert, D.P.; Heck, M.P.; Thorson, J.; Mintz, J.; Allai, T. Land-Cover and Climatic Controls on Water Temperature, Flow Permanence, and Fragmentation of Great Basin Stream Networks. Water 2020, 12, 1962. [Google Scholar] [CrossRef]
- Hoef, J.M.V.; Peterson, E.; Theobald, D. Spatial statistical models that use flow and stream distance. Environ. Ecol. Stat. 2006, 13, 449–464. [Google Scholar] [CrossRef]
- Hoef, J.M.V.; Peterson, E.E. A Moving Average Approach for Spatial Statistical Models of Stream Networks. J. Am. Stat. Assoc. 2010, 105, 6–18. [Google Scholar] [CrossRef]
- Barry, R.P.; Jay, M.V.H. Blackbox Kriging: Spatial Prediction without Specifying Variogram Models. J. Agric. Biol. Environ. Stat. 1996, 1, 297–322. [Google Scholar] [CrossRef]
- Neill, A.J.; Tetzlaff, D.; Strachan, N.J.C.; Hough, R.L.; Avery, L.M.; Watson, H.; Soulsby, C. Using spatial-stream-network models and long-term data to understand and predict dynamics of faecal contamination in a mixed land-use catchment. Sci. Total Environ. 2018, 612, 840–852. [Google Scholar] [CrossRef]
- Hunt, A.; Wu, M.; Hsu, T.-T.; Roberts-Lawler, N.; Miller, J.; Rossi, A.; Lee, L. Picking Up Where the TMDL Leaves Off: Using the Partnership Wild and Scenic River Framework for Collaborative River Restoration. Sustainability 2021, 13, 1878. [Google Scholar] [CrossRef]
- Stinnette, I.; Taylor, M.; Kerr, L.; Pirani, R.; Lipuma, S.; Lodge, J. State of the Estuary 2018; Hudson River Foundation: New York, NY, USA, 2018. [Google Scholar]
- New Jersey Department of Environmental Protection. Total Maximum Daily Loads for Fecal Coliform to Address 28 Streams in the Northwest Water Region; New Jersey Department of Environmental Protection: Camden, NJ, USA, 2003. [Google Scholar]
- Anderson, J.R.; Hardy, E.E.; Roach, J.T.; Witmer, R.E. A Land Use and Land Cover Classification System for Use with Remote Sensor Data; US Government Printing Office: Washington, DC, USA, 1976. [Google Scholar]
- Restoration, R.W. Protection Plan for the Musconetcong River Watershed from Hampton to Bloomsbury. In Water Quality Monitoring Data Report; Rutgers Cooperative Extension Water Resources Program: New Brunswick, NJ, USA, 2011. [Google Scholar]
- Garreta, V.; Monestiez, P.; Hoef, J.M.V. Spatial modelling and prediction on river networks: Up model, down model or hybrid? Environmetrics 2010, 21, 439–456. [Google Scholar] [CrossRef]
- Ver Hoef, J.M.; Peterson, E.; Clifford, D.; Shah, R. SSN: An R Package for Spatial Statistical Modeling on Stream Networks. J. Stat. Softw. 2014, 56, 1–45. [Google Scholar]
- Dubin, R.A. Spatial autocorrelation and neighborhood quality. Reg. Sci. Urban Econ. 1992, 22, 433–452. [Google Scholar] [CrossRef]
- Anselin, L.; Bera, A.K.; Florax, R.; Yoon, M.J. Simple diagnostic tests for spatial dependence. Reg. Sci. Urban Econ. 1996, 26, 77–104. [Google Scholar] [CrossRef]
- Paule-Mercado, M.; Ventura, J.; Memon, S.; Jahng, D.; Kang, J.-H.; Lee, C.-H. Monitoring and predicting the fecal indicator bacteria concentrations from agricultural, mixed land use and urban stormwater runoff. Sci. Total Environ. 2016, 550, 1171–1181. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.E. STARS: Spatial Tools for the Analysis of River Systems version 2.0. 6—A tutorial; Queensland University of Technology: Brisbane, Australia, 2017. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Peterson, E.E.; Merton, A.A.; Theobald, D.M.; Urquhart, N.S. Patterns of Spatial Autocorrelation in Stream Water Chemistry. Environ. Monit. Assess. 2006, 121, 571–596. [Google Scholar] [CrossRef] [PubMed]
- Cressie, N.; Lahiri, S. The Asymptotic Distribution of REML Estimators. J. Multivar. Anal. 1993, 45, 217–233. [Google Scholar] [CrossRef]
- Beleites, C.; Baumgartner, R.; Bowman, C.; Somorjai, R.; Steiner, G.; Salzer, R.; Sowa, M.G. Variance reduction in estimating classification error using sparse datasets. Chemom. Intell. Lab. Syst. 2005, 79, 91–100. [Google Scholar] [CrossRef]
- Detenbeck, N.E.; Morrison, A.C.; Abele, R.W.; Kopp, D.A. Spatial statistical network models for stream and river temperature in New England, USA. Water Resour. Res. 2016, 52, 6018–6040. [Google Scholar] [CrossRef]
- Post, C.J.; Cope, M.P.; Gerard, P.D.; Masto, N.M.; Vine, J.R.; Stiglitz, R.Y.; Hallstrom, J.O.; Newman, J.C.; Mikhailova, E.A. Monitoring spatial and temporal variation of dissolved oxygen and water temperature in the Savannah River using a sensor network. Environ. Monit. Assess. 2018, 190, 274. [Google Scholar] [CrossRef]
- Frieden, J.C.; Peterson, E.E.; Webb, J.A.; Negus, P.M. Improving the predictive power of spatial statistical models of stream macroinvertebrates using weighted autocovariance functions. Environ. Model. Softw. 2014, 60, 320–330. [Google Scholar] [CrossRef]
- Larsen, S.; Bruno, M.C.; Vaughan, I.P.; Zolezzi, G. Testing the River Continuum Concept with geostatistical stream-network models. Ecol. Complex. 2019, 39, 100773. [Google Scholar] [CrossRef]
- Holcomb, D.A.; Messier, K.P.; Serre, M.L.; Rowny, J.G.; Stewart, J.R. Geostatistical Prediction of Microbial Water Quality Throughout a Stream Network Using Meteorology, Land Cover, and Spatiotemporal Autocorrelation. Environ. Sci. Technol. 2018, 52, 7775–7784. [Google Scholar] [CrossRef] [PubMed]
- Franklin, R.B.; Mills, A.L. The Spatial Distribution of Microbes in the Environment; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Brokamp, C.; Beck, A.F.; Muglia, L.; Ryan, P. Combined sewer overflow events and childhood emergency department visits: A case-crossover study. Sci. Total Environ. 2017, 607–608, 1180–1187. [Google Scholar] [CrossRef] [PubMed]
- Borchardt, M.A.; Chyou, P.-H.; DeVries, E.O.; Belongia, E.A. Septic system density and infectious diarrhea in a defined population of children. Environ. Health Perspect. 2003, 111, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Crowther, J.; Kay, D.; Wyer, M.D. Faecal-indicator concentrations in waters draining lowland pastoral catchments in the UK: Relationships with land use and farming practices. Water Res. 2002, 36, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Burkholder, J.; Libra, B.; Weyer, P.; Heathcote, S.; Kolpin, D.; Thorne, P.S.; Wichman, M. Impacts of Waste from Concentrated Animal Feeding Operations on Water Quality. Environ. Health Perspect. 2007, 115, 308–312. [Google Scholar] [CrossRef]
- Harris, C.S.; Tertuliano, M.; Rajeev, S.; Vellidis, G.; Levy, K. Impact of storm runoff on Salmonella and Escherichia coli prevalence in irrigation ponds of fresh produce farms in southern Georgia. J. Appl. Microbiol. 2018, 124, 910–921. [Google Scholar] [CrossRef]
- Mitsch, W.J.; Gosselink, J.G. Wetlands; John Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar]
- Molleda, P.; Blanco, I.; Ansola, G.; de Luis, E. Removal of wastewater pathogen indicators in a constructed wetland in Leon, Spain. Ecol. Eng. 2008, 33, 252–257. [Google Scholar] [CrossRef]
- Hathaway, J.M.; Hunt, W.F.; Jadlocki, S. Indicator Bacteria Removal in Storm-Water Best Management Practices in Charlotte, North Carolina. J. Environ. Eng. 2009, 135, 1275–1285. [Google Scholar] [CrossRef]
- Hsu, T.-T.D.; Mitsch, W.J.; Martin, J.F.; Lee, J. Towards sustainable protection of public health: The role of an urban wetland as a frontline safeguard of pathogen and antibiotic resistance spread. Ecol. Eng. 2017, 108, 547–555. [Google Scholar] [CrossRef]
- Biswal, B.K.; Balasubramanian, R. Constructed Wetlands for Reclamation and Reuse of Wastewater and Urban Stormwater: A Review. Front. Environ. Sci. 2022, 10, 201. [Google Scholar] [CrossRef]
- Curriero, F.C.; Patz, J.A.; Rose, J.B.; Lele, S. The Association Between Extreme Precipitation and Waterborne Disease Outbreaks in the United States, 1948–1994. Am. J. Public Health 2001, 91, 1194–1199. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.M.; Charron, D.F.; Waltner-Toews, D.; Schuster, C.; Maarouf, A.R.; Holt, J.D. A role of high impact weather events in waterborne disease outbreaks in Canada, 1975–2001. Int. J. Environ. Health Res. 2006, 16, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Mac Kenzie, W.R.; Hoxie, N.J.; Proctor, M.E.; Gradus, M.S.; Blair, K.A.; Peterson, D.E.; Kazmierczak, J.J.; Addiss, D.G.; Fox, K.R.; Rose, J.B.; et al. A Massive Outbreak in Milwaukee of Cryptosporidium Infection Transmitted through the Public Water Supply. N. Engl. J. Med. 1994, 331, 161–167. [Google Scholar] [CrossRef] [PubMed]
- De Roos, A.J.; Kondo, M.C.; Robinson, L.F.; Rai, A.; Ryan, M.; Haas, C.N.; Lojo, J.; Fagliano, J.A. Heavy precipitation, drinking water source, and acute gastrointestinal illness in Philadelphia, 2015–2017. PLoS ONE 2020, 15, e0229258. [Google Scholar] [CrossRef]
- Dupigny-Giroux, L.A.; Mecray, E.L.; Lemcke-Stampone, M.D.; Hodgkins, G.A.; Lentz, E.E.; Mills, K.E.; Lane, E.D.; Miller, R.; Hollinger, D.Y.; Solecki, W.D.; et al. Northeast. In Impacts, Risks, and Adaptation in the United States: Fourth National Climate Assessment; U.S. Global Change Research Program: Washington, DC, USA, 2018; Volume II. [Google Scholar]



| Data | Sources/Methods |
|---|---|
| E. coli | EPA approved Hach mColiBlue24® |
| Urban land use | NJDEP, Bureau of Geographic Information Systems |
| Pasture land use | NJDEP, Bureau of Geographic Information Systems |
| Forest land use | NJDEP, Bureau of Geographic Information Systems |
| Wetland land use | NJDEP, Bureau of Geographic Information Systems |
| Precipitation | NJDEP, Department of Water Monitoring & Standards |
| Response Variable | Autocovariance | Coefficients for Explanatory Variables | Model Performance | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | Structure | Nugget | Partial Sill | Range (km) | % Urban | % Pasture | % Forest | % Wetland | Fixed Effect R2 | Random Effect R2 | Total R2 | LOOCV RMSPE | |
| log10 Geomean (overall) | OLS | Nugget only | 0.041 | NA | NA | * 0.018 | −0.003 | −0.006 | ^ −0.066 | 0.81 | NA | 0.81 | 1.234 |
| Euclidean Distance | Gaussian Euclidean + Nugget | 0.028 | 0.014 | 2.31 | * 0.018 | −0.003 | −0.005 | ^ −0.068 | 0.83 | 0.06 | 0.89 | 1.110 | |
| Stream Distance | LinearSill Tailup + Nugget | 0.000 | 0.040 | 2.87 | * 0.018 | −0.003 | −0.006 | ^ −0.068 | 0.81 | 0.19 | 1.00 | 1.226 | |
| log10 Geomean (storm) | OLS | Nugget only | 0.054 | NA | NA | * 0.021 | 0.014 | −0.008 | −0.026 | 0.83 | NA | 0.83 | 1.396 |
| Euclidean Distance | Gaussian Euclidean + Nugget | 0.033 | 0.005 | 3.11 | * 0.022 | 0.016 | −0.008 | −0.022 | 0.84 | 0.02 | 0.86 | 1.346 | |
| Stream Distance | LinearSill Tailup + Nugget | 0.000 | 0.038 | 3.37 | * 0.021 | * 0.019 | −0.009 | −0.024 | 0.83 | 0.17 | 1.00 | 1.290 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, T.-T.D.; Yu, D.; Wu, M. Predicting Fecal Indicator Bacteria Using Spatial Stream Network Models in A Mixed-Land-Use Suburban Watershed in New Jersey, USA. Int. J. Environ. Res. Public Health 2023, 20, 4743. https://doi.org/10.3390/ijerph20064743
Hsu T-TD, Yu D, Wu M. Predicting Fecal Indicator Bacteria Using Spatial Stream Network Models in A Mixed-Land-Use Suburban Watershed in New Jersey, USA. International Journal of Environmental Research and Public Health. 2023; 20(6):4743. https://doi.org/10.3390/ijerph20064743
Chicago/Turabian StyleHsu, Tsung-Ta David, Danlin Yu, and Meiyin Wu. 2023. "Predicting Fecal Indicator Bacteria Using Spatial Stream Network Models in A Mixed-Land-Use Suburban Watershed in New Jersey, USA" International Journal of Environmental Research and Public Health 20, no. 6: 4743. https://doi.org/10.3390/ijerph20064743
APA StyleHsu, T.-T. D., Yu, D., & Wu, M. (2023). Predicting Fecal Indicator Bacteria Using Spatial Stream Network Models in A Mixed-Land-Use Suburban Watershed in New Jersey, USA. International Journal of Environmental Research and Public Health, 20(6), 4743. https://doi.org/10.3390/ijerph20064743







