Molecular Effects of Physical Activity and Body Composition: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Study Selection Criteria
2.3. Search Strategy
2.4. Study Quality Assessment
2.5. Analysis
3. Results
3.1. Study Selection
3.2. Participant Characteristics
3.3. Study Design
3.4. Study Quality Assessment Results
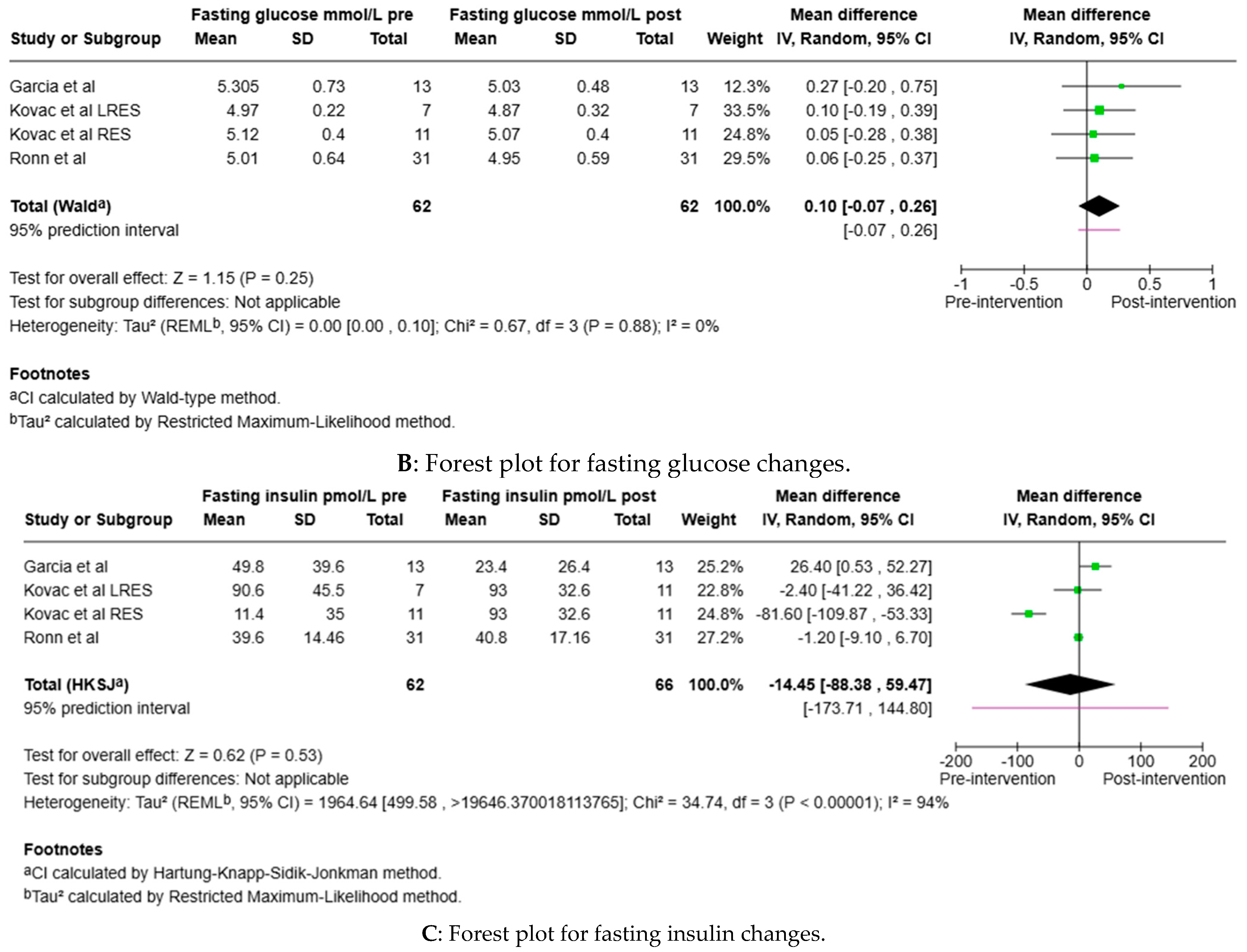
3.5. Clinical Metrics
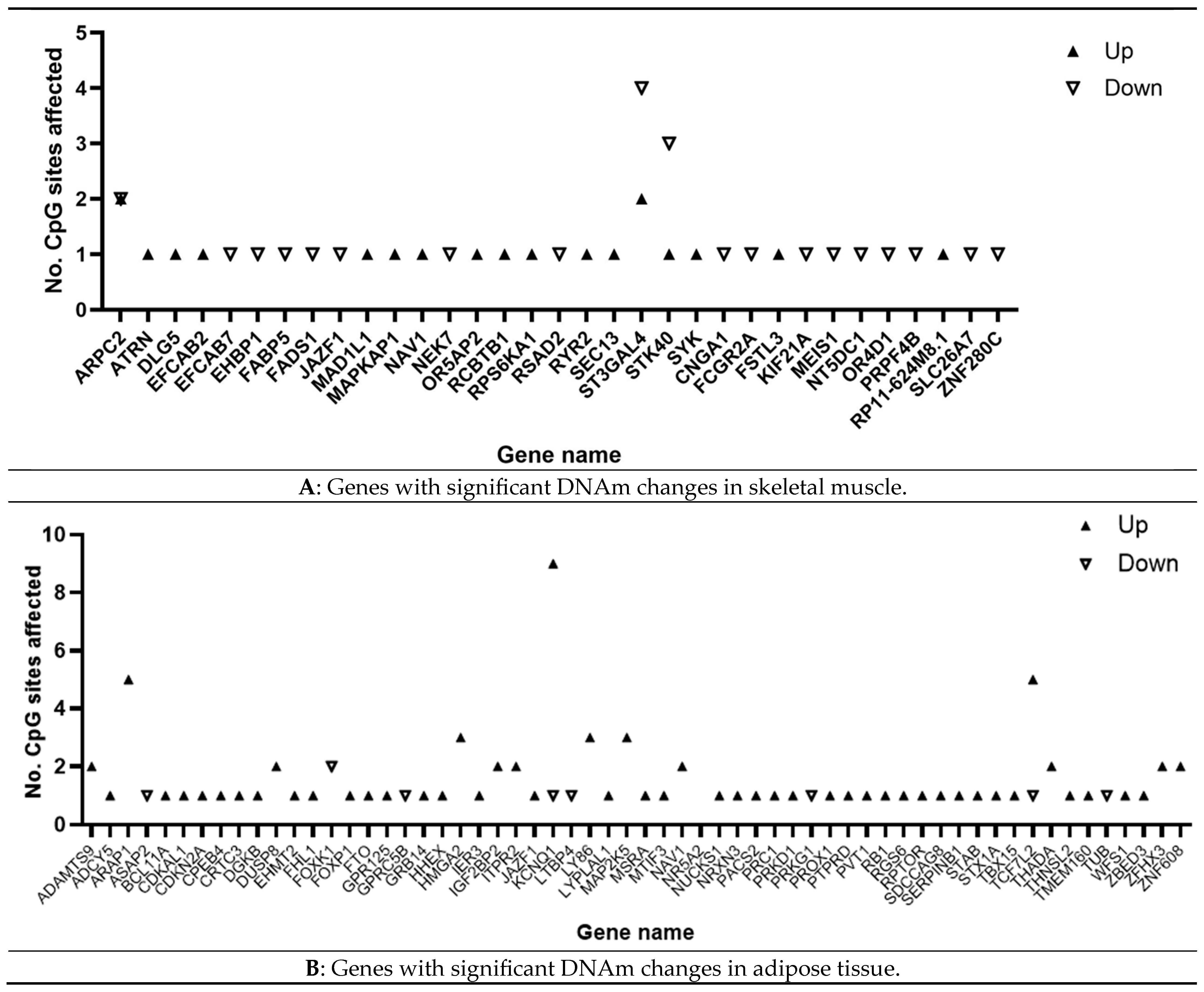
3.6. Epigenomic Meta-Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AT | Adipose tissue |
| BC | Body composition |
| BMI | Body mass index |
| CpG | Cytosine–phosphate–guanine |
| CVD | Cardiovascular disease |
| DNA | Deoxyribonucleic acid |
| DNAm | DNA methylation |
| EWAS | Epigenome-wide association studies |
| FDR | False discovery rate |
| GWAS | Genome-wide association studies |
| LRES | Low responders to exercise |
| MET | Metabolic equivalents |
| NICE | National Institute for Health and Care Excellence |
| PA | Physical activity |
| RES | High responders to exercise |
| RNA | Ribonucleic acid |
| SB | Sedentary behaviour |
| SKM | Skeletal muscle |
| T2D | Type II diabetes |
| WHO | World Health Organisation |
References
- Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 2 February 2025).
- Overweight and Obesity Management. Available online: https://www.nice.org.uk/guidance/ng246 (accessed on 14 February 2025).
- Pan, D.Z.; Miao, Z.; Comenho, C.; Rajkumar, S.; Koka, A.; Lee, S.H.T.; Alvarez, M.; Kaminska, D.; Ko, A.; Sinsheimer, J.S.; et al. Identification of TBX15 as an Adipose Master Trans Regulator of Abdominal Obesity Genes. Genome Med. 2021, 13, 123. [Google Scholar] [CrossRef] [PubMed]
- Kornmueller, K.; Amri, E.-Z.; Scheideler, M.; Prassl, R. Delivery of miRNAs to the Adipose Organ for Metabolic Health. Adv. Drug Deliv. Rev. 2022, 181, 114110. [Google Scholar] [CrossRef] [PubMed]
- Rubino, F.; Cummings, D.E.; Eckel, R.H.; Cohen, R.V.; Wilding, J.P.H.; Brown, W.A.; Stanford, F.C.; Batterham, R.L.; Farooqi, I.S.; Farpour-Lambert, N.J.; et al. Definition and Diagnostic Criteria of Clinical Obesity. Lancet Diabetes Endocrinol. 2025, 13, p221–p262. [Google Scholar] [CrossRef] [PubMed]
- Physical Activity. Available online: https://www.who.int/news-room/fact-sheets/detail/physical-activity (accessed on 2 February 2025).
- Dasso, N.A. How Is Exercise Different from Physical Activity? A Concept Analysis: DASSO. Nurs. Forum 2019, 54, 45–52. [Google Scholar] [CrossRef]
- Physical Activity Guidelines: Adults and Older Adults. Available online: https://www.gov.uk/government/publications/physical-activity-guidelines-adults-and-older-adults (accessed on 2 February 2025).
- Craig, C.L.; Marshall, A.L.; Sjöström, M.; Bauman, A.E.; Booth, M.L.; Ainsworth, B.E.; Pratt, M.; Ekelund, U.; Yngve, A.; Sallis, J.F.; et al. International Physical Activity Questionnaire: 12-Country Reliability and Validity. Med. Sci. Sports Exerc. 2003, 35, 1381–1395. [Google Scholar] [CrossRef]
- Edwardson, C.L.; Gorely, T.; Davies, M.J.; Gray, L.J.; Khunti, K.; Wilmot, E.G.; Yates, T.; Biddle, S.J.H. Association of Sedentary Behaviour with Metabolic Syndrome: A Meta-Analysis. PLoS ONE 2012, 7, e34916. [Google Scholar] [CrossRef]
- Engin, B.; Willis, S.A.; Malaikah, S.; Sargeant, J.A.; Biddle, G.J.H.; Razieh, C.; Argyridou, S.; Edwardson, C.L.; Jelleyman, C.; Stensel, D.J.; et al. Sedentary Time Is Independently Related to Adipose Tissue Insulin Resistance in Adults With or at Risk of Type 2 Diabetes. Med. Sci. Sports Exerc. 2023, 55, 1548–1554. [Google Scholar] [CrossRef]
- Henson, J.; De Craemer, M.; Yates, T. Sedentary Behaviour and Disease Risk. BMC Public Health 2023, 23, 2048. [Google Scholar] [CrossRef]
- Lavin, K.M.; Coen, P.M.; Baptista, L.C.; Bell, M.B.; Drummer, D.; Harper, S.A.; Lixandrão, M.E.; McAdam, J.S.; O’Bryan, S.M.; Ramos, S.; et al. State of Knowledge on Molecular Adaptations to Exercise in Humans: Historical Perspectives and Future Directions. In Comprehensive Physiology; Prakash, Y.S., Ed.; Wiley: Hoboken, NJ, USA, 2022; pp. 3193–3279. ISBN 978-0-470-65071-4. [Google Scholar]
- Noone, J.; Mucinski, J.M.; DeLany, J.P.; Sparks, L.M.; Goodpaster, B.H. Understanding the Variation in Exercise Responses to Guide Personalized Physical Activity Prescriptions. Cell Metab. 2024, 36, 702–724. [Google Scholar] [CrossRef]
- Rask-Andersen, M.; Karlsson, T.; Ek, W.E.; Johansson, Å. Genome-Wide Association Study of Body Fat Distribution Identifies Adiposity Loci and Sex-Specific Genetic Effects. Nat. Commun. 2019, 10, 339. [Google Scholar] [CrossRef]
- Keller, M.; Svensson, S.I.A.; Rohde-Zimmermann, K.; Kovacs, P.; Böttcher, Y. Genetics and Epigenetics in Obesity: What Do We Know so Far? Curr. Obes. Rep. 2023, 12, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Światowy, W.J.; Drzewiecka, H.; Kliber, M.; Sąsiadek, M.; Karpiński, P.; Pławski, A.; Jagodziński, P.P. Physical Activity and DNA Methylation in Humans. Int. J. Mol. Sci. 2021, 22, 12989. [Google Scholar] [CrossRef] [PubMed]
- Wahl, S.; Drong, A.; Lehne, B.; Loh, M.; Scott, W.R.; Kunze, S.; Tsai, P.-C.; Ried, J.S.; Zhang, W.; Yang, Y.; et al. Epigenome-Wide Association Study of Body Mass Index, and the Adverse Outcomes of Adiposity. Nature 2017, 541, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Aurich, S.; Müller, L.; Kovacs, P.; Keller, M. Implication of DNA Methylation during Lifestyle Mediated Weight Loss. Front. Endocrinol. 2023, 14, 1181002. [Google Scholar] [CrossRef]
- Voisin, S.; Eynon, N.; Yan, X.; Bishop, D.J. Exercise Training and DNA Methylation in Humans. Acta Physiol. 2015, 213, 39–59. [Google Scholar] [CrossRef]
- Ulrey, C.L.; Liu, L.; Andrews, L.G.; Tollefsbol, T.O. The Impact of Metabolism on DNA Methylation. Hum. Mol. Genet. 2005, 14, R139–R147. [Google Scholar] [CrossRef]
- Widmann, M.; Nieß, A.M.; Munz, B. Physical Exercise and Epigenetic Modifications in Skeletal Muscle. Sports Med. 2019, 49, 509–523. [Google Scholar] [CrossRef]
- Spartano, N.L.; Wang, R.; Yang, Q.; Chernofsky, A.; Murabito, J.M.; Vasan, R.S.; Levy, D.; Beiser, A.S.; Seshadri, S. Association of Accelerometer-Measured Physical Activity and Sedentary Time with Epigenetic Markers of Aging. Med. Sci. Sports Exerc. 2023, 55, 264–272. [Google Scholar] [CrossRef]
- Da Silva Rodrigues, G.; Noma, I.H.Y.; Noronha, N.Y.; Watanabe, L.M.; Da Silva Sobrinho, A.C.; De Lima, J.G.R.; Sae-Lee, C.; Benjamim, C.J.R.; Nonino, C.B.; Bueno, C.R. Eight Weeks of Physical Training Decreases 2 Years of DNA Methylation Age of Sedentary Women. Res. Q. Exerc. Sport 2024, 95, 405–415. [Google Scholar] [CrossRef]
- Day, S.E.; Coletta, R.L.; Kim, J.Y.; Campbell, L.E.; Benjamin, T.R.; Roust, L.R.; De Filippis, E.A.; Dinu, V.; Shaibi, G.Q.; Mandarino, L.J.; et al. Next-Generation Sequencing Methylation Profiling of Subjects with Obesity Identifies Novel Gene Changes. Clin. Epigenetics 2016, 8, 77. [Google Scholar] [CrossRef]
- Nishida, Y.; Hara, M.; Ohmomo, H.; Ono, K.; Shimizu, A.; Horita, M.; Shimanoe, C.; Taguchi, N.; Higaki, Y.; Tanaka, K. Epigenome-Wide Association Study Identified VTI1A DNA Methylation Associated With Accelerometer-Assessed Physical Activity. Med. Sci. Sports Exerc. 2022, 54, 1879–1888. [Google Scholar] [CrossRef] [PubMed]
- Willmer, T.; Oosthuizen, A.; Dias, S.; Mendham, A.E.; Goedecke, J.H.; Pheiffer, C. A Pilot Investigation of Genetic and Epigenetic Variation of FKBP5 and Response to Exercise Intervention in African Women with Obesity. Sci. Rep. 2022, 12, 11771. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Goldberg, J.; Vaccarino, V. Promoter Methylation of Serotonin Transporter Gene Is Associated with Obesity Measures: A Monozygotic Twin Study. Int. J. Obes. 2013, 37, 140–145. [Google Scholar] [CrossRef]
- Duncan, G.E.; Avery, A.; Thorson, J.L.M.; Nilsson, E.E.; Beck, D.; Skinner, M.K. Epigenome-Wide Association Study of Physical Activity and Physiological Parameters in Discordant Monozygotic Twins. Sci. Rep. 2022, 12, 20166. [Google Scholar] [CrossRef] [PubMed]
- Vetr, N.G.; Gay, N.R.; MoTrPAC Study Group; Adkins, J.N.; Albertson, B.G.; Amar, D.; Amper, M.A.S.; Armenteros, J.J.A.; Ashley, E.; Avila-Pacheco, J.; et al. The Impact of Exercise on Gene Regulation in Association with Complex Trait Genetics. Nat. Commun. 2024, 15, 3346. [Google Scholar] [CrossRef]
- Nair, V.D.; Pincas, H.; Smith, G.R.; Zaslavsky, E.; Ge, Y.; Amper, M.A.S.; Vasoya, M.; Chikina, M.; Sun, Y.; Raja, A.N.; et al. Molecular Adaptations in Response to Exercise Training Are Associated with Tissue-Specific Transcriptomic and Epigenomic Signatures. Cell Genom. 2024, 4, 100421. [Google Scholar] [CrossRef]
- Bordoni, L.; Malinowska, A.M.; Petracci, I.; Chmurzynska, A.; Gabbianelli, R. Biological Age and Diet: Measuring the Impact of Lifestyle on a 6CpG-Epigenetic Clock. Nutr. Healthy Aging 2022, 7, 121–134. [Google Scholar] [CrossRef]
- Mahmoud, A.M. An Overview of Epigenetics in Obesity: The Role of Lifestyle and Therapeutic Interventions. Int. J. Mol. Sci. 2022, 23, 1341. [Google Scholar] [CrossRef] [PubMed]
- Kresovich, J.K.; Garval, E.L.; Martinez Lopez, A.M.; Xu, Z.; Niehoff, N.M.; White, A.J.; Sandler, D.P.; Taylor, J.A. Associations of Body Composition and Physical Activity Level With Multiple Measures of Epigenetic Age Acceleration. Am. J. Epidemiol. 2021, 190, 984–993. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Wirsching, J.; Graßmann, S.; Eichelmann, F.; Harms, L.M.; Schenk, M.; Barth, E.; Berndzen, A.; Olalekan, M.; Sarmini, L.; Zuberer, H.; et al. Development and Reliability Assessment of a New Quality Appraisal Tool for Cross-Sectional Studies Using Biomarker Data (BIOCROSS). BMC Med. Res. Methodol. 2018, 18, 122. [Google Scholar] [CrossRef] [PubMed]
- Review Manager (RevMan). Version (9.0.0). The Cochrane Collaboration, (April 14, 2025). Available online: https://revman.cochrane.org/info (accessed on 14 April 2025).
- Battram, T.; Yousefi, P.; Crawford, G.; Prince, C.; Sheikhali Babaei, M.; Sharp, G.; Hatcher, C.; Vega-Salas, M.J.; Khodabakhsh, S.; Whitehurst, O.; et al. The EWAS Catalog: A Database of Epigenome-Wide Association Studies. Wellcome Open Res. 2022, 7, 41. [Google Scholar] [CrossRef]
- Relton, C.L.; Gaunt, T.; McArdle, W.; Ho, K.; Duggirala, A.; Shihab, H.; Woodward, G.; Lyttleton, O.; Evans, D.M.; Reik, W.; et al. Data Resource Profile: Accessible Resource for Integrated Epigenomic Studies (ARIES). Int. J. Epidemiol. 2015, 44, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Min, J.L.; Hemani, G.; Hannon, E.; Dekkers, K.F.; Castillo-Fernandez, J.; Luijk, R.; Carnero-Montoro, E.; Lawson, D.J.; Burrows, K.; Suderman, M.; et al. Genomic and Phenotypic Insights from an Atlas of Genetic Effects on DNA Methylation. Nat. Genet. 2021, 53, 1311–1321. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Shi, H.; Ossip, D.J.; Mayo, N.L.; Lopez, D.A.; Block, R.C.; Post, W.S.; Bertoni, A.G.; Ding, J.; Chen, S.; Yan, C.; et al. Role of DNA Methylation on the Association between Physical Activity and Cardiovascular Diseases: Results from the Longitudinal Multi-Ethnic Study of Atherosclerosis (MESA) Cohort. BMC Genom. 2021, 22, 790. [Google Scholar] [CrossRef]
- Hibler, E.; Huang, L.; Andrade, J.; Spring, B. Impact of a Diet and Activity Health Promotion Intervention on Regional Patterns of DNA Methylation. Clin. Epigenetics 2019, 11, 133. [Google Scholar] [CrossRef]
- Keller, M.; Yaskolka Meir, A.; Bernhart, S.H.; Gepner, Y.; Shelef, I.; Schwarzfuchs, D.; Tsaban, G.; Zelicha, H.; Hopp, L.; Müller, L.; et al. DNA Methylation Signature in Blood Mirrors Successful Weight-Loss during Lifestyle Interventions: The CENTRAL Trial. Genome Med. 2020, 12, 97. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Milagro, F.I.; Mansego, M.L.; Mourão, D.M.; Martínez, J.A.; Bressan, J. LINE-1 Methylation Is Positively Associated with Healthier Lifestyle but Inversely Related to Body Fat Mass in Healthy Young Individuals. Epigenetics 2016, 11, 49–60. [Google Scholar] [CrossRef]
- Fernández-Sanlés, A.; Sayols-Baixeras, S.; Castro De Moura, M.; Esteller, M.; Subirana, I.; Torres-Cuevas, S.; Pérez-Fernández, S.; Aslibekyan, S.; Marrugat, J.; Elosua, R. Physical Activity and Genome-Wide DNA Methylation: The REgistre GIroní Del COR Study. Med. Sci. Sports Exerc. 2020, 52, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L.A.; Zapata-Bustos, R.; Day, S.E.; Campos, B.; Hamzaoui, Y.; Wu, L.; Leon, A.D.; Krentzel, J.; Coletta, R.L.; De Filippis, E.; et al. Can Exercise Training Alter Human Skeletal Muscle DNA Methylation? Metabolites 2022, 12, 222. [Google Scholar] [CrossRef] [PubMed]
- Kovac, L.; Goj, T.; Ouni, M.; Irmler, M.; Jähnert, M.; Beckers, J.; Hrabé De Angelis, M.; Peter, A.; Moller, A.; Birkenfeld, A.L.; et al. Skeletal Muscle Gene Expression Signatures of Obese High and Low Responders to Endurance Exercise Training. J. Clin. Endocrinol. Metab. 2024, 109, 1318–1327. [Google Scholar] [CrossRef] [PubMed]
- Landen, S.; Jacques, M.; Hiam, D.; Alvarez-Romero, J.; Schittenhelm, R.B.; Shah, A.D.; Huang, C.; Steele, J.R.; Harvey, N.R.; Haupt, L.M.; et al. Sex Differences in Muscle Protein Expression and DNA Methylation in Response to Exercise Training. Biol. Sex Differ. 2023, 14, 56. [Google Scholar] [CrossRef]
- Rönn, T.; Volkov, P.; Davegårdh, C.; Dayeh, T.; Hall, E.; Olsson, A.H.; Nilsson, E.; Tornberg, Å.; Dekker Nitert, M.; Eriksson, K.-F.; et al. A Six Months Exercise Intervention Influences the Genome-Wide DNA Methylation Pattern in Human Adipose Tissue. PLoS Genet. 2013, 9, e1003572. [Google Scholar] [CrossRef]
- Sexton, C.L.; Godwin, J.S.; McIntosh, M.C.; Ruple, B.A.; Osburn, S.C.; Hollingsworth, B.R.; Kontos, N.J.; Agostinelli, P.J.; Kavazis, A.N.; Ziegenfuss, T.N.; et al. Skeletal Muscle DNA Methylation and mRNA Responses to a Bout of Higher versus Lower Load Resistance Exercise in Previously Trained Men. Cells 2023, 12, 263. [Google Scholar] [CrossRef]
- Landen, S.; Jacques, M.; Hiam, D.; Alvarez-Romero, J.; Harvey, N.R.; Haupt, L.M.; Griffiths, L.R.; Ashton, K.J.; Lamon, S.; Voisin, S.; et al. Skeletal Muscle Methylome and Transcriptome Integration Reveals Profound Sex Differences Related to Muscle Function and Substrate Metabolism. Clin. Epigenetics 2021, 13, 202. [Google Scholar] [CrossRef]
- Da Silva Rodrigues, G.; Noronha, N.Y.; Noma, I.H.Y.; De Lima, J.G.R.; Da Silva Sobrinho, A.C.; De Souza Pinhel, M.A.; De Almeida, M.L.; Watanabe, L.M.; Nonino, C.B.; Júnior, C.R.B. 14-Week Exercise Training Modifies the DNA Methylation Levels at Gene Sites in Non-Alzheimer’s Disease Women Aged 50 to 70 Years. Exp. Gerontol. 2024, 186, 112362. [Google Scholar] [CrossRef]
- Barry, V.W.; Baruth, M.; Beets, M.W.; Durstine, J.L.; Liu, J.; Blair, S.N. Fitness vs. Fatness on All-Cause Mortality: A Meta-Analysis. Prog. Cardiovasc. Dis. 2014, 56, 382–390. [Google Scholar] [CrossRef]
- Bullich-Vilarrubias, C.; Romaní-Pérez, M.; López-Almela, I.; Rubio, T.; García, C.J.; Tomás-Barberán, F.A.; Sanz, Y. Nav1.8-Expressing Neurons Control Daily Oscillations of Food Intake, Body Weight and Gut Microbiota in Mice. Commun. Biol. 2024, 7, 219. [Google Scholar] [CrossRef]
- Udit, S.; Burton, M.; Rutkowski, J.M.; Lee, S.; Bookout, A.L.; Scherer, P.E.; Elmquist, J.K.; Gautron, L. Na v 1.8 Neurons Are Involved in Limiting Acute Phase Responses to Dietary Fat. Mol. Metab. 2017, 6, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Romaní-Pérez, M.; Bullich-Vilarrubias, C.; López-Almela, I.; Sanz, Y. The Ablation of Sensory Neurons Expressing the Nav1.8 Sodium Channel Improves Glucose Homeostasis and Amplifies the GLP-1 Signaling in Obese Female Mice. Mol. Nutr. Food Res. 2024, 68, 2300474. [Google Scholar] [CrossRef] [PubMed]
- Elsadek, M.F. Effect of a Genetic Variant in the JAZF1 Gene among Obesity Population. J. King Saud Univ.-Sci. 2022, 34, 102112. [Google Scholar] [CrossRef]
- Liao, Z.; Wang, Y.; Qi, X.; Xiao, X. JAZF1, a Relevant Metabolic Regulator in Type 2 Diabetes. Diabetes Metab. Res. Rev. 2019, 35, e3148. [Google Scholar] [CrossRef]
- Jang, W.Y.; Bae, K.B.; Kim, S.H.; Yu, D.H.; Kim, H.J.; Ji, Y.R.; Park, S.J.; Park, S.J.; Kang, M.-C.; Jeong, J.I.; et al. Overexpression of Jazf1 Reduces Body Weight Gain and Regulates Lipid Metabolism in High Fat Diet. Biochem. Biophys. Res. Commun. 2014, 444, 296–301. [Google Scholar] [CrossRef]
- Klemp, I.; Hoffmann, A.; Müller, L.; Hagemann, T.; Horn, K.; Rohde-Zimmermann, K.; Tönjes, A.; Thiery, J.; Löffler, M.; Burkhardt, R.; et al. DNA Methylation Patterns Reflect Individual’s Lifestyle Independent of Obesity. Clin. Transl. Med. 2022, 12, e851. [Google Scholar] [CrossRef]
- Miranda Furtado, C.L.; Hansen, M.; Kogure, G.S.; Ribeiro, V.B.; Taylor, N.; Racy Soares, M.; Ferriani, R.A.; Aston, K.I.; Jenkins, T.; Dos Reis, R.M. Resistance and Aerobic Training Increases Genome-Wide DNA Methylation in Women with Polycystic Ovary Syndrome. Epigenetics 2024, 19, 2305082. [Google Scholar] [CrossRef]
- Schenk, A.; Koliamitra, C.; Bauer, C.J.; Schier, R.; Schweiger, M.R.; Bloch, W.; Zimmer, P. Impact of Acute Aerobic Exercise on Genome-Wide DNA-Methylation in Natural Killer Cells—A Pilot Study. Genes 2019, 10, 380. [Google Scholar] [CrossRef]
- Sailani, M.R.; Halling, J.F.; Møller, H.D.; Lee, H.; Plomgaard, P.; Pilegaard, H.; Snyder, M.P.; Regenberg, B. Lifelong Physical Activity Is Associated with Promoter Hypomethylation of Genes Involved in Metabolism, Myogenesis, Contractile Properties and Oxidative Stress Resistance in Aged Human Skeletal Muscle. Sci. Rep. 2019, 9, 3272. [Google Scholar] [CrossRef]
- Knigge, A.; Klöting, N.; Schön, M.R.; Dietrich, A.; Fasshauer, M.; Gärtner, D.; Lohmann, T.; Dreßler, M.; Stumvoll, M.; Kovacs, P.; et al. ADCY5 Gene Expression in Adipose Tissue Is Related to Obesity in Men and Mice. PLoS ONE 2015, 10, e0120742. [Google Scholar] [CrossRef]
- Pell, N.; Garcia-Pras, E.; Gallego, J.; Naranjo-Suarez, S.; Balvey, A.; Suñer, C.; Fernandez-Alfara, M.; Chanes, V.; Carbo, J.; Ramirez-Pedraza, M.; et al. Targeting the Cytoplasmic Polyadenylation Element-Binding Protein CPEB4 Protects against Diet-Induced Obesity and Microbiome Dysbiosis. Mol. Metab. 2021, 54, 101388. [Google Scholar] [CrossRef] [PubMed]
- Prats-Puig, A.; Soriano-Rodríguez, P.; Oliveras, G.; Carreras-Badosa, G.; Espuña, S.; Díaz-Roldán, F.; De Zegher, F.; Ibáñez, L.; Bassols, J.; Puig, T.; et al. Soluble CRTC3: A Newly Identified Protein Released by Adipose Tissue That Is Associated with Childhood Obesity. Clin. Chem. 2016, 62, 476–484. [Google Scholar] [CrossRef]
- Jollet, M.; Tramontana, F.; Jiang, L.Q.; Borg, M.L.; Savikj, M.; Kuefner, M.S.; Massart, J.; De Castro Barbosa, T.; Mannerås-Holm, L.; Checa, A.; et al. Diacylglycerol Kinase Delta Overexpression Improves Glucose Clearance and Protects against the Development of Obesity. Metabolism 2024, 158, 155939. [Google Scholar] [CrossRef]
- Able, A.A.; Richard, A.J.; Stephens, J.M. TNFα Effects on Adipocytes Are Influenced by the Presence of Lysine Methyltransferases, G9a (EHMT2) and GLP (EHMT1). Biology 2023, 12, 674. [Google Scholar] [CrossRef]
- Liu, P.; Huang, S.; Ling, S.; Xu, S.; Wang, F.; Zhang, W.; Zhou, R.; He, L.; Xia, X.; Yao, Z.; et al. Foxp1 Controls Brown/Beige Adipocyte Differentiation and Thermogenesis through Regulating Β3-AR Desensitization. Nat. Commun. 2019, 10, 5070. [Google Scholar] [CrossRef] [PubMed]
- The GIANT Consortium; Wood, A.R.; Tyrrell, J.; Beaumont, R.; Jones, S.E.; Tuke, M.A.; Ruth, K.S.; Yaghootkar, H.; Freathy, R.M.; Murray, A.; et al. Variants in the FTO and CDKAL1 Loci Have Recessive Effects on Risk of Obesity and Type 2 Diabetes, Respectively. Diabetologia 2016, 59, 1214–1221. [Google Scholar] [CrossRef]
- Xi, Y.; Shen, W.; Ma, L.; Zhao, M.; Zheng, J.; Bu, S.; Hino, S.; Nakao, M. HMGA2 Promotes Adipogenesis by Activating C/EBPβ-Mediated Expression of PPARγ. Biochem. Biophys. Res. Commun. 2016, 472, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Ravaud, C.; Esteve, D.; Villageois, P.; Bouloumie, A.; Dani, C.; Ladoux, A. IER3 Promotes Expansion of Adipose Progenitor Cells in Response to Changes in Distinct Microenvironmental Effectors. Stem Cells 2015, 33, 2564–2573. [Google Scholar] [CrossRef]
- Lei, X.; Callaway, M.; Zhou, H.; Yang, Y.; Chen, W. Obesity Associated Lyplal1 Gene Is Regulated in Diet Induced Obesity but Not Required for Adipocyte Differentiation. Mol. Cell. Endocrinol. 2015, 411, 207–213. [Google Scholar] [CrossRef]
- Lu, C.; Wang, H.-J.; Song, J.-Y.; Wang, S.; Li, X.-Y.; Huang, T.; Wang, H. Fine Mapping of the MAP2K5 Region Identified Rs7175517 as a Causal Variant Related to BMI in China and the United Kingdom Populations. Front. Genet. 2022, 13, 838685. [Google Scholar] [CrossRef]
- Albuquerque, D.; Nóbrega, C.; Rodríguez-López, R.; Manco, L. Association Study of Common Polymorphisms in MSRA, TFAP2B, MC4R, NRXN3, PPARGC1A, TMEM18, SEC16B, HOXB5 and OLFM4 Genes with Obesity-Related Traits among Portuguese Children. J. Hum. Genet. 2014, 59, 307–313. [Google Scholar] [CrossRef]
- Huang, M.; Coral, D.; Ardalani, H.; Spegel, P.; Saadat, A.; Claussnitzer, M.; Mulder, H.; Franks, P.W.; Kalamajski, S. Identification of a Weight Loss-Associated Causal eQTL in MTIF3 and the Effects of MTIF3 Deficiency on Human Adipocyte Function. eLife 2023, 12, e84168. [Google Scholar] [CrossRef]
- Heard-Costa, N.L.; Zillikens, M.C.; Monda, K.L.; Johansson, Å.; Harris, T.B.; Fu, M.; Haritunians, T.; Feitosa, M.F.; Aspelund, T.; Eiriksdottir, G.; et al. NRXN3 Is a Novel Locus for Waist Circumference: A Genome-Wide Association Study from the CHARGE Consortium. PLoS Genet. 2009, 5, e1000539. [Google Scholar] [CrossRef]
- Yang, Q.-Y.; Liang, J.-F.; Rogers, C.J.; Zhao, J.-X.; Zhu, M.-J.; Du, M. Maternal Obesity Induces Epigenetic Modifications to Facilitate Zfp423 Expression and Enhance Adipogenic Differentiation in Fetal Mice. Diabetes 2013, 62, 3727–3735. [Google Scholar] [CrossRef]
- Li, G.; Xing, Z.; Wang, W.; Luo, W.; Ma, Z.; Wu, Z.; Chen, H.; Li, Y.; Wang, C.; Zeng, F.; et al. Adipose-Specific Knockout of Protein Kinase D1 Suppresses de Novo Lipogenesis in Mice via SREBP1c-Dependent Signaling. Exp. Cell Res. 2021, 401, 112548. [Google Scholar] [CrossRef]
- Escobedo, N.; Proulx, S.T.; Karaman, S.; Dillard, M.E.; Johnson, N.; Detmar, M.; Oliver, G. Restoration of Lymphatic Function Rescues Obesity in Prox1-Haploinsufficient Mice. JCI Insight 2016, 1, e85096. [Google Scholar] [CrossRef]
- Parrillo, L.; Spinelli, R.; Longo, M.; Desiderio, A.; Mirra, P.; Nigro, C.; Fiory, F.; Hedjazifar, S.; Mutarelli, M.; Carissimo, A.; et al. Altered PTPRD DNA Methylation Associates with Restricted Adipogenesis in Healthy First-Degree Relatives of Type 2 Diabetes Subjects. Epigenomics 2020, 12, 873–888. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, D.; Qin, Z.; Li, J.; Shen, Z. The Role and Possible Mechanism of Long Noncoding RNA PVT1 in Modulating 3T3-L1 Preadipocyte Proliferation and Differentiation. IUBMB Life 2020, 72, 1460–1467. [Google Scholar] [CrossRef]
- Moreno-Navarrete, J.M.; Petrov, P.; Serrano, M.; Ortega, F.; García-Ruiz, E.; Oliver, P.; Ribot, J.; Ricart, W.; Palou, A.; Bonet, M.L.; et al. Decreased RB1 mRNA, Protein, and Activity Reflect Obesity-Induced Altered Adipogenic Capacity in Human Adipose Tissue. Diabetes 2013, 62, 1923–1931. [Google Scholar] [CrossRef]
- Han, J.-H.; Jang, K.-W.; Myung, C.-S. Garcinia cambogia Attenuates Adipogenesis by Affecting CEBPB and SQSTM1/P62-Mediated Selective Autophagic Degradation of KLF3 through RPS6KA1 and STAT3 Suppression. Autophagy 2022, 18, 518–539. [Google Scholar] [CrossRef]
- Scherag, A.; Kleber, M.; Boes, T.; Kolbe, A.; Ruth, A.; Grallert, H.; Illig, T.; Heid, I.M.; The GIANT Consortium; Toschke, A.M.; et al. SDCCAG8 Obesity Alleles and Reduced Weight Loss After a Lifestyle Intervention in Overweight Children and Adolescents. Obesity 2012, 20, 466–470. [Google Scholar] [CrossRef]
- Nahon, J.E.; Hoekstra, M.; Van Hulst, S.; Manta, C.; Goerdt, S.; Geerling, J.J.; Géraud, C.; Van Eck, M. Hematopoietic Stabilin-1 Deficiency Does Not Influence Atherosclerosis Susceptibility in LDL Receptor Knockout Mice. Atherosclerosis 2019, 281, 47–55. [Google Scholar] [CrossRef]
- Yu, H.; He, K.; Wang, L.; Hu, J.; Gu, J.; Zhou, C.; Lu, R.; Jin, Y. Stk40 Represses Adipogenesis through Translational Control of CCAAT/Enhancer-Binding Proteins. J. Cell Sci. 2015, 128, 2881–2890. [Google Scholar] [CrossRef]
- Knoll, M.; Winther, S.; Natarajan, A.; Yang, H.; Jiang, M.; Thiru, P.; Shahsafaei, A.; Chavarria, T.E.; Lamming, D.W.; Sun, L.; et al. SYK Kinase Mediates Brown Fat Differentiation and Activation. Nat. Commun. 2017, 8, 2115. [Google Scholar] [CrossRef]
- Verma, M.; Loh, N.Y.; Sabaratnam, R.; Vasan, S.K.; Van Dam, A.D.; Todorčević, M.; Neville, M.J.; Toledo, E.; Karpe, F.; Christodoulides, C. TCF7L2 Plays a Complex Role in Human Adipose Progenitor Biology, Which Might Contribute to Genetic Susceptibility to Type 2 Diabetes. Metabolism 2022, 133, 155240. [Google Scholar] [CrossRef]
- Nfor, O.N.; Wu, M.-F.; Lee, C.-T.; Wang, L.; Liu, W.-H.; Tantoh, D.M.; Hsu, S.-Y.; Lee, K.-J.; Ho, C.-C.; Debnath, T.; et al. Body Mass Index Modulates the Association between CDKAL1 Rs10946398 Variant and Type 2 Diabetes among Taiwanese Women. Sci. Rep. 2018, 8, 13235. [Google Scholar] [CrossRef]
- Wang, T.; Ma, X.; Peng, D.; Zhang, R.; Sun, X.; Chen, M.; Yan, J.; Wang, S.; Yan, D.; He, Z.; et al. Effects of Obesity Related Genetic Variations on Visceral and Subcutaneous Fat Distribution in a Chinese Population. Sci. Rep. 2016, 6, 20691. [Google Scholar] [CrossRef]
- Hong, J.; Shi, J.; Qi, L.; Cui, B.; Gu, W.; Zhang, Y.; Li, L.; Xu, M.; Wang, L.; Zhai, Y.; et al. Genetic Susceptibility, Birth Weight and Obesity Risk in Young Chinese. Int. J. Obes. 2013, 37, 673–677. [Google Scholar] [CrossRef]
- Yang, S.-A. Association Study between ZFHX3 Gene Polymorphisms and Obesity in Korean Population. J. Exerc. Rehabil. 2017, 13, 491–494. [Google Scholar] [CrossRef]
- Ntalla, I.; Panoutsopoulou, K.; Vlachou, P.; Southam, L.; William Rayner, N.; Zeggini, E.; Dedoussis, G.V. Replication of Established Common Genetic Variants for Adult BMI and Childhood Obesity in Greek Adolescents: The TEENAGE Study. Ann. Hum. Genet. 2013, 77, 268–274. [Google Scholar] [CrossRef]
- Nies, V.J.M.; Struik, D.; Wolfs, M.G.M.; Rensen, S.S.; Szalowska, E.; Unmehopa, U.A.; Fluiter, K.; Van Der Meer, T.P.; Hajmousa, G.; Buurman, W.A.; et al. TUB Gene Expression in Hypothalamus and Adipose Tissue and Its Association with Obesity in Humans. Int. J. Obes. 2018, 42, 376–383. [Google Scholar] [CrossRef]
- Graae, A.-S.; Grarup, N.; Ribel-Madsen, R.; Lystbæk, S.H.; Boesgaard, T.; Staiger, H.; Fritsche, A.; Wellner, N.; Sulek, K.; Kjolby, M.; et al. ADAMTS9 Regulates Skeletal Muscle Insulin Sensitivity Through Extracellular Matrix Alterations. Diabetes 2019, 68, 502–514. [Google Scholar] [CrossRef]
- Bailetti, D.; Sentinelli, F.; Prudente, S.; Cimini, F.A.; Barchetta, I.; Totaro, M.; Di Costanzo, A.; Barbonetti, A.; Leonetti, F.; Cavallo, M.G.; et al. Deep Resequencing of 9 Candidate Genes Identifies a Role for ARAP1 and IGF2BP2 in Modulating Insulin Secretion Adjusted for Insulin Resistance in Obese Southern Europeans. Int. J. Mol. Sci. 2022, 23, 1221. [Google Scholar] [CrossRef]
- Tang, L.; Wang, L.; Ye, H.; Xu, X.; Hong, Q.; Wang, H.; Xu, L.; Bu, S.; Zhang, L.; Cheng, J.; et al. BCL11A Gene DNA Methylation Contributes to the Risk of Type 2 Diabetes in Males. Exp. Ther. Med. 2014, 8, 459–463. [Google Scholar] [CrossRef]
- Kaneko, K.; Katagiri, H. Dual-specificity Phosphatase 8: A Gatekeeper in Hypothalamic Control of Glucose Metabolism in Males. J. Diabetes Investig. 2021, 12, 1138–1140. [Google Scholar] [CrossRef]
- Teran-Garcia, M.; Rankinen, T.; Rice, T.; Leon, A.S.; Rao, D.C.; Skinner, J.S.; Bouchard, C. Variations in the Four and a Half LIM Domains 1 Gene (FHL1) Are Associated with Fasting Insulin and Insulin Sensitivity Responses to Regular Exercise. Diabetologia 2007, 50, 1858–1866. [Google Scholar] [CrossRef]
- Atanes, P.; Ashik, T.; Persaud, S.J. Obesity-Induced Changes in Human Islet G Protein-Coupled Receptor Expression: Implications for Metabolic Regulation. Pharmacol. Ther. 2021, 228, 107928. [Google Scholar] [CrossRef]
- Ding, X.; Iyer, R.; Novotny, C.; Metzger, D.; Zhou, H.H.; Smith, G.I.; Yoshino, M.; Yoshino, J.; Klein, S.; Swaminath, G.; et al. Inhibition of Grb14, a Negative Modulator of Insulin Signaling, Improves Glucose Homeostasis without Causing Cardiac Dysfunction. Sci. Rep. 2020, 10, 3417. [Google Scholar] [CrossRef]
- Guney, E.; Arruda, A.P.; Parlakgul, G.; Cagampan, E.; Min, N.; Lee, G.Y.; Greene, L.; Tsaousidou, E.; Inouye, K.; Han, M.S.; et al. Aberrant Ca2+ Signaling by IP3 Rs in Adipocytes Links Inflammation to Metabolic Dysregulation in Obesity. Sci. Signal. 2021, 14, eabf2059. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, C.; Chen, Y.; Yuan, Z.; Yu, T.; Zhang, W.; Tang, F.; Gu, J.; Xu, Q.; Chi, X.; et al. A Variant in KCNQ1 Gene Predicts Metabolic Syndrome among Northern Urban Han Chinese Women. BMC Med. Genet. 2018, 19, 153. [Google Scholar] [CrossRef]
- Su, S.; Zhu, H.; Xu, X.; Wang, X.; Dong, Y.; Kapuku, G.; Treiber, F.; Gutin, B.; Harshfield, G.; Snieder, H.; et al. DNA Methylation of the LY86 Gene Is Associated With Obesity, Insulin Resistance, and Inflammation. Twin Res. Hum. Genet. 2014, 17, 183–191. [Google Scholar] [CrossRef]
- Brazill, J.M.; Shin, D.; Magee, K.; Majumdar, A.; Shen, I.R.; Cavalli, V.; Scheller, E.L. Knockout of TSC2 in Nav1.8+ Neurons Predisposes to the Onset of Normal Weight Obesity. Mol. Metab. 2023, 68, 101664. [Google Scholar] [CrossRef]
- Qiu, B.; Shi, X.; Wong, E.T.; Lim, J.; Bezzi, M.; Low, D.; Zhou, Q.; Akıncılar, S.C.; Lakshmanan, M.; Swa, H.L.F.; et al. NUCKS Is a Positive Transcriptional Regulator of Insulin Signaling. Cell Rep. 2014, 7, 1876–1886. [Google Scholar] [CrossRef]
- Li, Q.; Li, Z.; Guo, S.; Li, S.; Yao, M.; Li, Y.; Luo, X. Increased Circulating serpinB1 Levels in Children with Overweight/Obesity Are Associated with Obesity-Related Parameters: A Cross-sectional Study. BMC Pediatr. 2024, 24, 762. [Google Scholar] [CrossRef]
- Romeo, S.; Sentinelli, F.; Cavallo, M.G.; Leonetti, F.; Fallarino, M.; Mariotti, S.; Baroni, M.G. Search for Genetic Variants of the SYNTAXIN 1A (STX1A) Gene: The −352 A>T Variant in the STX1A Promoter Associates with Impaired Glucose Metabolism in an Italian Obese Population. Int. J. Obes. 2008, 32, 413–420. [Google Scholar] [CrossRef]
- Ivask, M.; Volke, V.; Raasmaja, A.; Kõks, S. High-Fat Diet Associated Sensitization to Metabolic Stress in Wfs1 Heterozygous Mice. Mol. Genet. Metab. 2021, 134, 203–211. [Google Scholar] [CrossRef]
- Jia, Y.; Yuan, L.; Hu, W.; Luo, Y.; Suo, L.; Yang, M.; Chen, S.; Wang, Y.; Liu, H.; Yang, G.; et al. Zinc-finger BED Domain-containing 3 (Zbed3) Is a Novel Secreted Protein Associated with Insulin Resistance in Humans. J. Intern. Med. 2014, 275, 522–533. [Google Scholar] [CrossRef]
- Tsuda, N.; Kumadaki, S.; Higashi, C.; Ozawa, M.; Shinozaki, M.; Kato, Y.; Hoshida, K.; Kikuchi, S.; Nakano, Y.; Ogawa, Y.; et al. Intestine-Targeted DGAT1 Inhibition Improves Obesity and Insulin Resistance without Skin Aberrations in Mice. PLoS ONE 2014, 9, e112027. [Google Scholar] [CrossRef]
- Chung, J.Y.; Hong, J.; Kim, H.-J.; Song, Y.; Yong, S.-B.; Lee, J.; Kim, Y.-H. White Adipocyte-Targeted Dual Gene Silencing of FABP4/5 for Anti-Obesity, Anti-Inflammation and Reversal of Insulin Resistance: Efficacy and Comparison of Administration Routes. Biomaterials 2021, 279, 121209. [Google Scholar] [CrossRef]
- Hirabayashi, Y.; Kim, Y.-J. Roles of GPRC5 Family Proteins: Focusing on GPRC5B and Lipid-Mediated Signalling. J. Biochem. 2020, 167, 541–547. [Google Scholar] [CrossRef]
- Rautenberg, E.K.; Hamzaoui, Y.; Coletta, D.K. Mini-Review: Mitochondrial DNA Methylation in Type 2 Diabetes and Obesity. Front. Endocrinol. 2022, 13, 968268. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Rundek, T.; Beecham, A.; Hudson, B.; Blanton, S.H.; Zhao, H.; Sacco, R.L.; Dong, C. Genome-Wide Interaction Study Identifies RCBTB1 as a Modifier for Smoking Effect on Carotid Intima-Media Thickness. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 219–225. [Google Scholar] [CrossRef]
- Li, J.; Zhou, L.; Ouyang, X.; He, P. Transcription Factor-7-Like-2 (TCF7L2) in Atherosclerosis: A Potential Biomarker and Therapeutic Target. Front. Cardiovasc. Med. 2021, 8, 701279. [Google Scholar] [CrossRef] [PubMed]
- Trang, K.; Grant, S.F.A. Genetics and Epigenetics in the Obesity Phenotyping Scenario. Rev. Endocr. Metab. Disord. 2023, 24, 775–793. [Google Scholar] [CrossRef] [PubMed]
- Szpakowicz, A.; Szum-Jakubowska, A.; Lisowska, A.; Dubatówka, M.; Raczkowski, A.; Czajkowski, M.; Szczerbiński, Ł.; Chlabicz, M.; Krętowski, A.; Kamiński, K.A. The FCGR2A Is Associated with the Presence of Atherosclerotic Plaques in the Carotid Arteries—A Case-Control Study. J. Clin. Med. 2023, 12, 6480. [Google Scholar] [CrossRef]
- Freundt, G.V.; Von Samson-Himmelstjerna, F.A.; Nitz, J.-T.; Luedde, M.; Waltenberger, J.; Wieland, T.; Frey, N.; Preusch, M.; Hippe, H.-J. The Orphan Receptor GPRC5B Activates Pro-Inflammatory Signaling in the Vascular Wall via Fyn and NFκB. Biochem. Biophys. Res. Commun. 2022, 592, 60–66. [Google Scholar] [CrossRef]
- Zakharkin, S.O.; Belay, A.T.; Fernandez, J.R.; De Luca, V.; Kennedy, J.L.; Sokolowski, M.B.; Allison, D.B. Lack of Association between Polymorphism of the Human Cyclic GMP-Dependent Protein Kinase Gene and Obesity. Int. J. Obes. 2005, 29, 872–874. [Google Scholar] [CrossRef]



| PICOS Element | Criteria |
|---|---|
| Population | Human population with no underlying health conditions, aged 18–65 years, non-smokers, pregnant or lactating excluded |
| Intervention | Assessment of DNA methylation resulting from: (1) PA levels; or (2) effects of PA programme |
| Comparison | (1) Control group for population study or (2) non-exercising group or participant baseline as control for PA programme |
| Outcome | DNA hypermethylation or hypomethylation in CpG sites |
| Study design | Population study or PA intervention study |
| Citation | Title | Country of Origin | Study Characteristics | Study Population | Study Numbers | Tissue Type | Publication | Risk of Bias Score (Table S3) |
|---|---|---|---|---|---|---|---|---|
| [47] | Physical activity and genome-wide DNA methylation: the REgistre GIroni del COR study. | Spain | Population study: validation of meta-analysis using PA questionnaires and blood sample analysis. | Existing cohort (REGICOR) | 619; 5% female | Blood | American College of Sports Medicine | 17 |
| [48] | Can exercise training alter human skeletal muscle DNA methylation? | US | Exercise intervention: 8 weeks endurance training. | Sedentary healthy adults | 13; 61% female | Skeletal muscle | Metabolites | 15 |
| [49] | Skeletal muscle gene expression signatures of obese high and low responders to endurance exercise training. | Germany | Exercise intervention: 8 weeks endurance training. | Healthy overweight adults | 18; 63% female | Skeletal muscle | Journal of Clinical Endocrinology and Metabolism | 17 |
| [50] | Sex differences in muscle protein expression and DNA methylation in response to exercise training. | Australia | Exercise intervention: 4 weeks endurance training. | Healthy adults | 78; 36% female | Skeletal muscle | BMC | 16 |
| [51] | A six-months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. | Sweden | Exercise intervention: 6 months endurance training. | Healthy middle-aged males | 31; 0% female | Adipose tissue | PLOS Genetics | 17 |
| [52] | Skeletal muscle DNA methylation and mRNA responses to a bout of higher versus lower load resistance exercise in previously trained men. | US | Exercise intervention: resistance load testing, not time-constrained. | Active young males | 11; 0% female | Skeletal muscle | Cells | 16 |
| Study/ Characteristic | [47] | [48] | RES [49] | LRE [49] | [50] * | [51] | [52] | Total/ Average |
|---|---|---|---|---|---|---|---|---|
| Number of participants | 619 | 13 | 11 | 7 | 78 | 31 | 11 | 770 total |
| Mean age (yrs) | 63.10 (11.70) | 34.60 (11.10) | 28.60 (4.72) | 27.60 (3.96) | 33.50 (7.50) | 37.30 (4.40) | 23.00 (4.00) | 35.39 (6.76) |
| % female | 49.90 | 61.00 | 54.50 | 71.40 | 35.90 | 0.00 | 0.00 | 38.96 |
| Height (m) | NS | NS | 1.72 (0.10) | 1.71 (0.09) | NS | NS | 1.80 (0.07) | 1.74 (0.09) |
| Weight (kg) | NS | 87.50 (24.10) | 91.80 (17.10) | 96.90 (17.30) | NS | 91.80 (11.00) | 86.00 (12.00) | 90.80 (16.30) |
| BMI (kg/m2) | 26.90 (4.00) | 30.70 (7.40) | 30.80 (3.65) | 33.30 (5.84) | NS | 28.20 (2.90) | 27.00 (3.00) | 29.4 (4.47) |
| Waist circumference (cm) | NS | NS | NS | NS | NS | 97.70 (8.60) | NS | 97.70 (8.60) |
| Waist-to-hip ratio | NS | NS | 0.90 (0.05) | 0.87 (0.05) | NS | 0.93 (0.05) | NS | 0.90 (0.05) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chambers, J.; Roscoe, C.M.P.; Chidley, C.; Wisniewska, A.; Duggirala, A. Molecular Effects of Physical Activity and Body Composition: A Systematic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2025, 22, 637. https://doi.org/10.3390/ijerph22040637
Chambers J, Roscoe CMP, Chidley C, Wisniewska A, Duggirala A. Molecular Effects of Physical Activity and Body Composition: A Systematic Review and Meta-Analysis. International Journal of Environmental Research and Public Health. 2025; 22(4):637. https://doi.org/10.3390/ijerph22040637
Chicago/Turabian StyleChambers, Jenni, Clare M. P. Roscoe, Corinna Chidley, Agnieszka Wisniewska, and Aparna Duggirala. 2025. "Molecular Effects of Physical Activity and Body Composition: A Systematic Review and Meta-Analysis" International Journal of Environmental Research and Public Health 22, no. 4: 637. https://doi.org/10.3390/ijerph22040637
APA StyleChambers, J., Roscoe, C. M. P., Chidley, C., Wisniewska, A., & Duggirala, A. (2025). Molecular Effects of Physical Activity and Body Composition: A Systematic Review and Meta-Analysis. International Journal of Environmental Research and Public Health, 22(4), 637. https://doi.org/10.3390/ijerph22040637







