Analysis of the Presence of the Virulence and Regulation Genes from Staphylococcus aureus (S. aureus) in Coagulase Negative Staphylococci and the Influence of the Staphylococcal Cross-Talk on Their Functions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material Isolation and Identification
2.1.1. Tested Strains
2.1.2. DNA and RNA Isolation
2.2. Analysis of the Virulence
2.2.1. Phenotypic Analysis of Haemolytic Activity of Tested Isolates
2.2.2. Phenotypic Analysis of Biofilm Formation
2.2.3. Detection of Genes Encoded Virulence Factors
2.3. Antimicrobial Susceptibility Testing
2.4. Agr Detection and Classification
2.5. Cross-Inhibition of the Biofilm Formation and the Influence on δ-Toxin Activity among Agr Groups
2.5.1. Supernatant Preparation
2.5.2. Analysis of Cross-Inhibition of Biofilm Formation
2.5.3. Analysis of the Influence on δ-Toxin Activity
2.6. Statistical analysis
3. Results
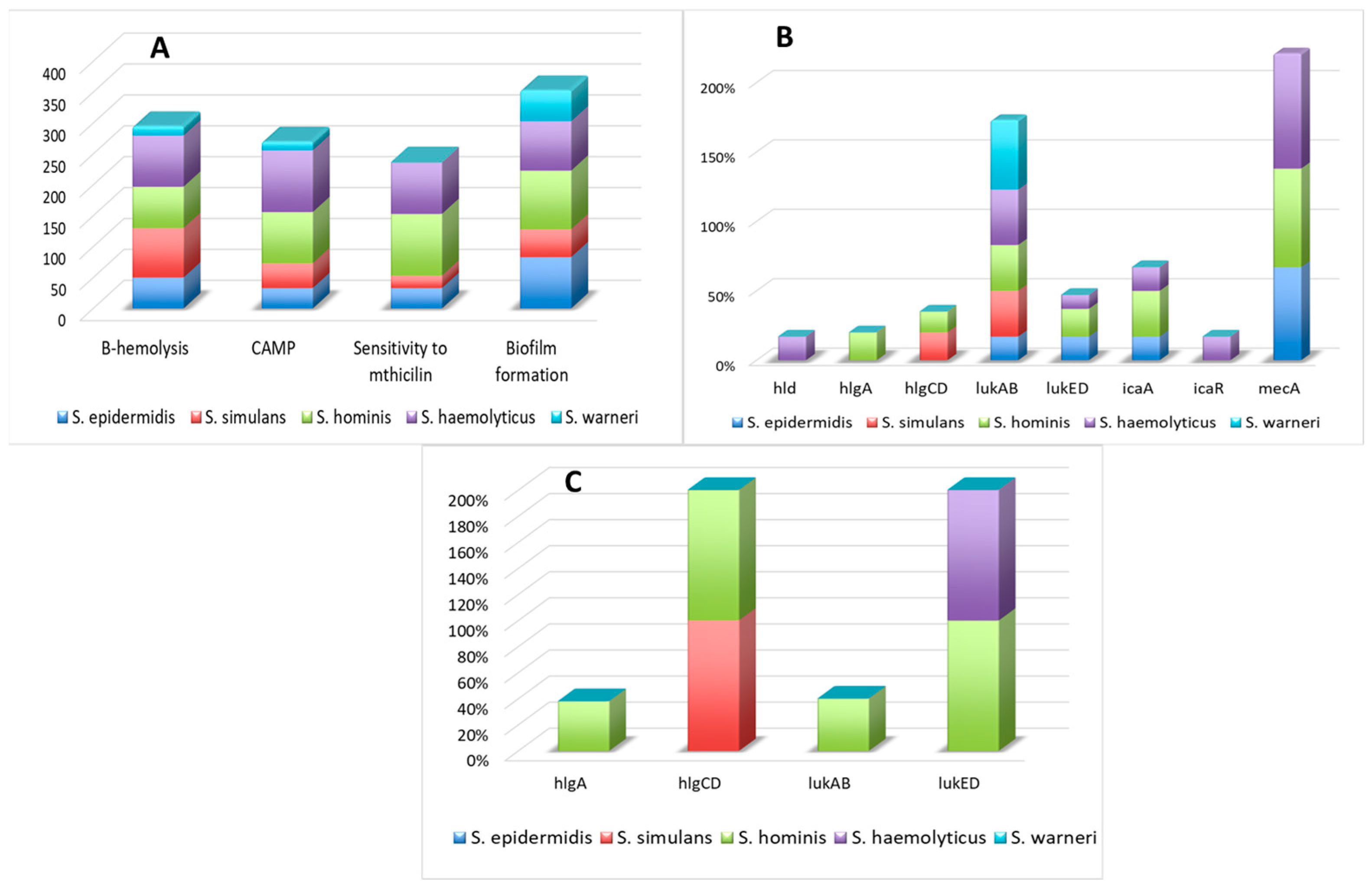
3.1. The Presence of Virulence Factors, mecA Gene, and Susceptibility to Methicillin
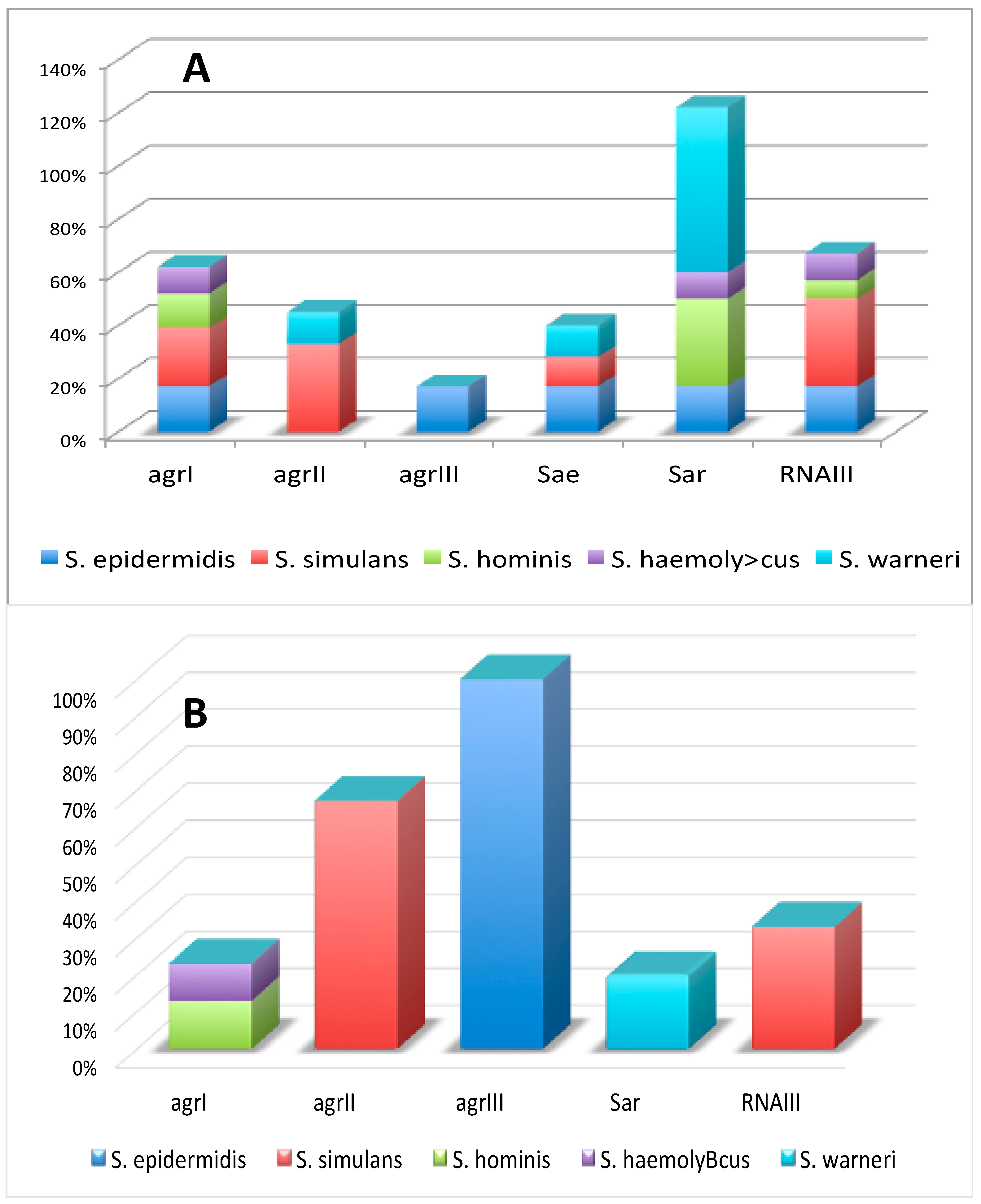
3.2. The Presence of Regulatory Factors
3.3. Statistical Analysis Materials: Będzie Ficant Associa
3.4. Activity of Genes Encoding Virulence and Regulatory Factors
3.5. The Cross-Talk Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yarwood, J.M.; Schlievert, P.M. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 2003, 112, 1620. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, A. Increasing virulence factors. Nat. Rev. Microbiol. 2018, 16, 658. [Google Scholar] [CrossRef] [PubMed]
- Spaan, A.N.; Vrieling, M.; Wallet, P.; Badiou, C.; Reyes-Robles, T.; Ohneck, E.A.; Yvonne Benito, Y.; de Haas, C.J.C.; Day, C.J.; Jennings, M.P.; et al. The staphylococcal toxins γ-haemolysin AB and CB differentially target phagocytes by employing specific chemokine receptors. Nat. Commun. 2014, 11, 5438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenesch, F.; Lin, G.; Henry, T. Staphylococcus aureus haemolysins, bicomponent leukocidins, and cytolytic peptides: A redundant arsenal of membrane-damaging virulence factors? Front. Cell. Infect. Microbial. 2012, 2, 12. [Google Scholar]
- Ventura, C.L.; Malachowa, N.; Hammer, C.H.; Nardone, G.A.; Robinson, M.A.; Kobayashi, S.D.; DeLeo, F.R. Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS ONE 2010, 5, e11634. [Google Scholar] [CrossRef]
- Kawada-Matsuo, M.; Yoshida, Y.; Nakamura, N.; Komatsuzawa, H. Role of two-component systems in the resistance of Staphylococcus aureus to antibacterial agents. Virulence 2011, 2, 427. [Google Scholar] [CrossRef] [Green Version]
- Novick, R.P. Autoinduction and Signal Transduction in the Regulation of Staphylococcal Virulence. Mol. Microbiol. 2003, 48, 1429. [Google Scholar] [CrossRef]
- Divyakolu, S.; Chikkala, R.; Ratnakar, K.S.; Sritharan, V. Haemolysins of Staphylococcus aureus—An Update on Their Biology, Role in Pathogenesis and as Targets for Anti-Virulence Therapy. Adv. Inf. Dis. 2019, 9, 80. [Google Scholar]
- Liu, Q.; Cho, H.; Yeo, W.S.; Bae, T. The extracytoplasmic linker peptide of the sensor protein SaeS tunes the kinase activity required for staphylococcal virulence in response to host signals. PLoS Pathog. 2015, 11, e1004799. [Google Scholar] [CrossRef]
- Lebeau, C.; Vandenesch, F.; Greenland, T.; Novick, R.P.; Etienne, J. Coagulase expression in Staphylococcus aureus is positively and negatively modulated by an agr-dependent mechanism. J. Bacteriol. 1994, 17, 5534. [Google Scholar] [CrossRef] [Green Version]
- Chovanová, R.; Mikulášová, M.; Vaverková, S. In vitro antibacterial and antibiotic resistance modifying effect of bioactive plant extracts on methicillin-resistant Staphylococcus epidermidis. Int. J. Microbiol. 2013, 9, 760969. [Google Scholar]
- Aldman, M.H.; Rasmussen, M.; Olaison, L.; Påhlman, L.I. Endocarditis due to Staphylococcus lugdunensis-A retrospective national registry-based study. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 40, 1103. [Google Scholar] [CrossRef] [PubMed]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ott, E.; Saathoff, S.; Graf, K.; Schwab, F.; Chaberny, I.F. The prevalence of nosocomial and community acquired infections in a university hospital: An observational study. Dtsch. Aerzteblatt Int. 2013, 110, 533. [Google Scholar]
- Papan, C.; Karremann, M.; Weis, M.; Petzold, A.; Zahn, K.; Schroten, H.; Weichert, S.; Tenenbaum, T. A 28-Day-Old Boy with Multifocal Osteomyelitis Mimicking Non-Accidental Injury. Klin. Padiatr. 2021, 233, 91. [Google Scholar] [CrossRef]
- Eichel, V.; Papan, C.; Mutters, N.T. Update Hygiene: Prevention of Vascular Catheter-Associated Infections in Premature and Newborn Infants. Klin. Padiatr. 2019, 231, 177–182. [Google Scholar]
- Marincola, G.; Liong, O.; Schoen, C.; Abouelfetouh, A.; Hamdy, A.; Wencker, F.D.R.; Marciniak, T.; Becker, K.; Köck, R.; Ziebuhr, W. Antimicrobial Resistance Profiles of Coagulase-Negative Staphylococci in Community-Based Healthy Individuals in Germany. Front. Public Health 2021, 17, 684456. [Google Scholar] [CrossRef]
- Deyno, S.; Fekadu, S.; Seyfe, S. Prevalence and antimicrobial resistance of coagulase negative staphylococci clinical isolates from Ethiopia: A meta-analysis. BMC Microbiol. 2018, 18, 43. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Kim, H.K.; Rauch, S.; Schneewind, O.; Missiakas, D. Pathogenic conversion of coagulase-negative staphylococci. Microbes Infect. 2017, 19, 101. [Google Scholar] [CrossRef] [Green Version]
- Szemraj, M.; Grazul, M.; Balcerczak, E.; Szewczyk, E.M. Staphylococcal species less frequently isolated from human clinical specimens-are they a threat for hospital patients? BMC Infect. Dis. 2020, 20, 128. [Google Scholar] [CrossRef] [Green Version]
- Hirotaki, S.; Sasaki, T.; Kuwahara-Arai, K.; Hiramatsu, K. Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR. J. Clin. Microbiol. 2011, 49, 3627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hébert, G.A.; Hancock, G.A. Synergistic haemolysis exhibited by species of Staphylococci. J. Clin. Microbiol. 1985, 22, 409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Tool, G.A. Microtiter dish biofilm formation assay. JoVE 2011, 47, 2437. [Google Scholar]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis 2007, 115, 891. [Google Scholar] [CrossRef] [PubMed]
- Monday, S.R.; Bohach, G.A. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 1999, 37, 3411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarraud, S.; Mougel, C.; Thioulouse, J.; Lina, G.; Meugnier, H.; Forey, F.; Nesme, X.; Etienne, J.; Vandenesch, F. Relationships between Staphylococcus aureus Genetic Background, Virulence Factors, agr Groups (Alleles), and Human Disease. Infect. Immun. 2002, 70, 631. [Google Scholar] [CrossRef] [Green Version]
- Ünal, N.; Çinar, O. Detection of stapylococcal enterotoxin, methicillin-resistant and Panton–Valentine leukocidin genes in coagulase-negative staphylococci isolated from cows and ewes with subclinical mastitis. Trop. Anim. Health Prod. 2012, 44, 369. [Google Scholar] [CrossRef]
- Zhang, K.; McClure, J.A.; Elsayed, S.; Louie, T.; Conly, J.M. Novel Multiplex PCR Assay for Characterization and Concomitant Subtyping of Staphylococcal Cassette Chromosome. J. Clin. Microbiol. 2005, 43, 5026. [Google Scholar] [CrossRef] [Green Version]
- Soares, B.S.; Melo, D.A.; Motta, C.C.; Marques, V.F.; Barreto, N.B.; Coelho, S.M.O.; Coelho, S.; Souza, M.M.S. Characterization of virulence and antibiotic profile and agr typing of Staphylococcus aureus from milk of subclinical mastitis bovine in State of Rio de Janeiro. Arq. Bras. Med. Vet. Zootec. 2017, 69, 843–850. [Google Scholar] [CrossRef] [Green Version]
- Frebourg, N.B.; Lefebvre, S.; Baert, S.; Lemeland, J.F. PCR-Based Assay for Discrimination between Invasive and Contaminating Staphylococcus epidermidis Strains. J. Clin. Microbiol. 2000, 38, 877. [Google Scholar] [CrossRef] [Green Version]
- Giraudo, A.; Calzolari, A.; Cataldi, A.; Bogni, C.; Nagel, R. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 1999, 177, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993, 12, 3967. [Google Scholar] [CrossRef]
- Lina, G.; Boutite, F.; Tristan, A.; Bes, M.; Etienne, J.; Vandenesch, F. Bacterial competition for human nasal cavity colonization: Role of staphylococcal agr alleles. Appl. Environ. Microbiol. 2003, 69, 18. [Google Scholar] [CrossRef] [Green Version]
- Gilot, P.; Lina, G.; Cochard, T.; Poutrel, B. Analysis of the Genetic Variability of Genes Encoding the RNA III-Activating Components Agr and TRAP in a Population of Staphylococcus aureus Strains Isolated from Cows with Mastitis. J. Clin. Microbiol. 2002, 40, 4060. [Google Scholar] [CrossRef] [Green Version]
- Attia, A.S.; Benson, M.A.; Stauff, D.L.; Torres, V.J.; Skaar, E.P. Membrane Damage Elicits an Immunomodulatory Program in Staphylococcus aureus. PLoS Pathog. 2010, 6, e1000802. [Google Scholar] [CrossRef] [Green Version]
- Benson, M.A.; Ohneck, E.A.; Ryan, C.; Alonzo, F.; Smith, H.; Narechania, A.; Kolokotronis, S.O.; Satola, S.W.; Uhlemann, A.C.; Sebra, R.; et al. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol. Microbiol. 2014, 93, 664. [Google Scholar] [CrossRef] [Green Version]
- Alonzo, F.; Benson, M.A.; Chen, J.; Novick, R.P.; Shopsin, B.; Torres, V.J. Staphylococcus aureus leukocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol. Microbiol. 2012, 83, 423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezar, I.F.; Mashruwala, A.A.; Boy, J.M.; Stock, A.M. Drug-like Fragments Inhibit agr-Mediated Virulence Expression in Staphylococcus aureus. Sci. Rep. 2019, 9, 6786. [Google Scholar] [CrossRef] [Green Version]
- Kessel, C.I. Molecular Subtyping of Staphylococcus aureus Isolates from the u.p. Community for the Presence of Toxin-Encoding Genes, Nmu Commons All Nmu. Master’s Thesis, Northern Michigan University, Marquette, MI, USA, 2017; p. 133. [Google Scholar]
- Francois, P.; Koessler, T.; Huyghe, A.; Harbarth, S.; Bento, M.; Lew, D.; Etienne, J.; Pittet, D.; Schrenzel, J. Rapid Staphylococcus aureus agr Type Determination by a Novel Multiplex Real-Time Quantitative PCR Assay. J. Clin. Microbiol. 2006, 44, 1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, X.; Qin, N.; Wu, C.; Sheng, J.; Yang, R.; Zheng, B.; Ma, Z.; Liu, L.; Peng, X.; Jia, A. Transcriptome analysis of the biofilm formed by methicillin-susceptible Staphylococcus aureus. Sci. Rep. 2015, 5, 11997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, Z.; Seleem, M.N.; Hussain, H.I.; Huang, L.; Hao, H.; Yuan, Z. Comparative virulence studies and transcriptome analysis of Staphylococcus aureus strains isolated from animals. Sci. Rep. 2016, 6, 35442. [Google Scholar] [CrossRef]
- Cho, H.; Jeong, D.W.; Liu, Q.; Yeo, W.S.; Vogl, T.; Skaar, E.P.; Chazin, W.J.; Bae, T. Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Staphylococcus aureus Infections. PLoS Pathog. 2015, 11, e1005026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-García, S.; Ortiz-García, C.I.; Cruz-Aguilar, M.; Zenteno, J.C.; Murrieta-Coxca, J.M.; Pérez-Tapia, S.M.; Rodríguez-Martínez, S.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. Competition/antagonism associations of biofilm formation among Staphylococcus epidermidis Agr groups I, II, and III. J. Microbiol. 2019, 57, 143. [Google Scholar] [CrossRef] [PubMed]
- May, L.; Klein, E.Y.; Rothman, R.E.; Laxminarayan, R. Trends in antibiotic resistance in coagulase-negative staphylococci in the United States, 1999 to 2012. Antimicrob. Agents Chemother. 2014, 58, 1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Robles, T.; Torres, V.J. Staphylococcus aureus Pore-Forming Toxin. Curr. Top. Microbiol. Immunol. 2016, 409, 121. [Google Scholar]
- Goerke, C.; Köller, J.; Wolz, C. Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 171. [Google Scholar] [CrossRef] [Green Version]
- Huseby, M.; Shi, K.; Brown, C.K.; Digre, J.; Mengistu, F.; Seo, K.S.; Bohach, G.A.; Schlievert, P.M.; Ohlendorf, D.H.; Earhart, C.A. Structure and Biological Activities of β Toxin from Staphylococcus aureus. J. Bacteriol. 2007, 189, 8719. [Google Scholar] [CrossRef] [Green Version]
- Traber, K.; Novick, R. A Slipped-Mispairing Mutation in AgrA of Laboratory Strains and Clinical Isolates Results in Delayed Activation of agr and Failure to Translate δ- and α-haemolysins. Mol. Microbiol. 2006, 59, 1519. [Google Scholar] [CrossRef]
- Nakamura, Y.; Oscherwitz, J.; Cease, K.B.; Chan, S.M.; Munoz-Planillo, R.; Hasegawa, M.; Villaruz, A.E.; Cheung, G.Y.C.; McGavin, M.J.; Travers, J.B.; et al. Staphylococcus δ-Toxin Induces Allergic Skin Disease by Activating Mast Cells. Nature 2013, 503, 397. [Google Scholar] [CrossRef] [Green Version]
- Burnside, K.; Lembo, A.; de los Reyes, M.; Iliuk, A.; BinhTran, N.T.; Connelly, J.E.; Lin, W.J.; Schmidt, B.Z.; Richardson, A.R.; Fang, F.C.; et al. Regulation of haemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS ONE 2010, 5, e11071. [Google Scholar] [CrossRef]
- Cooney, J.; Kienle, Z.; Foster, T.J.; O’Toole, P.W. The gammahaemolysin locus of Staphylococcus aureus comprises three linked genes, two of which are identical to the genes for the F and S components of leukocidin. Infect. Immun. 1993, 61, 768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiger, T.; Goerke, C.; Mainiero, M.; Kraus, D.; Wolz, C. The virulence regulator Sae of Staphylococcus aureus: Promoter activities and response to phagocytosiFs-related signals. J. Bacteriol. 2008, 190, 3419. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, A.J.; Lindsay, J.A. Staphylococcus aureus innate immune evasion is lineage-specific: A bioinfomatics study. Infect. Genet. Evol. 2013, 19, 7. [Google Scholar] [CrossRef] [PubMed]
- Yoong, P.; Torres, V.J. The effects of Staphylococcus aureus leukotoxins on the host: Cell lysis and beyond. Curr. Opin. Microbiol. 2013, 16, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, T.; Bae, T.; Schneewind, O.; Takeuchi, F.; Hiramatsu, K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: Polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 2008, 190, 300. [Google Scholar] [CrossRef] [Green Version]
- Gravet, A.; Couppie, P.; Meunier, O.; Clyti, E.; Moreau, B.; Pradinaud, R.; Monteil, H.; Prevost, G. Staphylococcus aureus isolated in cases of impetigo produces both epidermolysin A or B and LukE-LukD in 78% of 131 retrospective and prospective cases. J. Clin. Microbiol. 2001, 39, 4349. [Google Scholar] [CrossRef] [Green Version]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Yeh, A.J.; Cheung, G.Y.; Villaruz, A.E.; Tan, V.Y.; Joo, H.S.; Chatterjee, S.S.; Yu, Y.; Otto, M. Basis of virulence in a panton-valentine leukocidin-negative community-associated methicillin-resistant Staphylococcus aureus strain. J. Infect. Dis. 2015, 2011, 472. [Google Scholar] [CrossRef] [Green Version]
- Udo, E.E.; Al-Bustan, M.A.; Jacob, L.E.; Chugh, T.D. Enterotoxin production by coagulase-negative staphylococci in restaurant workers from Kuwait City may be a potential cause of food poisoning. J. Med. Microbiol. 1999, 48, 819. [Google Scholar] [CrossRef]
- Vasconcelos, N.G.; Pereira, V.C.; Araujo, J.P.; da Cunha, M.d.L.R.S. Molecular detection of enterotoxins E, G, H and I in Staphylococcus aureus and coagulase-negative staphylococci isolated from clinical samples of newborns in Brazil. J. Appl. Microbiol. 2011, 111, 749. [Google Scholar] [CrossRef]
- Nasaj, M.; Saeidi, Z.; Asghari, B.; Roshanaei, G.; Arabestani, M.R. Identifcation of haemolysin encodin genes and their association with antimicrobial resistance pattern among clinical isolates of coagulase-negative Staphylococci. BMC Res. Notes 2020, 13, 68. [Google Scholar] [CrossRef] [Green Version]
- da Silva Sdos, S.; Cidral, T.A.; Soares, M.J.; de Melo, M.C. Enterotoxin-Encoding Genes in Staphylococcus spp. from Food Handlers in a University Restaurant. Foodborne Pathog. Dis. 2015, 12, 921. [Google Scholar] [CrossRef] [PubMed]
- MacFadden, D.R.; McGough, S.F.; Fisman, D.; Santillana, M.; Brownstein, J.S. Antibiotic resistance increases with local temperature. Nat. Clim. Chang. 2018, 8, 510. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.C.; Shaw, H.; Rhodes, V.; Hart, A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 2016, 7, 1728. [Google Scholar] [CrossRef] [Green Version]
- Klingenberg, C.; Aarag, E.; Ronnestad, A.; Sollid, J.E.; Abrahamsen, T.G.; Kjeldsen, G.; Flaegstad, T. Coagulase-negative staphylococcal sepsis in neonates. Association between antibiotic resistance, biofilm formation and the host inflammatory response. Pediatr Infect. Dis. J. 2005, 24, 817–822. [Google Scholar] [CrossRef]
- Hira, V.; Sluijter, M.; Goessens, W.H.; Ott, A.; de Groot, R.; Hermans, P.W.; Kornelisse, R.F. Coagulase-negative staphylococcal skin carriage among neonatal intensive care unit personnel: From population to infection. J. Clin. Microbiol. 2010, 48, 3876. [Google Scholar] [CrossRef] [Green Version]
- Kord, M.; Ardebili, A.; Jamalan, M.; Jahanbakhsh, R.; Behnampour, N.; Ghaemia, E.A. Evaluation of Biofilm Formation and Presence of Ica Genes in Staphylococcus epidermidis Clinical Isolates. Osong Public Health Res. Perspect. 2018, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- den Reijer, P.M.; Haisma, E.M.; Lemmens-den Toom, N.A.; Willemse, J.; Koning, R.A.; Demmers, J.A.; Dekkers, D.H.W.; Rijkers, E.; El Ghalbzouri, A.; Nibbering, P.H.; et al. Detection of alpha-toxin and other virulence factors in biofilms of Staphylococcus aureus on polystyrene and a human epidermal model. PLoS ONE 2016, 11, e0145722. [Google Scholar]
- Ma, X.X.; Wang, E.H.; Liu, Y.; Luo, E.J. Antibiotic susceptibility of coagulase-negative staphylococci (CoNS): Emergence of teicoplanin-non-susceptible CoNS strains with inducible resistance to vancomycin. J. Med. Microbiol. 2011, 60, 1661. [Google Scholar] [CrossRef]
- Chlebowicz, M.A.; Nganou, K.; Kozytska, S.; Arends, J.P.; Engelmann, S.; Grundmann, H.; Ohlsen, K.; van Dijl, J.M. Recombination between ccrC genes in a type V (5C2&5) Staphylococcal Cassette Chromosome mec (SCCmec) of Staphylococcus aureus ST398 leads to conversion from methicillin resistance to methicillin susceptibility in vivo. Antimicrob. Agents Chemother. 2010, 54, 783. [Google Scholar]
- Kriegeskorte, A.; Idelevich, E.A.; Schlattmann, A.; Layer, F.; Strommenger, B.; Denis, O.; Paterson, G.K.; Holmes, M.A.; Werner, G.; Becker, K. Comparison of different phenotypic approaches to screen and detect mecC-harboring methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2018, 56, e00826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Alvarez, L. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 2011, 11, 595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wushouer, H.; Zhang, Z.X.; Wang, J.H.; Ji, P.; Zhu, Q.F.; Aishan, R.; Shi, L.W. Trends and relationship between antimicrobial resistance and antibiotic use in Xinjiang Uyghur Autonomous Region, China: Based on a 3 year surveillance data, 2014–2016. J. Infect. Public Health 2018, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Kateete, D.P.; Asiimwe, B.B.; Mayanja, R.; Najjuka, C.F.; Rutebemberwa, E. Species and drug susceptibility profiles of staphylococci isolated from healthy children in Eastern Uganda. PLoS ONE 2020, 15, e0229026. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Lin, J.; Li, Y.; Zhang, W.; Zhou, J.; Han, Z.; Yao, Z. Insights into the epidemiology of methicillin-resistant coagulase-negative Staphylococci carriage in community-based drug users. J. Infect. Public Health 2020, 13, 1742. [Google Scholar] [CrossRef]
- Budri, P.E.; Shore, A.C.; Coleman, D.C.; Kinnevey, P.M.; Humpreys, H.; Fitzgerald-Hughes, D. Observational cross-sectional study of nasal staphylococcal species of medical students of diverse geographical origin, prior to healthcare exposure: Prevalence of SCCmec, fusC, fusB and the arginine catabolite mobile element (ACME) in the absence of selective antibiotic pressure. BMJ Open 2018, 8, e020391. [Google Scholar]
- Cavanagh, J.P.; Wolden, R.; Heise, P.; Esaiassen, E.; Klingenberg, C.; Aarag Fredheim, E.G. Antimicrobial susceptibility and body site distribution of community isolates of coagulase-negative staphylococci. Apmis 2016, 124, 973. [Google Scholar] [CrossRef] [Green Version]
- Bronner, S.; Monteil, H.; Prevost, G. Regulation of virulence determinants in Staphylococcus aureus: Complexity and applications. FEMS Microbiol. Rev. 2004, 28, 183. [Google Scholar] [CrossRef] [Green Version]
- Viedma, E.; Perez-Montarelo, D.; Villa, J.; Munoz-Gallego, I.; Larrosa, N.; Fernandez-Hidalgo, N.; Gavaldà, J.; Almirante, B.; Chaves, F. Sub-inhibitory concentrations of oxacillin modify the expression of agr locus in Staphylococcus aureus clinical strains belonging to different clonal complexes. BMC Infect. Dis. 2018, 16, 177. [Google Scholar] [CrossRef] [Green Version]
- Gray, B.; Hall, P.; Gresham, H. Targeting agrand agr-like quorum sensing systems for development of common therapeutics to treat multiple Gram-positive bacterial infections. Sensors 2013, 13, 5130. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Li, S.R.; Jiang, B.; Hu, X.M.; Li, S. Therapeutic Targeting of the Staphylococcus aureus Accessory Gene Regulator (agr) System. Front. Microbiol. 2018, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Guan, M.; Jiang, X.F.; Yuan, F.Y.; Xu, M.; Zhang, W.Z.; Lu, Y. Genetic polymorphism of the accessory gene regulator (agr) locus in Staphylococcus epidermidis and its association with pathogenicity. J. Med. Microbiol. 2004, 53, 545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Chiueh, W.Y.; Sun, J.R.; Tsao, S.M.; Lu, J.J. Molecular typing and phenotype characterization of methicillin-resistant Staphylococcus aureus isolates from blood in Taiwan. PLoS ONE 2012, 7, e30394. [Google Scholar] [CrossRef] [PubMed]
- Neetu, J.P.; Murugan, S. Genotyping of methicillin resistant Staphylococcus aureus from tertiary care hospitals in Coimbatore, South India. J. Glob. Infect. Dis. 2016, 8, 68. [Google Scholar] [PubMed]
- Nichol, K.A.; Adam, H.J.; Hussain, Z.; Mulvey, M.R.; McCracken, M.; Mataseje, L.F.; Thompson, K.; Kost, S.; Lagacé-Wiens, P.R.S.; Hoban, D.J.; et al. Comparison of community-associated and health care-associated methicillin-resistant Staphylococcus aureus in Canada: Results of the CANWARD 2007–2009 study. Diagn. Microbiol. Infect. Dis. 2011, 69, 320. [Google Scholar] [CrossRef]
- Mohsenzadeh, M.; Ghazvini, K.; Azimian, A. Frequency of specific agr groups and antibiotic resistance in Staphylococcus aureus isolated from bovine mastitis in the northeast of Iran. Vet. Res. Forum. 2015, 6, 295. [Google Scholar] [PubMed]
- Wright, J.S.; Traber, K.E.; Corrigan, R.; Benson, S.A.; Musser, J.M.; Novick, R.P. The agr radiation: An early event in the evolution of staphylococci. J. Bacteriol. 2005, 187, 5585. [Google Scholar] [CrossRef] [Green Version]
- Tegmark, K.; Morfeldt, E.; Arvidson, S. Regulation of agr-dependent virulence genes in Staphylococcus aureus by RNAIII from coagulase-negative staphylococci. J. Bacteriol. 1998, 180, 3181. [Google Scholar] [CrossRef] [Green Version]
- Van Wamel, W.J.; Van Rossum, G.; Verhoef, J.; Vandenbroucke-Grauls, C.M.; Fluit, A.C. Cloning and characterization of an accessory gene regulator (agr)-like locus from Staphylococcus epidermidis. FEMS Microbiol. Lett. 1998, 163, 1. [Google Scholar] [CrossRef]
- Khoramrooz, S.S.; Mansouri, F.; Marashifard, M.; Hosseini, S.A.A.M.; Chenarestane-Olia, F.A.; Ganavehei, B.; Gharibpour, F.; Shahbazi, A.; Mirzaii, M.; Darban-Sarokhalil, D. Detection of biofilm related genes, classical enterotoxin genes and agr typing among Staphylococcus aureus isolated from bovine with subclinical mastitis in southwest of Iran. Microb. Pathog. 2016, 97, 45. [Google Scholar] [CrossRef]
- Ikonomidis, A.; Vasdeki, A.; Kristo, I.; Maniatis, A.N.; Tsakris, A.; Malizos, K.N.; Pournaras, S. Association of biofilm formation and methicillin-resistance with accessory gene regulator (agr) loci in Greek Staphylococcus aureus clones. Microb. Pathog. 2009, 47, 341. [Google Scholar] [CrossRef]
- Khelissa, S.O.; Jama, C.; Abdallah, M.; Boukherroub, R.; Faille, C.; Chihib, N.E. Efect of incubation duration, growth temperature, and abiotic surface type on cell surface properties, adhesion and pathogenicity of biofilm detached Staphylococcus aureus cells. AMB Express. 2017, 7, 191. [Google Scholar] [CrossRef]
- Todd, O.A.; Fidel, P.L.; Harro, J.M.; Hilliard, J.J.; Tkaczyk, C.; Sellman, B.R.; Noverr, M.C.; Peters, B.M. Candida albicans Augments Staphylococcus aureus Virulence by Engaging the Staphylococcal agr Quorum Sensing System. mBio 2019, 10, e00910-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, M.; Echner, H.; Voelter, W.; Gotz, F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 2001, 69, 1957. [Google Scholar] [CrossRef] [Green Version]
- LaSarre, B.; Federle, M.J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73. [Google Scholar] [CrossRef] [Green Version]
- Canovas, J.; Baldry, M.; Bojer, M.S.; Andersen, P.S.; Gless, B.H.; Grzeskowiak, P.K.; Stegger, M.; Damborg, P.; Olsen, C.A.; Ingmer, H. Cross-Talk between Staphylococcus aureus and Other Staphylococcal Species via the agr Quorum Sensing System. Front. Microbiol. 2016, 8, 1949. [Google Scholar] [CrossRef]
- Ji, G.; Pei, W.; Zhang, L.; Qiu, R.; Lin, J.; Benito, Y.; Lina, G.; Novick, R.P. Staphylococcus intermedius Produces a Functional agr Autoinducing Peptide Containing a Cyclic Lactone. J. Bacteriol. 2005, 187, 3139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toledo-Silva, B.; Nogueira de Souza, F.; Piepers, S.; Mertens, K.; Haesebrouck, F.; De Vliegher, S. Metabolites of bovine-associated non-aureus staphylococci influence expression of Staphylococcus aureus agr-related genes in vitro. Vet. Res. 2021, 52, 62. [Google Scholar] [CrossRef]
- Ferreira, F.A.; Rodrigues Souza, R.; de Sousa Moraes, B.; de Amorim Ferreira, A.M.; Américo, M.A.; Longo Fracalanzza, S.E.; dos Santos Silva Couceiro, J.N.; Sá Figueiredo, A.M. Impact of agr dysfunction on virulence profiles and infections associated with a novel methicillin-resistant Staphylococcus aureus (MRSA) variant of the lineage ST1-SCCmec IV. BMC Microbiol. 2013, 13, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, L.; Yang, L.; Parsons, C.; Findlay, V.J.; Molin, S.; Qin, Z. Staphylococcus epidermidis recovered from indwelling catheters exhibit enhanced biofilm dispersal and “selfrenewal” through downregulation of agr. BMC Microbiol. 2012, 12, 102. [Google Scholar] [CrossRef] [Green Version]
- Periasamy, S.; Joo, H.S.; Duong, A.C.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, G.Y.C.; Duong, A.C.; Otto, M. Direct and synergistic haemolysis caused by Staphylococcus phenol-soluble modulins: Implications for diagnosis and pathogenesis. Microbes Infect. 2012, 14, 380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suryadevara, M.; Clark, A.E.; Wolk, D.M.; Carman, A.; Rosenbaum, P.F.; Shaw, J. Molecular characterization of invasive Staphylococcus aureus infection in central New York children: Importance of two clonal groups and inconsistent presence of selected virulence determinants. J. Pediatr. Infect. Dis. Soc. 2013, 2, 30. [Google Scholar] [CrossRef] [Green Version]
- Rudkin, J.K.; Edwards, A.M.; Bowden, M.G.; Brown, E.L.; Pozzi, C.; Waters, E.M.; Chan, W.C.; Williams, P.; O’Gara, J.P.; Massey, R.C. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J. Infect. Dis. 2012, 205, 798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Huber, C.; Stamm, I.; Ziebuhr, W.; Marincola, G.; Bischoff, M.; Strommenger, B.; Jaschkowitz, G.; Marciniak, T.; Cuny, C.; Witte, W.; et al. Silence as a way of niche adaptation: mecC-MRSA with variations in the accessory gene regulator (agr) functionality express kaleidoscopic phenotypes. Sci. Rep. 2020, 10, 14787. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.; Wang, R.; Khan, B.A.; Sturdevant, D.E.; Otto, M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 2011, 79, 1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLeo, F.R.; Otto, M.; Kreiswirth, B.N.; Chambers, H.F. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 2010, 375, 1557. [Google Scholar] [CrossRef] [Green Version]
| Lp. | Transcript | Author | Sequence | Size [bp] |
|---|---|---|---|---|
| 1. | sea | [25] | 5′-GCAGGGAACAGCTTTAGGCGT-3′ 5′-TCTGTAGAAGTATGAAACACG-3′ | 520 |
| 2. | seb | [25] | 5′-ATGTAATTTTGATATTCGCAGTG-3′ 5′-TGCAGGCATCATATCATACCA-3′ | 643 |
| 3. | sei | [25] | 5′-CAACTCGAATTTTCAACAGGTAC-3′ 5′-CAGGCAGTCCATCTCCTG-3′ | 465 |
| 4. | seg | [25] | 5′-CGTCTCCACCTGTTGAAGG-3′ 5′-CCAAGTGATTGTCTATTGTCG-3′ | 327 |
| 5. | Hla | [26] | 5′-CTTTCCAGCCTACTTTTTTATCAGT-3′ 5′-CTGATTACTATCCAAGAAATTCGATTG-3′ | 209 |
| 6. | Hlb | [26] | -GTTGATGAGTAGCTACCTTCAGT-3′ -GTGCACTTACTGACAATAGTGC-3′ | 309 |
| 7. | hld | [26] | 5′-TTAGTGAATTTGTTCACTGTGTCGA-3′ 5′-AAGAATTTTTATCTTAATTAAGGAAGGAGTG-3′ | 111 |
| 8. | hlg | [26] | 5′-CACCAAATGTATAGCCTAAAGTA-3′ 5′-GTCAYAGAGTCCATAATGCATTTAA-3′ | 535 |
| 9. | hlg2 | [26] | 5′-ATAGTCATTAGGATTAGGTTTCACAAAG-3′ 5′-GACATAGAGTCCATAATGCATTYGT-3′ | 390 |
| 10. | hlgCB | [26] | 5′-GCCAATCCGTTATTAGAAAATGC-3′ 5′-CCATAGAYGTAGCAACGGAT-3′ | 938 |
| 11. | lukED | [26] | 5′-TGAAAAAGGTTCAAAGTTGATACGAG-3′ 5′-TGTATTCGATAGCAAAAGCAGTGCA-3′ | 269 |
| 12. | lukAB | [26] | 5′-TCACTTCTCCACCATACTTC-3′ 5′-TATCAGCAGCAACGACTC-3′ | 638 |
| 13. | pvl | [27] | 5′-ATCATTAGGTAAAATGTCTGGACATGATCCA-3′ 5′-GCATCAASTGTATTGGATAGCAAAAGC-3′ | 433 |
| Lp. | Transcript | Author | Sequence | Size [bp] |
|---|---|---|---|---|
| 1. | sarA | [30] | 5′-TGG TCA CTT ATG CTG ACA GAT T-3′ | 313 |
| 5′-TTT GCT TCT GTG ATA CGG TTG-3′ | ||||
| 2. | Sae | [31] | 5′-TGT GGG GTT CAG GAA TTG TT-3′ | 680 |
| 5′-ATT GAT GAG AAG GAT GCC CA-3′ | ||||
| 3. | RNAIII | [32] | 5′-ATGATCACAGAGATGTGA-3′ | 514 |
| 5′-CTGAGTCCTAGGAAACTAACTC-3′ | ||||
| 4. | agrI | [33,34] | 5′-GTCACAAGTACTATAAGCTGCGAT-3′ | 439/441 |
| 5′-ATGCACATGGTGCACATGC-3′ | ||||
| 5. | agrII | [33,34] | 5′-TATTACTAATTGAAAAGTGGCCATAGC-3′ | 572/575 |
| 5′-ATGCACATGGTGCACATGC-3′ | ||||
| 6. | agrIII | [33,34] | 5′-GTAATGTAATAGCTTGTATAATAATACCCAG-3′ | 321/323 |
| 5′-ATGCACATGGTGCACATGC-3′ | ||||
| 7. | agrIV | [33,34] | 5′-ATGCACATGGTGCACATGC-3′ | 657/659 |
| 5′-CGATAATGCCGTAATACCCG3′ |
| Lp. | Transcript | Author | Sequence |
|---|---|---|---|
| 1. | 16S rRNA | [35] | 5′- TGAGATGTTGGGTTAAGTCCCGCA-3′ |
| 5′-CGGTTTCGCTGCCCTTTGTATTGT-3′ | |||
| 2. | hlgA | [36] | 5′-AATCGGAGGCAGTGGCTCATTCAA-3′ |
| 5′-GGACCAGTTGGGTCTTGTGCAAAT-3′ | |||
| 3. | hlgCB | [36] | 5′-TCGGTGGTAATTTCCAATCAGCCC-3′ |
| 5′-CGAATGAATTCGCTTTGACGCCC-3′ | |||
| 4. | lukED | [37] | 5′-GAAATGGGGCGTTACTCAAA-3′ |
| 5′-GAATGGCCAAATCATTCGTT-3′ | |||
| 5. | RNAIII | [38] | 5′-TTTATCTTAATTAAGGAAGGAGTGA-3′ |
| 5′-TGAATTTGTTCACTGTGTCG-3′ | |||
| 6. | lukAB | [39] | 5′-CGT GGA GCG TTA ACT GGA AAT A -3′ |
| 5′-ACA CCT TTA TGT GAC GTA GAT TGA -3′ | |||
| 7. | agrI | [40] | 5′-CCAGCTATAATTAGTGGTATTAAGTACAGTAAACT-3′ |
| 5′-AGGACGCGCTATCAAACATTTT-3′ | |||
| 5′-ATAGGAATTTCGACATTATC-3′ | |||
| 8. | agrII | [40] | 5′-CAATAGTAACAATTTTAGTGACCATGATCA-3′ |
| 5′-GCAGGATCAGTAGTGTATTTTCTTAAAGTT-3′ | |||
| 5′-TTGCAACAGTAGGTTTGTT-3′ | |||
| 9. | agrIII | [40] | 5′-CATTATAACAATTTCACACAGCGTGTT-3′ |
| 5′-GCAAGTGCATAAGAAATTGATACATACA-3′ | |||
| 5′-ATAGTTCTACCAATCTTTTTGG-3′ | |||
| 10. | hld | [41] | 5′-AAGAATTTTTATCTTAATTAAGGAAGGAGTG-3′ |
| 5′-TTAGTGAATTTGTTCACTGTGTGA-3′ | |||
| 11. | sarA | [42] | 5′-GTAATGAGCATGATGAAAGAACTGT-3′ |
| 5′-CGTTGTTTGCTTCAGTGATTCG-3′ | |||
| 12. | sae | [43] | 5′-CAACCATTGCGATTTCTTTACC-3′ |
| 5′-TTAGCTTTAGGTGCTTGTGG-3′ |
| Supernatant | Strain | CAMP | Β-Haemolysis | Biofilm |
|---|---|---|---|---|
| Without supernatant | S. epidermidis ATCC12228 * | + | + | + |
| S. aureus ATCC 25923 * | n/a | + | + | |
| S. simulans 01 * | - | + | ++ | |
| S. simulans 02 * | + | + | +++ | |
| S. epidermidis */** | - | + | +++ | |
| S. simulans 03 ** | + | + | +++ | |
| S. hominis 01 * | + | + | ++ | |
| S. haemolyticus * | + | + | ++ | |
| S. hominis 02 * | + | + | ++ | |
| S. simulans 04 ** | + | + | ++ | |
| S. simulans 03 | S. epidermidis ATCC12228 | + | + | + |
| S. simulans 01 | + | + | + | |
| S. simulans 02 | + | + | + | |
| S. epidermidis | + | + | + | |
| S. hominis 01 | S. epidermidis ATCC12228 | - | + | + |
| S. simulans 01 | - | + | + | |
| S. simulans 02 | - | + | + | |
| S. epidermidis | - | + | + | |
| S. haemolyticus | S. epidermidis ATCC12228 | + | + | + |
| S. simulans 01 | + | + | + | |
| S. simulans 02 | + | + | + | |
| S. epidermidis | + | + | + | |
| S. hominis 02 | S. epidermidis ATCC12228 | - | + | + |
| S. simulans 01 | - | + | + | |
| S. simulans 02 | - | + | + | |
| S. epidermidis | - | + | + | |
| S. simulans 04 | S. epidermidis ATCC12228 | + | + | + |
| S. simulans 01 | + | + | + | |
| S. simulans 02 | + | + | + | |
| S. epidermidis | + | + | + | |
| S. epidermidis | S. epidermidis ATCC12228 | + | + | + |
| S. simulans 01 | + | + | + | |
| S. simulans 02 | + | + | + | |
| S. aureus ATCC 25923 | S. epidermidis ATCC12228 | n/a | + | + |
| S. simulans 01 | n/a | + | + | |
| S. simulans 02 | n/a | + | + | |
| S. epidermidis | n/a | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grazul, M.; Balcerczak, E.; Sienkiewicz, M. Analysis of the Presence of the Virulence and Regulation Genes from Staphylococcus aureus (S. aureus) in Coagulase Negative Staphylococci and the Influence of the Staphylococcal Cross-Talk on Their Functions. Int. J. Environ. Res. Public Health 2023, 20, 5155. https://doi.org/10.3390/ijerph20065155
Grazul M, Balcerczak E, Sienkiewicz M. Analysis of the Presence of the Virulence and Regulation Genes from Staphylococcus aureus (S. aureus) in Coagulase Negative Staphylococci and the Influence of the Staphylococcal Cross-Talk on Their Functions. International Journal of Environmental Research and Public Health. 2023; 20(6):5155. https://doi.org/10.3390/ijerph20065155
Chicago/Turabian StyleGrazul, Magdalena, Ewa Balcerczak, and Monika Sienkiewicz. 2023. "Analysis of the Presence of the Virulence and Regulation Genes from Staphylococcus aureus (S. aureus) in Coagulase Negative Staphylococci and the Influence of the Staphylococcal Cross-Talk on Their Functions" International Journal of Environmental Research and Public Health 20, no. 6: 5155. https://doi.org/10.3390/ijerph20065155
APA StyleGrazul, M., Balcerczak, E., & Sienkiewicz, M. (2023). Analysis of the Presence of the Virulence and Regulation Genes from Staphylococcus aureus (S. aureus) in Coagulase Negative Staphylococci and the Influence of the Staphylococcal Cross-Talk on Their Functions. International Journal of Environmental Research and Public Health, 20(6), 5155. https://doi.org/10.3390/ijerph20065155






