Literary Identification of Differentially Hydroxymethylated DNA Regions for Type 2 Diabetes Mellitus: A Scoping Minireview
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy for Evidence
2.2. Evidence Selection Criteria
- (1)
- Original research studies that might include quantitative, qualitative, and mixed methods, observational studies, interventions, randomized trials, and experiments.
- (2)
- Studies that included type 2 diabetes patients as the primary subjects but might have animals in addition to human subjects.
- (3)
- Studies were published on PubMed prior to 1 August 2022, as the initial search was conducted between 1 June and 1 August 2022 for the required concept areas.
- (4)
- Studies are written in the English language.
- (5)
- Studies that addressed DNA hydroxymethylation and its relation to T2DM.
- (6)
- Studies that had comparisons to healthy or non-diabetic reference groups.
2.3. Evidence Assessment
2.4. Reviewer Protocol
- a.
- If the second reviewer withdrew after completing a section of the review, the completed portion would be included and compared to that of the other independent reviewer.
- b.
- If the second reviewer withdrew prior to completing a section of the review, the work in progress would be excluded, and an additional independent reviewer should be brought in.
- c.
- If an additional second reviewer was unable to be obtained within the constraints, the scoping review could be completed based on the results of a single reviewer.
2.5. Data Extraction
2.6. Summarizing and Reporting
2.7. Ethics and Dissemination
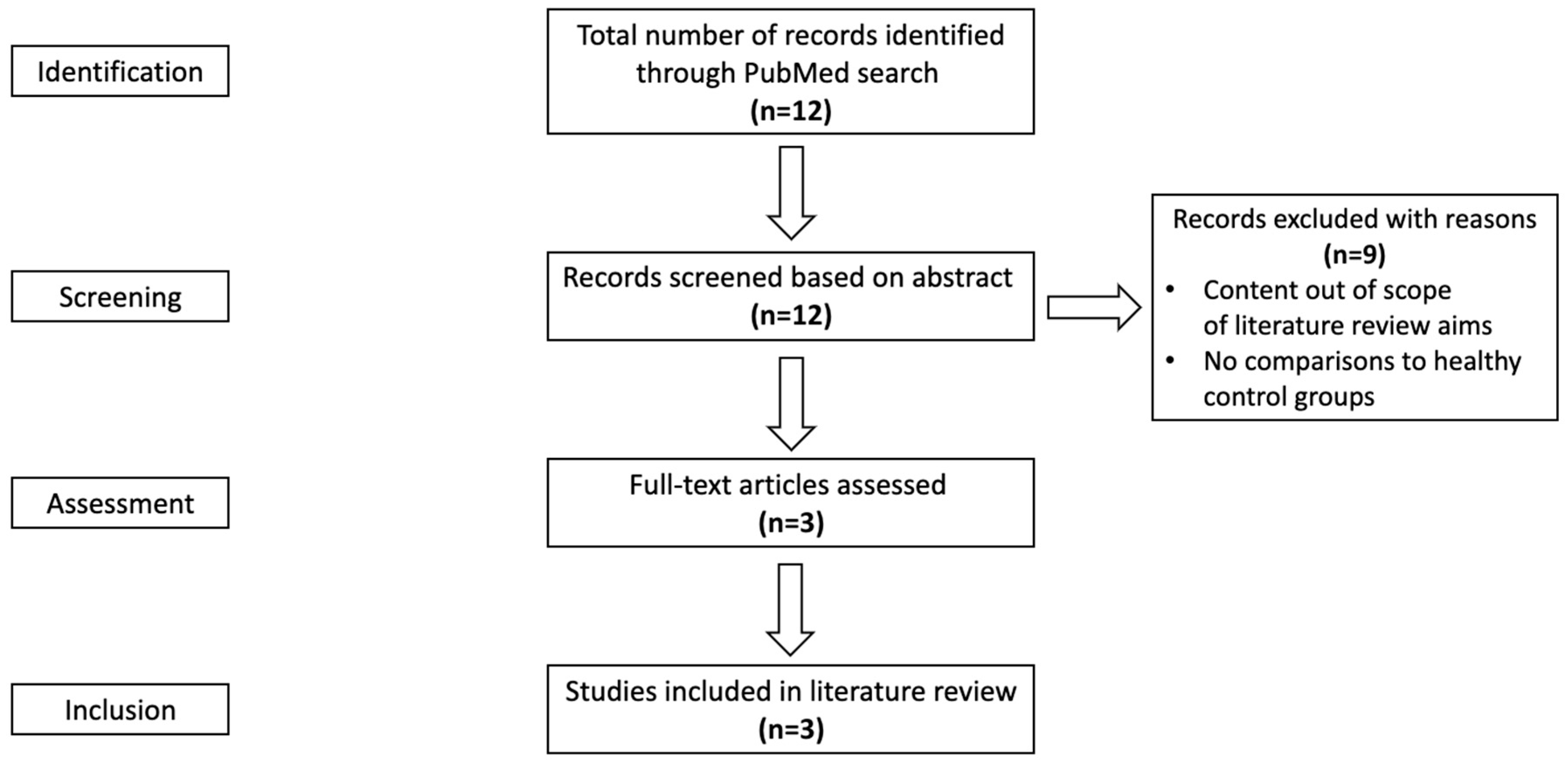
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stumvoll, M.; Goldstein, B.J.; van Haeften, T.W. Type 2 diabetes: Principles of pathogenesis and therapy. Lancet 2005, 365, 1333–1346. [Google Scholar] [CrossRef]
- Rix, I.; Nexøe-Larsen, C.; Bergmann, N.C.; Lund, A.; Knop, F.K. Glucagon Physiology. In Endotext; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Fowler, M.J. Microvascular and Macrovascular Complications of Diabetes. Clin. Diabetes 2008, 26, 77–82. [Google Scholar] [CrossRef]
- Dai, J.; Leung, M.; Guan, W.; Guo, H.T.; Krasnow, R.E.; Wang, T.J.; El-Rifai, W.; Zhao, Z.; Reed, T. Whole-Genome Differentially Hydroxymethylated DNA Regions among Twins Discordant for Cardiovascular Death. Genes 2021, 12, 1183. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Krasnow, R.E.; Liu, L.; Sawada, S.G.; Reed, T. The association between postload plasma glucose levels and 38-year mortality risk of coronary heart disease: The prospective NHLBI Twin Study. PLoS ONE 2013, 8, e69332. [Google Scholar] [CrossRef] [PubMed]
- ElSayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Hilliard, M.E.; Isaacs, D.; Johnson, E.L.; et al. 2. Classification and Diagnosis of Diabetes: Standards of Care in Diabetes—2023. Diabetes Care 2022, 46, S19–S40. [Google Scholar] [CrossRef] [PubMed]
- Association, A.D. Diagnosis and classification of diabetes mellitus. Diabetes Care 2014, 37 (Suppl. 1), S81–S90. [Google Scholar] [CrossRef] [PubMed]
- Eyth, E.; Basit, H.; Swift, C.J. Glucose Tolerance Test. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2023. [Google Scholar]
- Saudek, C.D.; Brick, J.C. The clinical use of hemoglobin A1c. J. Diabetes Sci. Technol. 2009, 3, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Farmer, A. Use of HbA1c in the diagnosis of diabetes. BMJ 2012, 345, e7293. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Meng, H.; Cao, Y.; Qin, J.; Song, X.; Zhang, Q.; Shi, Y.; Cao, L. DNA methylation, its mediators and genome integrity. Int. J. Biol. Sci. 2015, 11, 604–617. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Makleff, S.; Varshney, K.; Krishna, R.N.; Romero, L.; Fisher, J. Mental Health and Community Resilience among Vulnerable Populations Affected by Natural Hazards: Protocol for Scoping Reviews. Methods Protoc. 2022, 5, 88. [Google Scholar] [CrossRef]
- Nnyanzi, L.A.; Adisa, A.O.; Kanmodi, K.K.; Aladelusi, T.O.; Salami, A.A.; Amzat, J.; Angione, C.; Nwafor, J.N.; Uwambaye, P.; Okee, M.; et al. Status of Omics Research Capacity on Oral Cancer in Africa: A Systematic Scoping Review Protocol. BioMedInformatics 2023, 3, 327–338. [Google Scholar] [CrossRef]
- Falagas, M.E.; Pitsouni, E.I.; Malietzis, G.A.; Pappas, G. Comparison of PubMed, Scopus, Web of Science, and Google Scholar: Strengths and weaknesses. Faseb J. 2008, 22, 338–342. [Google Scholar] [CrossRef]
- Waffenschmidt, S.; Knelangen, M.; Sieben, W.; Bühn, S.; Pieper, D. Single screening versus conventional double screening for study selection in systematic reviews: A methodological systematic review. BMC Med. Res. Methodol. 2019, 19, 132. [Google Scholar] [CrossRef]
- Pinzón-Cortés, J.A.; Perna-Chaux, A.; Rojas-Villamizar, N.S.; Díaz-Basabe, A.; Polanía-Villanueva, D.C.; Jácome, M.F.; Mendivil, C.O.; Groot, H.; López-Segura, V. Effect of diabetes status and hyperglycemia on global DNA methylation and hydroxymethylation. Endocr. Connect. 2017, 6, 708–725. [Google Scholar] [CrossRef] [PubMed]
- Yuan, E.F.; Yang, Y.; Cheng, L.; Deng, X.; Chen, S.M.; Zhou, X.; Liu, S.M. Hyperglycemia affects global 5-methylcytosine and 5-hydroxymethylcytosine in blood genomic DNA through upregulation of SIRT6 and TETs. Clin. Epigenetics 2019, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, M.; Bacalini, M.G.; Barchetta, I.; Scalea, S.; Cimini, F.A.; Bertoccini, L.; Tagliatesta, S.; De Matteis, G.; Zardo, G.; Cavallo, M.G.; et al. Increased PARylation impacts the DNA methylation process in type 2 diabetes mellitus. Clin. Epigenetics 2021, 13, 114. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Chen, C.; Lu, X.; Song, Y.; Zhang, Z.; Zeng, C.; Chiu, R.; Li, L.; Xu, M.; He, C.; et al. Alterations of 5-hydroxymethylcytosines in circulating cell-free DNA reflect retinopathy in type 2 diabetes. Genomics 2021, 113, 79–87. [Google Scholar] [CrossRef]
- Yang, Y.; Zeng, C.; Lu, X.; Song, Y.; Nie, J.; Ran, R.; Zhang, Z.; He, C.; Zhang, W.; Liu, S.M. 5-Hydroxymethylcytosines in Circulating Cell-Free DNA Reveal Vascular Complications of Type 2 Diabetes. Clin. Chem. 2019, 65, 1414–1425. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, V.R.; Jarmasz, J.S.; Murugeshan, N.; Del Bigio, M.R.; Rastegar, M.; Davie, J.R. DNA modifications: Function and applications in normal and disease States. Biology 2014, 3, 670–723. [Google Scholar] [CrossRef] [PubMed]
- Nestor, C.E.; Lentini, A.; Hägg Nilsson, C.; Gawel, D.R.; Gustafsson, M.; Mattson, L.; Wang, H.; Rundquist, O.; Meehan, R.R.; Klocke, B.; et al. 5-Hydroxymethylcytosine Remodeling Precedes Lineage Specification during Differentiation of Human CD4(+) T Cells. Cell Rep. 2016, 16, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zeng, C.; Yang, K.; Xu, S.; Zhang, Z.; Cai, Q.; He, C.; Zhang, W.; Liu, S.M. Genome-wide Analysis Reflects Novel 5-Hydroxymethylcytosines Implicated in Diabetic Nephropathy and the Biomarker Potential. Extracell. Vesicles Circ. Nucl. Acids 2022, 3, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Pirola, C.J.; Scian, R.; Gianotti, T.F.; Dopazo, H.; Rohr, C.; Martino, J.S.; Castaño, G.O.; Sookoian, S. Epigenetic Modifications in the Biology of Nonalcoholic Fatty Liver Disease: The Role of DNA Hydroxymethylation and TET Proteins. Medicine 2015, 94, e1480. [Google Scholar] [CrossRef]
- Hathaway, Q.A.; Roth, S.M.; Pinti, M.V.; Sprando, D.C.; Kunovac, A.; Durr, A.J.; Cook, C.C.; Fink, G.K.; Cheuvront, T.B.; Grossman, J.H.; et al. Machine-learning to stratify diabetic patients using novel cardiac biomarkers and integrative genomics. Cardiovasc. Diabetol. 2019, 18, 78. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.A.; Torres, E.R.; Impey, S.; Stevens, J.F.; Raber, J. Apolipoprotein E4 and Insulin Resistance Interact to Impair Cognition and Alter the Epigenome and Metabolome. Sci. Rep. 2017, 7, 43701. [Google Scholar] [CrossRef] [PubMed]
- Spallotta, F.; Cencioni, C.; Atlante, S.; Garella, D.; Cocco, M.; Mori, M.; Mastrocola, R.; Kuenne, C.; Guenther, S.; Nanni, S.; et al. Stable Oxidative Cytosine Modifications Accumulate in Cardiac Mesenchymal Cells From Type2 Diabetes Patients: Rescue by α-Ketoglutarate and TET-TDG Functional Reactivation. Circ. Res. 2018, 122, 31–46. [Google Scholar] [CrossRef]
- Kang, M.; Park, S.; Park, S.H.; Lee, H.G.; Park, J.H. A Double-Edged Sword: The Two Faces of PARylation. Int. J. Mol. Sci. 2022, 23, 9826. [Google Scholar] [CrossRef]
- Ciccarone, F.; Zampieri, M.; Caiafa, P. PARP1 orchestrates epigenetic events setting up chromatin domains. Semin. Cell Dev. Biol. 2017, 63, 123–134. [Google Scholar] [CrossRef]
- Choi, E.H.; Park, S.J. TXNIP: A key protein in the cellular stress response pathway and a potential therapeutic target. Exp. Mol. Med. 2023, 55, 1348–1356. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.H.; Ansari, S.A.; Mensah-Brown, E.P.K.; Emerald, B.S. The role of DNA methylation in the pathogenesis of type 2 diabetes mellitus. Clin. Epigenetics 2020, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Rehman, K.; Akash, M.S.H.; Liaqat, A.; Kamal, S.; Qadir, M.I.; Rasul, A. Role of Interleukin-6 in Development of Insulin Resistance and Type 2 Diabetes Mellitus. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 229–236. [Google Scholar] [CrossRef]
- Klimczak, S.; Śliwińska, A. Epigenetic regulation of inflammation in insulin resistance. Semin. Cell Dev. Biol. 2024, 154, 185–192. [Google Scholar] [CrossRef]
- Wu, S.H.; Neale, M.C.; Acton, A.J., Jr.; Considine, R.V.; Krasnow, R.E.; Reed, T.; Dai, J. Genetic and environmental influences on the prospective correlation between systemic inflammation and coronary heart disease death in male twins. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 2168–2174. [Google Scholar] [CrossRef]
- Su, S.; Snieder, H.; Miller, A.H.; Ritchie, J.; Bremner, J.D.; Goldberg, J.; Dai, J.; Jones, L.; Murrah, N.V.; Zhao, J.; et al. Genetic and environmental influences on systemic markers of inflammation in middle-aged male twins. Atherosclerosis 2008, 200, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Miller, A.H.; Bremner, J.D.; Goldberg, J.; Jones, L.; Shallenberger, L.; Buckham, R.; Murrah, N.V.; Veledar, E.; Wilson, P.W.; et al. Adherence to the mediterranean diet is inversely associated with circulating interleukin-6 among middle-aged men: A twin study. Circulation 2008, 117, 169–175. [Google Scholar] [CrossRef]
- Tanti, J.F.; Ceppo, F.; Jager, J.; Berthou, F. Implication of inflammatory signaling pathways in obesity-induced insulin resistance. Front. Endocrinol. 2012, 3, 181. [Google Scholar] [CrossRef]
- Chambers, J.C.; Loh, M.; Lehne, B.; Drong, A.; Kriebel, J.; Motta, V.; Wahl, S.; Elliott, H.R.; Rota, F.; Scott, W.R.; et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: A nested case-control study. Lancet. Diabetes Endocrinol. 2015, 3, 526–534. [Google Scholar] [CrossRef]
- Dayeh, T.; Tuomi, T.; Almgren, P.; Perfilyev, A.; Jansson, P.A.; de Mello, V.D.; Pihlajamäki, J.; Vaag, A.; Groop, L.; Nilsson, E.; et al. DNA methylation of loci within ABCG1 and PHOSPHO1 in blood DNA is associated with future type 2 diabetes risk. Epigenetics 2016, 11, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Oishi, Y.; Spann, N.J.; Link, V.M.; Muse, E.D.; Strid, T.; Edillor, C.; Kolar, M.J.; Matsuzaka, T.; Hayakawa, S.; Tao, J.; et al. SREBP1 Contributes to Resolution of Pro-inflammatory TLR4 Signaling by Reprogramming Fatty Acid Metabolism. Cell Metab. 2017, 25, 412–427. [Google Scholar] [CrossRef] [PubMed]
- Minn, A.H.; Hafele, C.; Shalev, A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology 2005, 146, 2397–2405. [Google Scholar] [CrossRef] [PubMed]
| Search | Source Title | Author |
|---|---|---|
| MeSH | Alterations of 5-hydroxymethylcytosines in circulating cell-free DNA reflect retinopathy in type 2 diabetes. | Han et al. [23] |
| 5-Hydroxymethylcytosines in Circulating Cell-Free DNA Reveal Vascular Complications of Type 2 Diabetes. | Yang et al. [24] | |
| Hyperglycemia affects global 5-methylcytosine and 5-hydroxymethylcytosine in blood genomic DNA through upregulation of SIRT6 and TETs. ** | Yuan et al. [21] | |
| PubMed Keyword search | Alterations of 5-hydroxymethylcytosines in circulating cell-free DNA reflect retinopathy in type 2 diabetes. | Han et al. [23] |
| Increased PARylation impacts the DNA methylation process in type 2 diabetes mellitus. * | Zampieri et al. [22] | |
| 5-Hydroxymethylcytosines in Circulating Cell-Free DNA Reveal Vascular Complications of Type 2 Diabetes. | Yang et al. [24] | |
| Effect of diabetes status and hyperglycemia on global DNA methylation and hydroxymethylation. * | Pinzon-Cortes et al. [20] | |
| Hyperglycemia affects global 5-methylcytosine and 5-hydroxymethylcytosine in blood genomic DNA through upregulation of SIRT6 and TETs. ** | Yuan et al. [21] | |
| DNA modifications: function and applications in normal and disease States. | Liyanage et al. [25] | |
| 5-Hydroxymethylcytosine Remodeling Precedes Lineage Specification during Differentiation of Human CD4(+) T Cells. | Nestor et al. [26] | |
| Genome-wide Analysis Reflects Novel 5-Hydroxymethylcytosines Implicated in Diabetic Nephropathy and the Biomarker Potential. | Yang et al. [27] | |
| Epigenetic Modifications in the Biology of Nonalcoholic Fatty Liver Disease: The Role of DNA Hydroxymethylation and TET Proteins. | Pirola et al. [28] | |
| Machine-learning to stratify diabetic patients using novel cardiac biomarkers and integrative genomics. | Hathaway et al. [29] | |
| Apolipoprotein E4 and Insulin Resistance Interact to Impair Cognition and Alter the Epigenome and Metabolome. | Johnson et al. [30] | |
| Stable Oxidative Cytosine Modifications Accumulate in Cardiac Mesenchymal Cells From Type 2 Diabetes Patients: Rescue by α-Ketoglutarate and TET-TDG Functional Reactivation. | Spallota et al. [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luong, R.A.M.; Guan, W.; Vue, F.C.; Dai, J. Literary Identification of Differentially Hydroxymethylated DNA Regions for Type 2 Diabetes Mellitus: A Scoping Minireview. Int. J. Environ. Res. Public Health 2024, 21, 177. https://doi.org/10.3390/ijerph21020177
Luong RAM, Guan W, Vue FC, Dai J. Literary Identification of Differentially Hydroxymethylated DNA Regions for Type 2 Diabetes Mellitus: A Scoping Minireview. International Journal of Environmental Research and Public Health. 2024; 21(2):177. https://doi.org/10.3390/ijerph21020177
Chicago/Turabian StyleLuong, Ryan Anh Minh, Weihua Guan, Fue Chee Vue, and Jun Dai. 2024. "Literary Identification of Differentially Hydroxymethylated DNA Regions for Type 2 Diabetes Mellitus: A Scoping Minireview" International Journal of Environmental Research and Public Health 21, no. 2: 177. https://doi.org/10.3390/ijerph21020177





