Biochemical and Molecular Characteristics of a Novel Hyaluronic Acid Lyase from Citrobacter freundii
Abstract
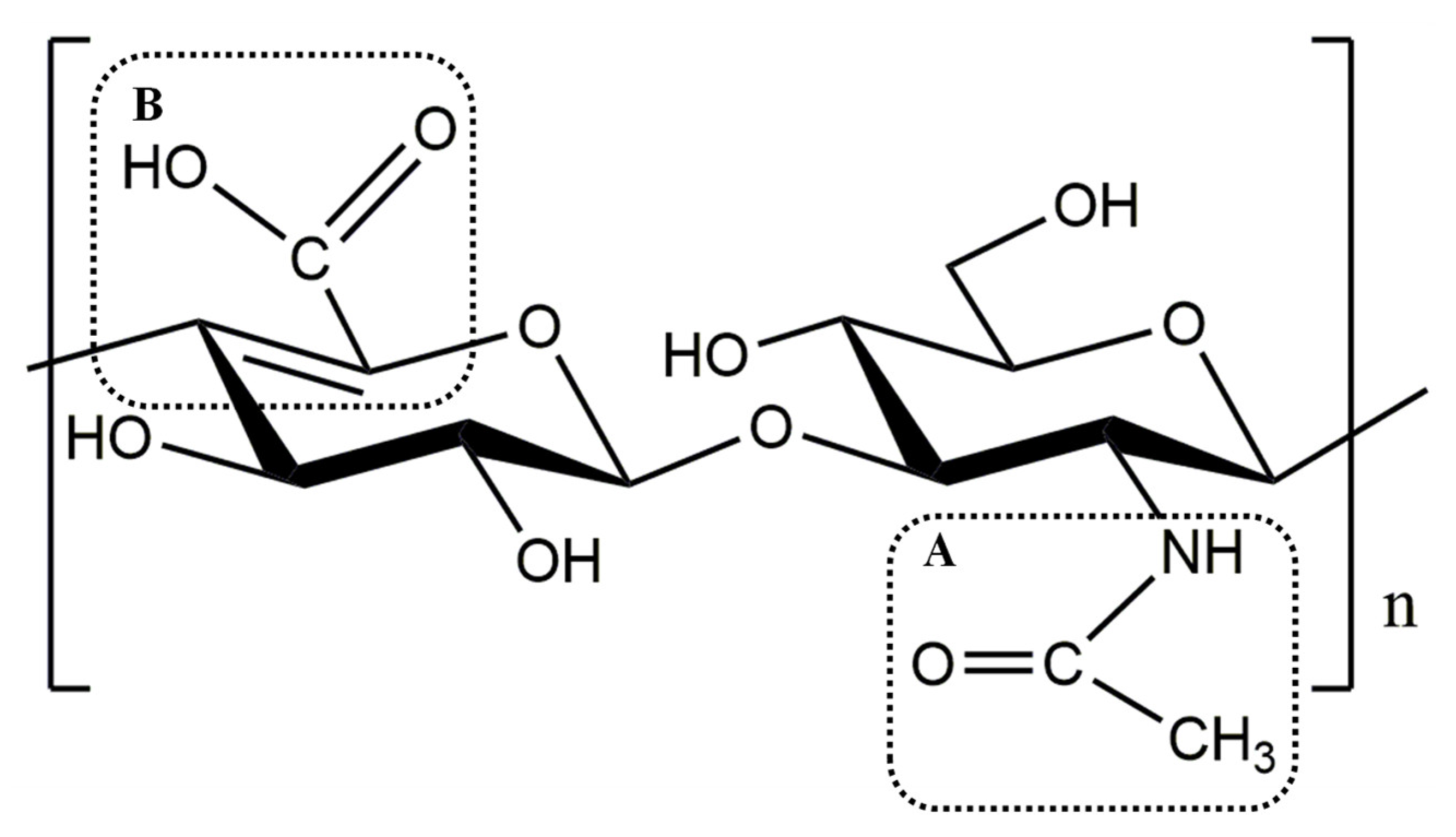
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain, Vectors, and Reagents
2.2. Gene Mining
2.3. Sequence Analyses
2.4. Heterologous Expression, Purifying, and Identifying Recombinant Hyaluronic Acid Lyase in E. coli
2.5. Enzyme Assay and Substrate Specificity
2.6. Biochemical Characterization of Recombinant Enzyme
2.7. Analysis of the Cleavage Products
2.8. Antioxidant Properties of Cleavage Products In Vitro
2.8.1. Assay of ABTS Radical Scavenging Activity
2.8.2. Assay of DPPH Radicals Scavenging Activity
2.8.3. Assay of Superoxide Anion Scavenging Activity
2.8.4. Assay of Hydroxyl Radical Scavenging Activity
2.9. Accession Number
2.10. Statistical Analysis
3. Results and Discussion
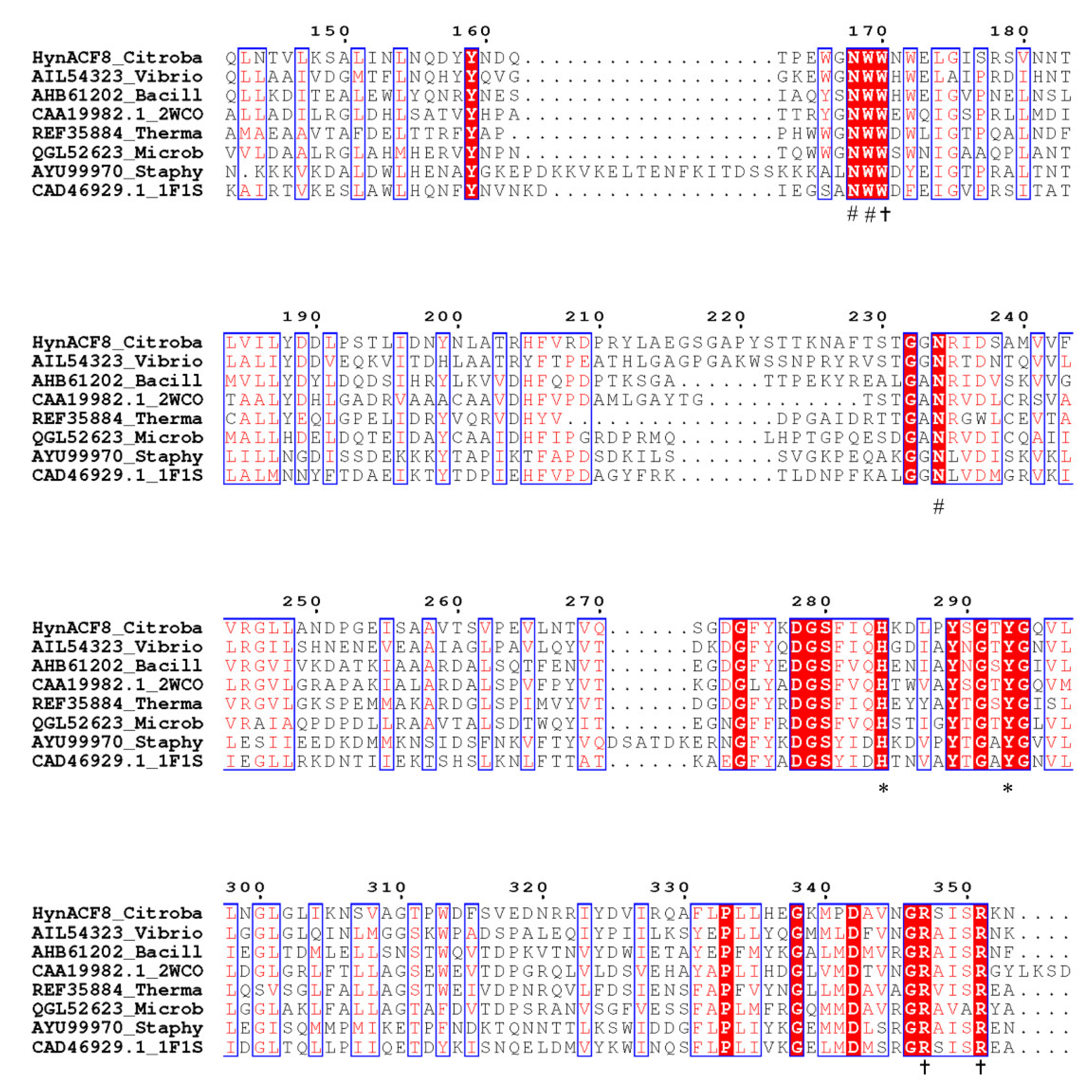
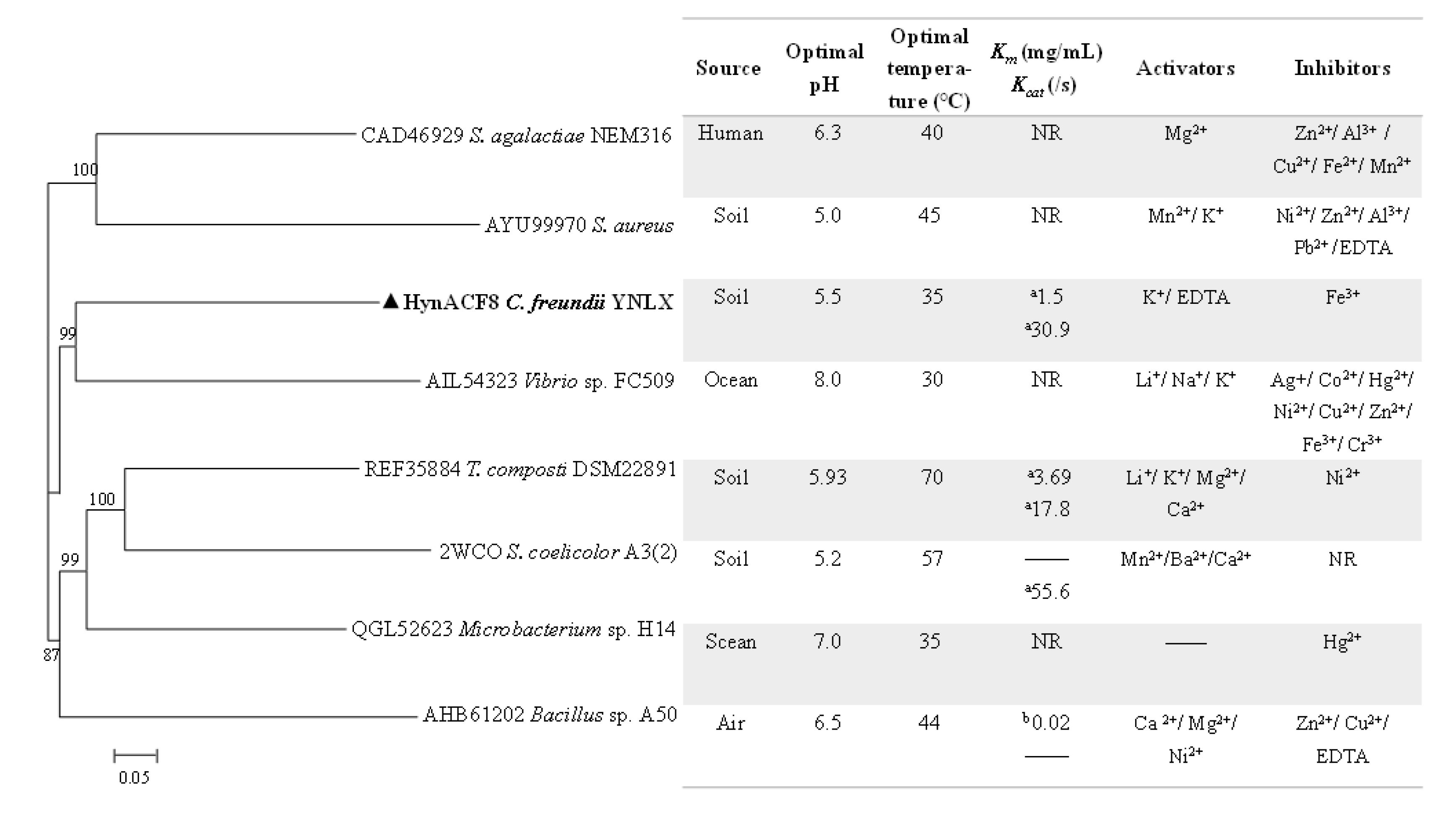
3.1. Genome Sequencing and Sequence Analyses
3.2. Expression and Purification of rHynACF8
3.3. Substrate Specificity
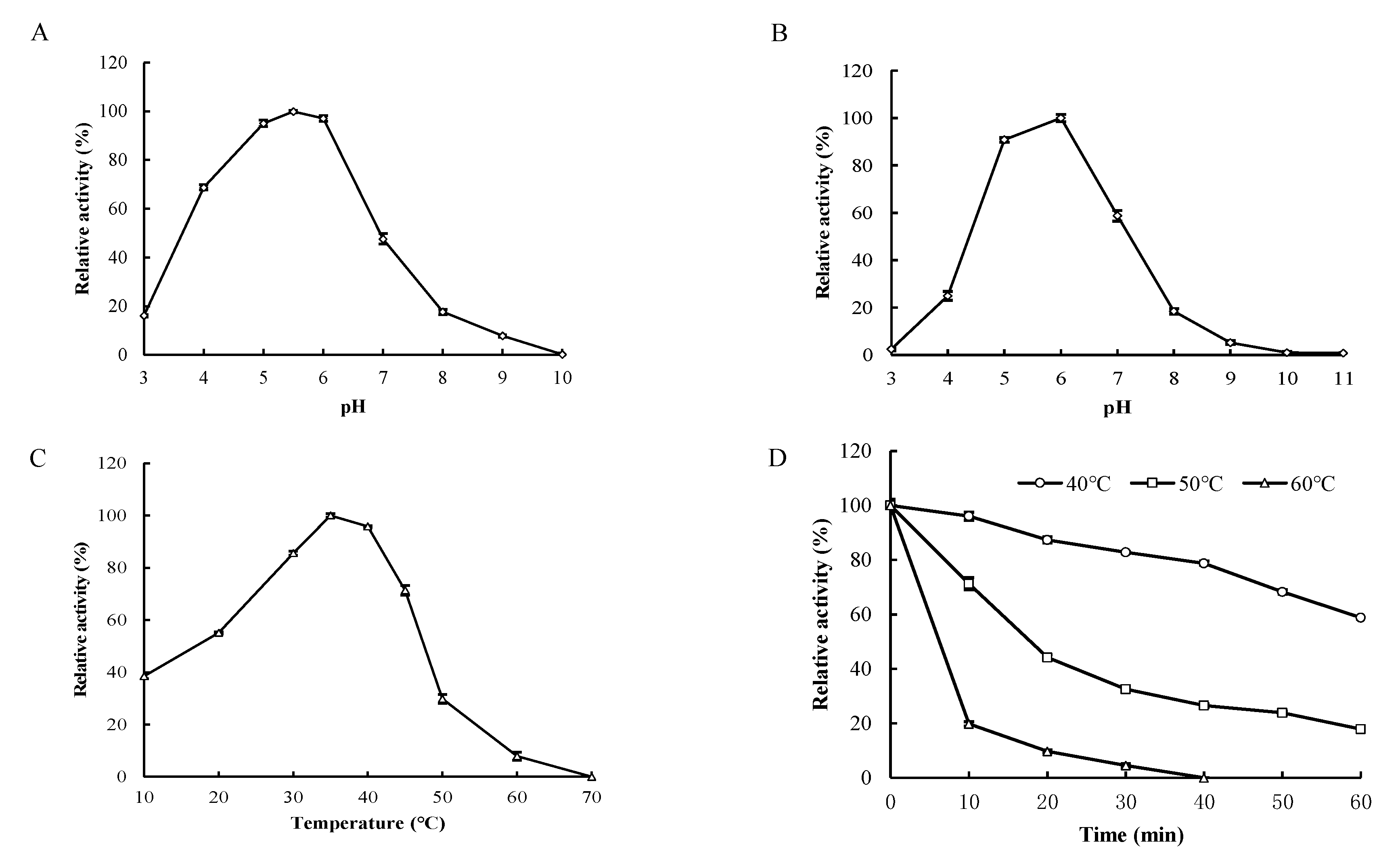
3.4. Biochemical Characterization
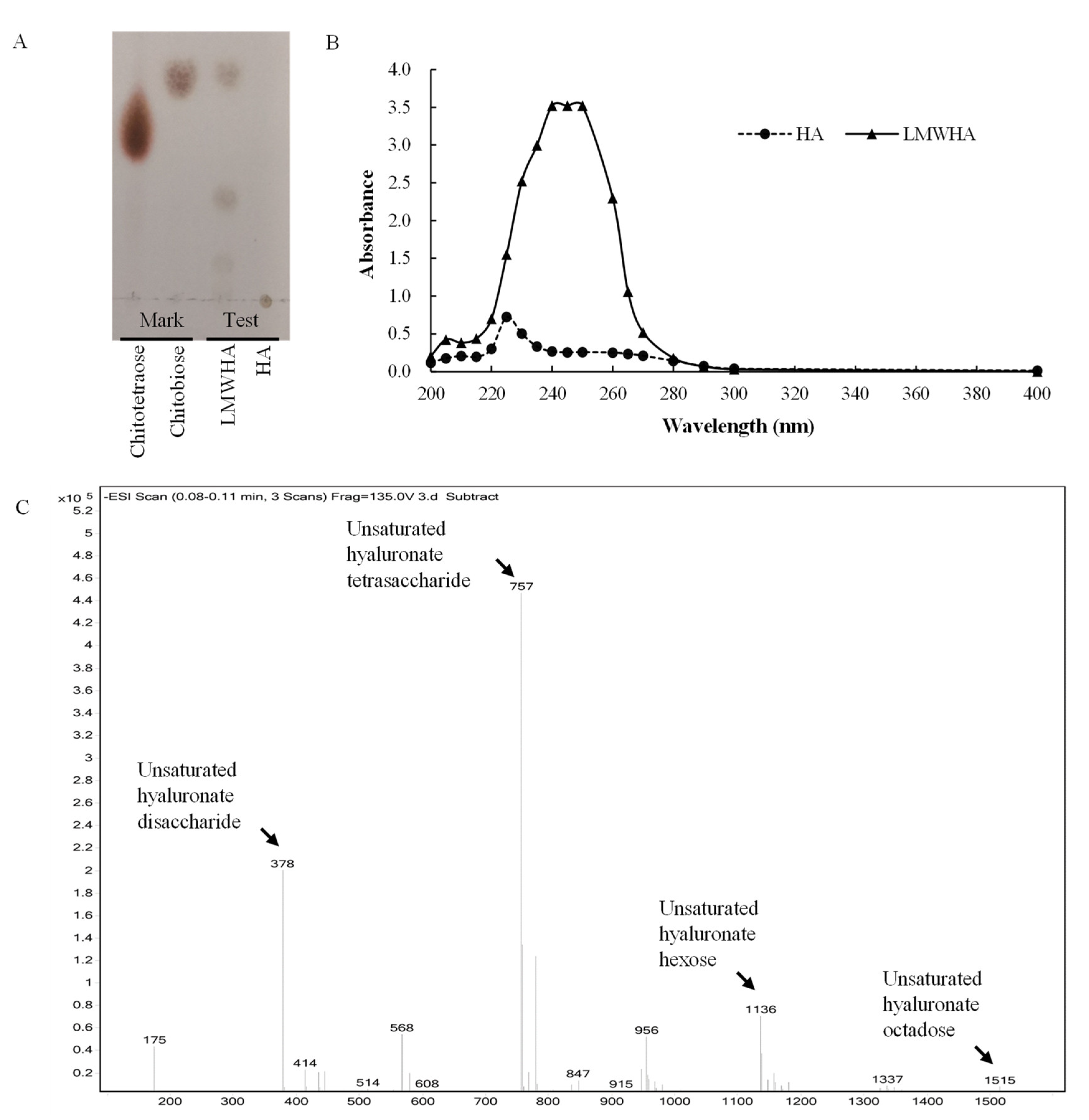
3.5. Degradation Pattern and Cleavage Products of rHynACF8
3.6. Antioxidant Properties of Cleavage Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Qiu, Y.; Ma, Y.; Huang, Y.; Li, S.; Xu, H.; Su, E. Current advances in the biosynthesis of hyaluronic acid with variable molecular weights. Carbohyd. Polym. 2021, 269, 118320–118333. [Google Scholar] [CrossRef] [PubMed]
- Cornelia, T.; Patrick, T.; Eva, T. Specific sizes of hyaluronan oligosaccharides stimulate fibroblast migration and excisional wound repair. PLoS ONE 2015, 9, e88479–e88489. [Google Scholar] [CrossRef] [Green Version]
- Snetkov, P.; Zakharova, K.; Morozkina, S.; Olekhnovich, R.; Uspenskaya, M. Hyaluronic acid: The influence of molecular weight on structural, physical, physico-chemical, and degradable properties of biopolymer. Polymers 2020, 12, 1800. [Google Scholar] [CrossRef] [PubMed]
- Ke, C.; Wang, D.; Sun, Y.; Qiao, D.; Ye, H.; Zeng, X. Immunostimulatory and antiangiogenic activities of low molecular weight hyaluronic acid. Food Chem. Toxicol. 2013, 58, 401–407. [Google Scholar] [CrossRef]
- Torigoe, K.; Tanaka, H.F.; Ohkochi, H.; Miyasaka, M.; Yamanokuchi, H.; Yoshidad, K.; Yoshida, T. Hyaluronan tetrasaccharide promotes regeneration of peripheral nerve: In vivo analysis by film model method. Brain Res. 2011, 1385, 87–92. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Li, F. Hyaluronidase and chondroitinase. Adv. Exp. Med. Biol. 2016, 2016, 75–87. [Google Scholar] [CrossRef]
- Duran-Reynals, F. Tissue permeability and the spreading factors in infection. Bacteriol. Rev. 1942, 6, 197–252. [Google Scholar] [CrossRef]
- El Safory, N.S.; Fazary, A.E.; Lee, C.K. Hyaluronidases, a group of glycosidases: Current and future perspectives. Carbohyd. Polym. 2010, 81, 165–181. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2021, 50, D571–D577. [Google Scholar] [CrossRef]
- Holm, V.A.; Nicolas, T.; Vincent, L.; Gurvan, M.; Mirjam, C.; Bernard, H.; Harry, B. A subfamily roadmap of the evolutionarily diverse glycoside hydrolase family 16 (GH16). J. Biol. Chem. 2019, 294, 15973–15986. [Google Scholar] [CrossRef] [Green Version]
- Lei, X.; Zhang, R.; Huang, Z.; Zhou, J. Research progress of hyaluronidases. Microbiol. China 2021, 48, 882–895. [Google Scholar] [CrossRef]
- Ozegowski, J.H.; Günther, E.; Reichardt, W. Purification and characterization of hyaluronidase from Streptococcus agalactiae. Zent. Bakteriol. 1994, 280, 497–506. [Google Scholar] [CrossRef]
- Xiaolong, C.; Xi, L.; Shiyun, M.; Qianru, C.; Zunxi, H.; Junpei, Z.; Rui, Z. Heterologous expression and characterization of the hyaluronic acid lyase HylS from Staphylococcus aureus. Biotecnol. Bull. 2022, 38, 1–12. [Google Scholar] [CrossRef]
- Sun, J.; Han, X.; Song, G.; Gong, Q.; Yu, W. Cloning, expression, and characterization of a new glycosaminoglycan lyase from Microbacterium sp. H14. Mar. Drugs 2019, 17, 681. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Shi, Y.; Sheng, J.; Wang, F. A novel hyaluronidase produced by Bacillus sp. A50. PLoS ONE 2014, 9, e94156–e94164. [Google Scholar] [CrossRef] [Green Version]
- Kurata, A.; Matsumoto, M.; Kobayashi, T.; Deguchi, S.; Kishimoto, N. Hyaluronate lyase of a deep-sea Bacillus niacini. Mar. Biotechnol. 2015, 17, 277–284. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, S.; Wu, H.; Li, Y.; Yu, W.; Han, F. Expression and characterization of a thermotolerant and pH-stable hyaluronate lyase from Thermasporomyces composti DSM22891. Protein Expres. Purif. 2021, 182, 105840–105848. [Google Scholar] [CrossRef]
- Elmabrouk, Z.H.; Vincent, F.; Zhang, M.; Smith, N.L.; Turkenburg, J.P.; Charnock, S.J.; Black, G.W.; Taylor, E.J. Crystal structures of a family 8 polysaccharide lyase reveal open and highly occluded substrate-binding cleft conformations. Proteins Struct. Funct. Bioinform. 2011, 79, 965–974. [Google Scholar] [CrossRef]
- Han, W.; Wang, W.; Zhao, M.; Sugahara, K.; Li, F. A novel eliminase from a marine bacterium that degrades hyaluronan and chondroitin sulfate. J. Biol. Chem. 2014, 289, 27886–27898. [Google Scholar] [CrossRef] [Green Version]
- Lei, X.; Zhou, J.; Huang, Z.; Cen, X.; Yang, L.; Zhang, R. Identification of a hyaluronidase producing strain and its enzymatic properties. Genomics Appl. Biol. 2021, 40, 1604–1610. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Gouet, P.; Robert, X.; Courcelle, E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003, 31, 3320–3323. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Han, X.W.; Xu, S.J.; Wei, C.Y.; Liu, X. Glycoside hydrolase family 39 β-xylosidase of Sphingomonas showing salt/ethanol/trypsin tolerance, low-pH/low-temperature activity, and transxylosylation activity. J. Agric. Food Chem. 2018, 66, 9465–9472. [Google Scholar] [CrossRef]
- Smith, N.L.; Taylor, E.J.; Lindsay, A.-M.; Charnock, S.J.; Turkenburg, J.P.; Dodson, E.J.; Davies, G.J.; Black, G.W. Structure of a group a streptococcal phage-encoded virulence factor reveals a catalytically active triple-stranded β-helix. Proc. Natl. Acad. Sci. USA 2005, 102, 17652–17657. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Li, X.; Cen, X.; Gao, Q.; Zhang, M.; Li, K.; Wu, Q.; Mu, Y.; Tang, X.; Zhou, J.; et al. Enzymatic preparation of manno-oligosaccharides from locust bean gum and palm kernel cake, and investigations into its prebiotic activity. Electron. J. Biotechnol. 2021, 49, 64–71. [Google Scholar] [CrossRef]
- Li, S.; Jedrzejas, M.J. Hyaluronan binding and degradation by Streptococcus agalactiae hyaluronate lyase. J. Biol. Chem. 2001, 276, 41407–41416. [Google Scholar] [CrossRef] [Green Version]
- Garron, M.L.; Cygler, M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 2010, 20, 1547–1573. [Google Scholar] [CrossRef] [Green Version]
- Rani, A.; Goyal, A. A new member of family 8 polysaccharide lyase chondroitin AC lyase (PsPL8A) from Pedobacter saltans displays endo- and exo-lytic catalysis. J. Mol. Catal. B Enzym. 2016, 134, 215–224. [Google Scholar] [CrossRef]
- Zhang, R.; Li, N.; Liu, Y.; Han, X.; Tu, T.; Shen, J.; Xu, S.; Wu, Q.; Zhou, J.; Huang, Z. Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 β-xylosidase: Activity and instability at high neutral salt concentrations. Food Chem. 2019, 301, 125266–125274. [Google Scholar] [CrossRef] [PubMed]
- Gulcin, İ. Antioxidants and antioxidant methods: An updated overview. Arch. Toxicol. 2020, 94, 651–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafsa, J.; Chaouch, M.A.; Charfeddine, B.; Rihouey, C.; Limem, K.; le Cerf, D.; Rouatbi, S.; Majdoub, H. Effect of ultrasonic degradation of hyaluronic acid extracted from rooster comb on antioxidant and antiglycation activities. Pharm. Biol. 2017, 55, 156–163. [Google Scholar] [CrossRef] [Green Version]
- Ke, C.; Sun, L.; Qiao, D.; Wang, D.; Zeng, X. Antioxidant acitivity of low molecular weight hyaluronic acid. Food Chem. Toxicol. 2011, 49, 2670–2675. [Google Scholar] [CrossRef]
- Choi, J.; Kim, J.; Kim, J.; Kweon, D.; Lee, J. Degradation of hyaluronic acid powder by electron beam irradiation, gamma ray irradiation, microwave irradiation and thermal treatment: A comparative study. Carbohyd. Polym. 2010, 79, 1080–1085. [Google Scholar] [CrossRef]
- Belcastro, M.; Marino, T.; Russo, N.; Toscano, M. Structural and electronic characterization of antioxidants from marine organisms. Theor. Chem. Acc. 2006, 115, 361–369. [Google Scholar] [CrossRef]
- Wang, J.; Hu, S.; Nie, S.; Yu, Q.; Xie, M. Reviews on mechanisms of In Vitro antioxidant activity of polysaccharides. Oxid. Med. Cell. Longev. 2016, 2016, 5692852–5692865. [Google Scholar] [CrossRef] [Green Version]

| Substance | Relative Activity (%) a | Substance | Relative Activity (%) a |
|---|---|---|---|
| none | 100.0 ± 0.8 | MgSO4 | 99.4 ± 2.2 |
| KCl | 119.9 ± 1.4 | MnSO4 | 98.8 ± 0.6 |
| LiCl | 109.4 ± 1.0 | ZnSO4 | 97.5 ± 0.9 |
| NaCl | 109.3 ± 1.1 | NiSO4 | 93.4 ± 0.5 |
| CaCl2 | 106.9 ± 0.6 | FeSO4 | 79.1 ± 0.4 |
| CoCl2 | 96.4 ± 0.7 | CuSO4 | 75.5 ± 1.3 |
| AlCl3 | 55.5 ± 1.1 | PbAc | 77.1 ± 1.9 |
| FeCl3 | 0 | EDTA | 116.3 ± 1.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Li, F.; Ma, J.; Li, M.; Lei, X.; Tang, X.; Wu, Q.; Huang, Z.; Zhang, R. Biochemical and Molecular Characteristics of a Novel Hyaluronic Acid Lyase from Citrobacter freundii. Foods 2022, 11, 1989. https://doi.org/10.3390/foods11131989
Li X, Li F, Ma J, Li M, Lei X, Tang X, Wu Q, Huang Z, Zhang R. Biochemical and Molecular Characteristics of a Novel Hyaluronic Acid Lyase from Citrobacter freundii. Foods. 2022; 11(13):1989. https://doi.org/10.3390/foods11131989
Chicago/Turabian StyleLi, Xinyue, Fang Li, Junhao Ma, Mingjun Li, Xi Lei, Xianghua Tang, Qian Wu, Zunxi Huang, and Rui Zhang. 2022. "Biochemical and Molecular Characteristics of a Novel Hyaluronic Acid Lyase from Citrobacter freundii" Foods 11, no. 13: 1989. https://doi.org/10.3390/foods11131989
APA StyleLi, X., Li, F., Ma, J., Li, M., Lei, X., Tang, X., Wu, Q., Huang, Z., & Zhang, R. (2022). Biochemical and Molecular Characteristics of a Novel Hyaluronic Acid Lyase from Citrobacter freundii. Foods, 11(13), 1989. https://doi.org/10.3390/foods11131989





