Artificial Intelligence in Colorectal Cancer Screening, Diagnosis and Treatment. A New Era
Abstract
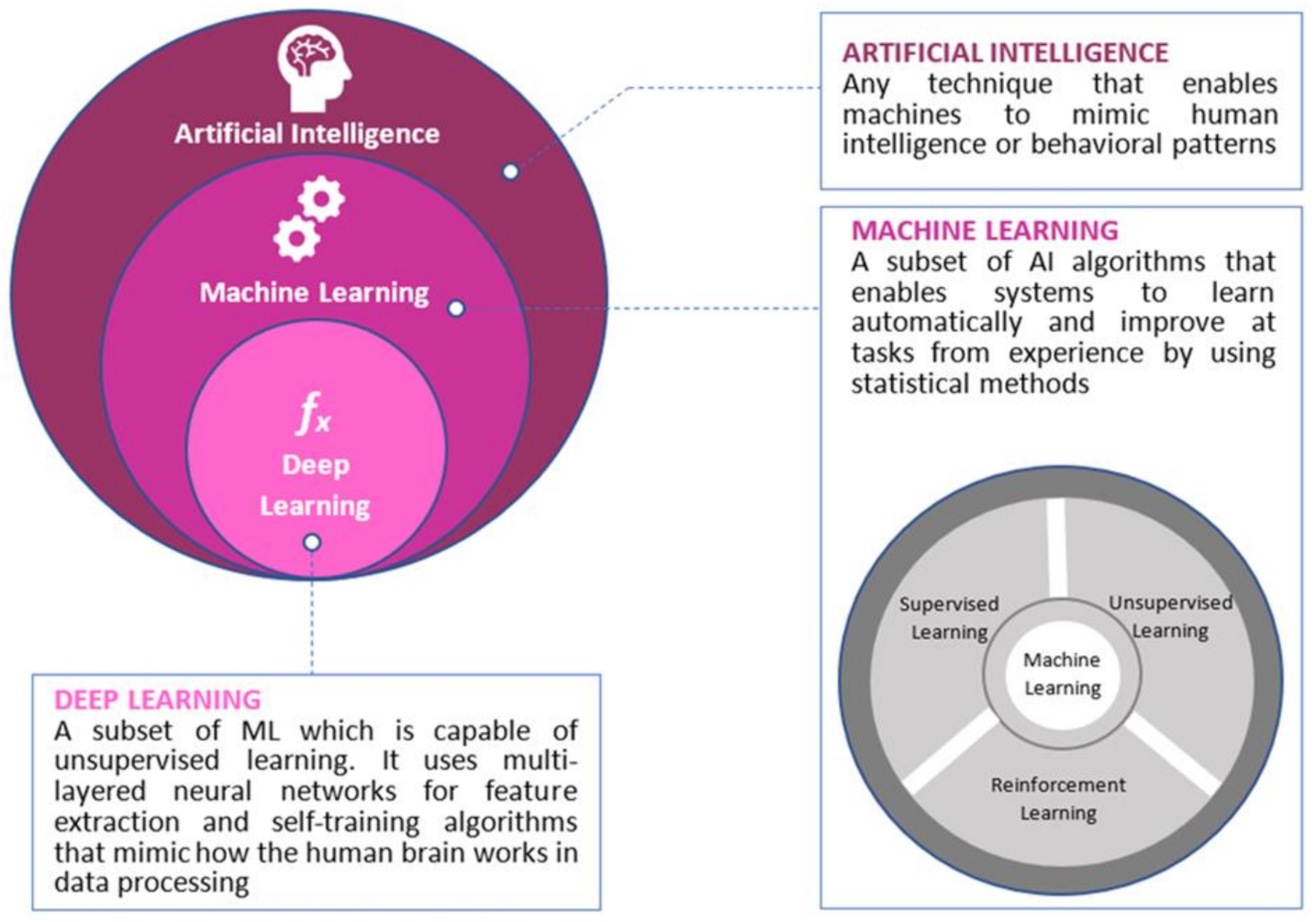
:1. Introduction
2. Artificial Intelligence, Colorectal Cancer and Genomics
3. Colorectal Cancer Screening
3.1. Colonoscopy
3.2. Virtual Colonoscopy
3.3. Capsule Endoscopy (CE)
3.4. Blood Tests
4. Polyp Detection
5. Polyp Characterization
5.1. Magnification Endoscopy with Narrow-Band Imaging (NBI)
5.2. Magnifying Chromoendoscopy
5.3. Endocytoscopy
5.4. Confocal Laser Endomicroscopy
5.5. Laser-Induced Fluorescence Spectroscopy (LIFS)
5.6. Autofluorescence Endoscopy (AFE)
5.7. White Light Endoscopy (WLE)
6. Treatment
6.1. Robotic-Assisted Surgery
6.2. Chemotherapy
7. Current Status of Precision Oncology in Colorectal Cancer
8. Solutions, Limitations and Future Directions
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Shalev-Shwartz, S.; Ben-David, S. Understanding Machine Learning: From Theory to Algorithms; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69, S36–S40. [Google Scholar] [CrossRef] [PubMed]
- Ruffle, J.K.; Farmer, A.D.; Aziz, Q. Artificial Intelligence-Assisted Gastroenterology—Promises and Pitfalls. Am. J. Gastroenterol. 2019, 114, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Kudo, S.; Mori, Y.; Misawa, M.; Takeda, K.; Kudo, T.; Itoh, H.; Oda, M.; Mori, K. Artificial intelligence and colonoscopy: Current status and future perspectives. Dig. Endosc. 2019, 31, 363–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, Y.; Kudo, S.-E.; Berzin, T.M.; Misawa, M.; Takeda, K. Computer-aided diagnosis for colonoscopy. Endoscopy 2017, 49, 813–819. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Rodríguez, R.M.; Rubio-Dorado-Manzanares, M.; Díaz-Pavón, J.M.; Reyes-Díaz, M.L.; Vazquez-Monchul, J.M.; Garcia-Cabrera, A.M.; Padillo, J.; De la Portilla, F. Learning Curve in Robotic Rectal Cancer Surgery: Current State of Affairs. Int. J. Colorectal Dis. 2016, 31, 1807–1815. [Google Scholar] [CrossRef]
- Krzyszczyk, P.; Acevedo, A.; Davidoff, E.J.; Timmins, L.M.; Marrero-Berrios, I.; Patel, M.; White, C.; Lowe, C.; Sherba, J.J.; Hartmanshenn, C.; et al. The growing role of precision and personalized medicine for cancer treatment. Technology 2018, 6, 79–100. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Niu, Z.; Bai, Y.; Tan, X. Cancer classification based on gene expression using neural networks. Genet. Mol. Res. 2015, 14, 17605–17611. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, M.; Zhu, H.; Xu, J. A 15-gene signature for prediction of colon cancer recurrence and prognosis based on SVM. Gene 2017, 604, 33–40. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Y.; Wang, Y.; Fan, Q. Detection of the BRAF V600E Mutation in Colorectal Cancer by NIR Spectroscopy in Conjunction with Counter Propagation Artificial Neural Network. Molecules 2019, 24, 2238. [Google Scholar] [CrossRef] [Green Version]
- Coppedè, F.; Grossi, E.; Lopomo, A.; Spisni, R.; Buscema, M.; Migliore, L.; Buscema, P.M. Application of artificial neural networks to link genetic and environmental factors to DNA methylation in colorectal cancer. Epigenomics 2015, 7, 175–186. [Google Scholar] [CrossRef]
- Wang, Q.; Wei, J.; Chen, Z.; Zhang, T.; Zhong, J.; Zhong, B.; Yang, P.; Li, W.; Cao, J. Establishment of multiple diagnosis models for colorectal cancer with artificial neural networks. Oncol. Lett. 2019, 17, 3314–3322. [Google Scholar] [CrossRef] [Green Version]
- Wan, N.; Weinberg, D.; Liu, T.-Y.; Niehaus, K.; Ariazi, E.A.; Delubac, D.; Kannan, A.; White, B.; Bailey, M.; Bertin, M.; et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer 2019, 19, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kel, A.; Boyarskikh, U.; Stegmaier, P.; Leskov, L.S.; Sokolov, A.V.; Yevshin, I.; Mandrik, N.; Stelmashenko, D.; Koschmann, J.; Kel-Margoulis, O.; et al. Walking pathways with positive feedback loops reveal DNA methylation biomarkers of colorectal cancer. BMC Bioinform. 2019, 20, 119. [Google Scholar] [CrossRef]
- Galamb, O.; Barták, B.K.; Kalmár, A.; Nagy, Z.B.; Szigeti, K.A.; Tulassay, Z.; Igaz, P.; Molnár, B. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J. Gastroenterol. 2019, 25, 5026–5048. [Google Scholar] [CrossRef]
- Chang, K.H.; Miller, N.; Kheirelseid, E.A.H.; Lemetre, C.; Ball, G.R.; Smith, M.J.; Regan, M.; McAnena, O.J.; Kerin, M.J. MicroRNA signature analysis in colorectal cancer: Identification of expression profiles in stage II tumors associated with aggressive disease. Int. J. Color. Dis. 2011, 26, 1415–1422. [Google Scholar] [CrossRef]
- Amirkhah, R.; Farazmand, A.; Gupta, S.K.; Ahmadi, H.; Wolkenhauer, O.; Schmitz, U. Naïve Bayes classifier predicts functional microRNA target interactions in colorectal cancer. Mol. BioSyst. 2015, 11, 2126–2134. [Google Scholar] [CrossRef] [Green Version]
- Herreros-Villanueva, M.; Duran-Sanchon, S.; Martín, A.C.; Pérez-Palacios, R.; Vila-Navarro, E.; Marcuello, M.; Diaz-Centeno, M.; Cubiella, J.; Diez, M.S.; Bujanda, L.; et al. Plasma MicroRNA Signature Validation for Early Detection of Colorectal Cancer. Clin. Transl. Gastroenterol. 2019, 10, e00003. [Google Scholar] [CrossRef]
- Afshar, S.; Afshar, S.; Warden, E.; Manochehri, H.; Saidijam, M. Application of Artificial Neural Network in miRNA Biomarker Selection and Precise Diagnosis of Colorectal Cancer. Iran. Biomed. J. 2019, 23, 175–183. [Google Scholar] [CrossRef] [Green Version]
- Xuan, P.; Dong, Y.; Guo, Y.; Zhang, T.; Liu, Y. Dual Convolutional Neural Network Based Method for Predicting Disease-Related miRNAs. Int. J. Mol. Sci. 2018, 19, 3732. [Google Scholar] [CrossRef] [Green Version]
- Gründner, J.; Prokosch, H.-U.; Stürzl, M.; Croner, R.; Christoph, J.; Toddenroth, D. Predicting Clinical Outcomes in Colorectal Cancer Using Machine Learning. Stud. Health Technol. Inform. 2018, 247, 101–105. [Google Scholar]
- Zhi, J.; Sun, J.; Wang, Z.; Ding, W. Support vector machine classifier for prediction of the metastasis of colorectal cancer. Int. J. Mol. Med. 2018, 41, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Ge, P.; Wang, W.; Li, L.; Zhang, G.; Gao, Z.; Tang, Z.; Dang, X.; Wu, Y. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of colorectal cancer. Biomed. Pharmacother. 2019, 118, 109228. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandel, J.S.; Bond, J.H.; Church, T.R.; Snover, D.C.; Bradley, G.M.; Schuman, L.M.; Ederer, F. Reducing Mortality from Colorectal Cancer by Screening for Fecal Occult Blood. N. Engl. J. Med. 1993, 328, 1365–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maida, M.; Macaluso, F.S.; Ianiro, G.; Mangiola, F.; Sinagra, E.; Hold, G.; Maida, C.; Cammarota, G.; Gasbarrini, A.; Scarpulla, G. Screening of colorectal cancer: Present and future. Expert Rev. Anticancer. Ther. 2017, 17, 1131–1146. [Google Scholar] [CrossRef] [PubMed]
- Lauby-Secretan, B.; Vilahur, N.; Bianchini, F.; Guha, N.; Straif, K.; International Agency for Research on Cancer Handbook Working Group. The IARC Perspective on Colorectal Cancer Screening. N. Engl. J. Med. 2018, 378, 1734–1740. [Google Scholar] [CrossRef]
- Morson, B. The Polyp-Cancer Sequence in the Large Bowel. Proc. R. Soc. Med. 1974, 67, 451–457. [Google Scholar] [CrossRef] [Green Version]
- Hewitson, P.; Glasziou, P.; Irwig, L.; Towler, B.; Watson, E. Screening for Colorectal Cancer Using the Faecal Occult Blood Test, Hemoccult. Cochrane Database Syst. Rev. 2007, 1, CD001216. [Google Scholar] [CrossRef] [PubMed]
- Doubeni, C.A.; Corley, D.A.; Quinn, V.P.; Jensen, C.D.; Zauber, A.G.; Goodman, M.; Johnson, J.R.; Mehta, S.J.; A Becerra, T.; Zhao, W.K.; et al. Effectiveness of screening colonoscopy in reducing the risk of death from right and left colon cancer: A large community-based study. Gut 2016, 67, 291–298. [Google Scholar] [CrossRef]
- Brenner, H.; Stock, C.; Hoffmeister, M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: Systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ 2014, 348, g2467. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.A.; Ba, K.S.A.; Brooks, D.; Fedewa, S.A.; Manassaram-Baptiste, D.; Saslow, D.; Wender, R.C. Cancer screening in the United States, 2019: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J. Clin. 2019, 69, 184–210. [Google Scholar] [CrossRef]
- Issa, I.A.; Noureddine, M. Colorectal cancer screening: An updated review of the available options. World J. Gastroenterol. 2017, 23, 5086–5096. [Google Scholar] [CrossRef]
- Nartowt, B.J.; Hart, G.R.; Muhammad, W.; Liang, Y.; Stark, G.F.; Deng, J. Robust Machine Learning for Colorectal Cancer Risk Prediction and Stratification. Front. Big Data 2020, 3, 3389. [Google Scholar] [CrossRef] [Green Version]
- Bressler, B.; Paszat, L.F.; Chen, Z.; Rothwell, D.M.; Vinden, C.; Rabeneck, L. Rates of New or Missed Colorectal Cancers After Colonoscopy and Their Risk Factors: A Population-Based Analysis. Gastroenterology 2007, 132, 96–102. [Google Scholar] [CrossRef]
- Morris, E.J.A.; Rutter, M.D.; Finan, P.J.; Thomas, J.D.; Valori, R. Post-colonoscopy colorectal cancer (PCCRC) rates vary considerably depending on the method used to calculate them: A retrospective observational population-based study of PCCRC in the English National Health Service. Gut 2014, 64, 1248–1256. [Google Scholar] [CrossRef]
- Kaminski, M.F.; Regula, J.; Kraszewska, E.; Polkowski, M.; Wojciechowska, U.; Didkowska, J.; Zwierko, M.; Rupinski, M.; Nowacki, M.P.; Butruk, E. Quality Indicators for Colonoscopy and the Risk of Interval Cancer. N. Engl. J. Med. 2010, 362, 1795–1803. [Google Scholar] [CrossRef] [Green Version]
- Corley, D.A.; Jensen, C.D.; Marks, A.R.; Zhao, W.K.; Lee, J.K.; Doubeni, C.A.; Zauber, A.G.; De Boer, J.; Fireman, B.H.; Schottinger, J.E.; et al. Adenoma Detection Rate and Risk of Colorectal Cancer and Death. N. Engl. J. Med. 2014, 370, 1298–1306. [Google Scholar] [CrossRef] [Green Version]
- Ahn, S.B.; Han, D.S.; Bae, J.H.; Byun, T.J.; Kim, J.P.; Eun, C.S. The Miss Rate for Colorectal Adenoma Determined by Quality-Adjusted, Back-to-Back Colonoscopies. Gut Liver 2012, 6, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Zheng, B.; Rieder, E.; Cassera, M.A.; Martinec, D.V.; Lee, G.; Panton, O.N.M.; Park, A.; Swanström, L.L. Quantifying Mental Workloads of Surgeons Performing Natural Orifice Transluminal Endoscopic Surgery (NOTES) Procedures. Surg. Endosc. 2012, 26, 1352–1358. [Google Scholar] [CrossRef]
- Kaminski, M.F.; Thomas-Gibson, S.; Bugajski, M.; Bretthauer, M.; Rees, C.J.; Dekker, E.; Hoff, G.; Jover, R.; Suchanek, S.; Ferlitsch, M.; et al. Performance measures for lower gastrointestinal endoscopy: A European Society of Gastrointestinal Endoscopy (ESGE) quality improvement initiative. United Eur. Gastroenterol. J. 2017, 5, 309–334. [Google Scholar] [CrossRef]
- Spadaccini, M.; Frazzoni, L.; Vanella, G.; East, J.; Radaelli, F.; Spada, C.; Fuccio, L.; Benamouzig, R.; Bisschops, R.; Bretthauer, M.; et al. Efficacy and Tolerability of High- vs Low-Volume Split-Dose Bowel Cleansing Regimens for Colonoscopy: A Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2020, 18, 1454–1465.e14. [Google Scholar] [CrossRef] [PubMed]
- Van Rijn, J.C.; Reitsma, J.B.; Stoker, J.; Bossuyt, P.M.; Van Deventer, S.J.; Dekker, E. Polyp Miss Rate Determined by Tandem Colonoscopy: A Systematic Review. Am. J. Gastroenterol. 2006, 101, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Park, D.I.; Lee, S.-H.; Hwangbo, Y.; Eun, C.S.; Han, D.S.; Cha, J.M.; Lee, B.-I.; Shin, J.E. Participation by experienced endoscopy nurses increases the detection rate of colon polyps during a screening colonoscopy: A multicenter, prospective, randomized study. Gastrointest. Endosc. 2011, 74, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Aslanian, H.R.; Shieh, F.K.; Chan, F.W.; Ciarleglio, M.M.; Deng, Y.; Rogart, J.N.; Jamidar, P.A.; Siddiqui, U.D. Nurse Observation During Colonoscopy Increases Polyp Detection: A Randomized Prospective Study. Am. J. Gastroenterol. 2013, 108, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Nogueira-Rodríguez, A.; Domínguez-Carbajales, R.; López-Fernández, H.; Iglesias, Á.; Cubiella, J.; Fdez-Riverola, F.; Reboiro-Jato, M.; Glez-Peña, D. Deep Neural Networks Approaches for Detecting and Classifying Colorectal Polyps. Neurocomputing 2021, 423, 721–734. [Google Scholar] [CrossRef]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Berzin, T.M.; Brown, J.R.G.; Bharadwaj, S.; Becq, A.; Xiao, X.; Liu, P.; Li, L.; Song, Y.; Zhang, D.; et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: A prospective randomised controlled study. Gut 2019, 68, 1813–1819. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Kudo, S.-E.; Misawa, M.; Saito, Y.; Ikematsu, H.; Hotta, K.; Ohtsuka, K.; Urushibara, F.; Kataoka, S.; Ogawa, Y.; et al. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps during Colonoscopy: A Prospective Study. Ann. Intern. Med. 2018, 169, 357–366. [Google Scholar] [CrossRef]
- Vining, D.J.; Gelfand, D.W.; Bechtold, R.E.; Scharling, E.S.; Grishaw, E.K.; Shifrin, R.Y. Technical Feasibility of Colon Imaging with Helical CT and Virtual Reality. AJR Am. J. Roentgenol. 1994, 162, 104. [Google Scholar]
- Song, B.; Zhang, G.; Lu, H.; Wang, H.; Zhu, W.; Pickhardt, P.J.; Liang, Z. Volumetric texture features from higher-order images for diagnosis of colon lesions via CT colonography. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 1021–1031. [Google Scholar] [CrossRef]
- Grosu, S.; Wesp, P.; Graser, A.; Maurus, S.; Schulz, C.; Knösel, T.; Cyran, C.C.; Ricke, J.; Ingrisch, M.; Kazmierczak, P.M. Machine Learning–based Differentiation of Benign and Premalignant Colorectal Polyps Detected with CT Colonography in an Asymptomatic Screening Population: A Proof-of-Concept Study. Radiology 2021, 202363. [Google Scholar] [CrossRef]
- The Paris Endoscopic Classification of Superficial Neoplastic Lesions: Esophagus, Stomach, and Colon: November 30 to December 1, 2002. Gastrointest. Endosc. 2003, 58 (Suppl. 6), S3–S43. [CrossRef]
- Endoscopic Classification Review Group Update on the Paris Classification of Superficial Neoplastic Lesions in the Digestive Tract. Endoscopy 2005, 37, 570–578. [CrossRef]
- O’brien, M.J.; Winawer, S.J.; Zauber, A.G.; Bushey, M.T.; Sternberg, S.S.; Gottlieb, L.S.; Bond, J.H.; Waye, J.D.; Schapiro, M.; National Polyp Study Workgroup. Flat Adenomas in the National Polyp Study: Is There Increased Risk for High-Grade Dysplasia Initially or during Surveillance? Clin. Gastroenterol. Hepatol. 2004, 2, 905–911. [Google Scholar] [CrossRef]
- Taylor, S.A.; Iinuma, G.; Saito, Y.; Zhang, J.; Halligan, S. CT colonography: Computer-aided detection of morphologically flat T1 colonic carcinoma. Eur. Radiol. 2008, 18, 1666–1673. [Google Scholar] [CrossRef]
- Eliakim, R.; Fireman, Z.; Gralnek, I.M.; Yassin, K.; Waterman, M.; Kopelman, Y.; Lachter, J.; Koslowsky, B.; Adler, S.N. Evaluation of the PillCam Colon capsule in the detection of colonic pathology: Results of the first multicenter, prospective, comparative study. Endoscopy 2006, 38, 963–970. [Google Scholar] [CrossRef]
- Van Gossum, A.; Munoz-Navas, M.; Fernandez-Urien, I.; Carretero, C.; Gay, G.; Delvaux, M.; Lapalus, M.G.; Ponchon, T.; Neuhaus, H.; Philipper, M.; et al. Capsule Endoscopy versus Colonoscopy for the Detection of Polyps and Cancer. N. Engl. J. Med. 2009, 361, 264–270. [Google Scholar] [CrossRef] [Green Version]
- Blanes-Vidal, V.; Baatrup, G.; Nadimi, E.S. Addressing priority challenges in the detection and assessment of colorectal polyps from capsule endoscopy and colonoscopy in colorectal cancer screening using machine learning. Acta Oncol. 2019, 58, S29–S36. [Google Scholar] [CrossRef]
- Hosoe, N.; Kamiya, K.J.L.L.; Hayashi, Y.; Sujino, T.; Ogata, H.; Kanai, T. Current status of colon capsule endoscopy. Dig. Endosc. 2020. [Google Scholar] [CrossRef]
- Soares, F.; Becker, K.; Anzanello, M.J. A hierarchical classifier based on human blood plasma fluorescence for non-invasive colorectal cancer screening. Artif. Intell. Med. 2017, 82, 1–10. [Google Scholar] [CrossRef]
- Hornbrook, M.C.; Goshen, R.; Choman, E.; O’Keeffe-Rosetti, M.; Kinar, Y.; Liles, E.G.; Rust, K.C. Early Colorectal Cancer Detected by Machine Learning Model Using Gender, Age, and Complete Blood Count Data. Dig. Dis. Sci. 2017, 62, 2719–2727. [Google Scholar] [CrossRef] [Green Version]
- Kinar, Y.; Kalkstein, N.; Akiva, P.; Levin, B.; Half, E.E.; Goldshtein, I.; Chodick, G.; Shalev, V. Development and validation of a predictive model for detection of colorectal cancer in primary care by analysis of complete blood counts: A binational retrospective study. J. Am. Med. Inform. Assoc. 2016, 23, 879–890. [Google Scholar] [CrossRef] [Green Version]
- Spell, D.W.; Jones, D.V.; Harper, W.F.; Bessman, J.D. The value of a complete blood count in predicting cancer of the colon. Cancer Detect. Prev. 2004, 28, 37–42. [Google Scholar] [CrossRef]
- Kinar, Y.; Akiva, P.; Choman, E.; Kariv, R.; Shalev, V.; Levin, B.; Narod, S.A.; Goshen, R. Performance analysis of a machine learning flagging system used to identify a group of individuals at a high risk for colorectal cancer. PLoS ONE 2017, 12, e0171759. [Google Scholar] [CrossRef]
- Hilsden, R.J.; Heitman, S.J.; Mizrahi, B.; Narod, S.A.; Goshen, R. Prediction of findings at screening colonoscopy using a machine learning algorithm based on complete blood counts (ColonFlag). PLoS ONE 2018, 13, e0207848. [Google Scholar] [CrossRef]
- Gupta, P.; Gulzar, Z.; Hsieh, B.; Lim, A.; Watson, D.; Mei, R. Analytical validation of the CellMax platform for early detection of cancer by enumeration of rare circulating tumor cells. J. Circ. Biomark. 2019, 8. [Google Scholar] [CrossRef]
- Ivancic, M.M.; Megna, B.W.; Sverchkov, Y.; Craven, M.; Reichelderfer, M.; Pickhardt, P.J.; Sussman, M.R.; Kennedy, G.D. Noninvasive Detection of Colorectal Carcinomas Using Serum Protein Biomarkers. J. Surg. Res. 2020, 246, 160–169. [Google Scholar] [CrossRef]
- Clercq, C.M.; Bouwens, M.W.; Rondagh, E.J.; Bakker, C.M.; Keulen, E.T.; Ridder, R.J.; Winkens, B.; Masclee, A.A.; Sanduleanu, S. Postcolonoscopy Colorectal Cancers Are Preventable: A Population-Based Study. Gut 2014, 63, 957–963. [Google Scholar] [CrossRef]
- Ahmad, O.F.; Soares, A.S.; Mazomenos, E.; Brandao, P.; Vega, R.; Seward, E.; Stoyanov, D.; Chand, M.; Lovat, L.B. Artificial Intelligence and Computer-Aided Diagnosis in Colonoscopy: Current Evidence and Future Directions. Lancet Gastroenterol. Hepatol. 2019, 4, 71–80. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Esparrach, G.; Bernal, J.; López-Cerón, M.; Córdova, H.; Sánchez-Montes, C.; Rodríguez de Miguel, C.; Sánchez, F.J. Exploring the Clinical Potential of an Automatic Colonic Polyp Detection Method Based on the Creation of Energy Maps. Endoscopy 2016, 48, 837–842. [Google Scholar] [CrossRef]
- Geetha, K.; Rajan, C. Automatic Colorectal Polyp Detection in Colonoscopy Video Frames. Asian Pac. J. Cancer Prev. 2016, 17, 4869–4873. [Google Scholar]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Integrating Online and Offline Three-Dimensional Deep Learning for Automated Polyp Detection in Colonoscopy Videos. IEEE J. Biomed. Health Inform. 2016, 21, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zheng, Y.; Mak, T.W.C.; Yu, R.; Wong, S.H.; Lau, J.Y.W.; Poon, C.C.Y. Automatic Detection and Classification of Colorectal Polyps by Transferring Low-Level CNN Features From Nonmedical Domain. IEEE J. Biomed. Health Inform. 2017, 21, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Billah, M.; Waheed, S.; Rahman, M.M. An Automatic Gastrointestinal Polyp Detection System in Video Endoscopy Using Fusion of Color Wavelet and Convolutional Neural Network Features. Int. J. Biomed. Imaging 2017, 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Misawa, M.; Kudo, S.-E.; Mori, Y.; Cho, T.; Kataoka, S.; Yamauchi, A.; Ogawa, Y.; Maeda, Y.; Takeda, K.; Ichimasa, K.; et al. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology 2018, 154, 2027–2029.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urban, G.; Tripathi, P.; Alkayali, T.; Mittal, M.; Jalali, F.; Karnes, W.; Baldi, P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology 2018, 155, 1069–1078.e8. [Google Scholar] [CrossRef]
- Figueiredo, P.N.; Figueiredo, I.N.; Pinto, L.; Kumar, S.; Tsai, Y.-H.R.; Mamonov, A.V. Polyp detection with computer-aided diagnosis in white light colonoscopy: Comparison of three different methods. Endosc. Int. Open 2019, 7, E209–E215. [Google Scholar] [CrossRef]
- Klare, P.; Sander, C.; Prinzen, M.; Haller, B.; Nowack, S.; Abdelhafez, M.; Poszler, A.; Brown, H.; Wilhelm, D.; Schmid, R.M.; et al. Automated polyp detection in the colorectum: A prospective study (with videos). Gastrointest. Endosc. 2019, 89, 576–582.e1. [Google Scholar] [CrossRef]
- Yamada, M.; Saito, Y.; Imaoka, H.; Saiko, M.; Yamada, S.; Kondo, H.; Takamaru, H.; Sakamoto, T.; Sese, J.; Kuchiba, A.; et al. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Liu, W.-N.; Zhang, Y.-Y.; Bian, X.-Q.; Wang, L.-J.; Yang, Q.; Zhang, X.-D. Study on detection rate of polyps and adenomas in artificial-intelligence-aided colonoscopy. Saudi J. Gastroenterol. 2020, 26, 13–19. [Google Scholar] [CrossRef]
- Su, J.-R.; Li, Z.; Shao, X.-J.; Ji, C.-R.; Ji, R.; Zhou, R.-C.; Li, G.-C.; Liu, G.-Q.; He, Y.-S.; Zuo, X.-L.; et al. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: A prospective randomized controlled study (with videos). Gastrointest. Endosc. 2020, 91, 415–424.e4. [Google Scholar] [CrossRef]
- Ozawa, T.; Ishihara, S.; Fujishiro, M.; Kumagai, Y.; Shichijo, S.; Tada, T. Automated endoscopic detection and classification of colorectal polyps using convolutional neural networks. Ther. Adv. Gastroenterol. 2020, 13. [Google Scholar] [CrossRef] [Green Version]
- Gong, D.; Wu, L.; Zhang, J.; Mu, G.; Shen, L.; Liu, J.; Wang, Z.; Zhou, W.; An, P.; Huang, X.; et al. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): A randomised controlled study. Lancet Gastroenterol. Hepatol. 2020, 5, 352–361. [Google Scholar] [CrossRef]
- Wang, P.; Liu, X.; Berzin, T.M.; Brown, J.R.G.; Liu, P.; Zhou, C.; Lei, L.; Li, L.; Guo, Z.; Lei, S.; et al. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): A double-blind randomised study. Lancet Gastroenterol. Hepatol. 2020, 5, 343–351. [Google Scholar] [CrossRef]
- Hassan, C.; Wallace, M.B.; Sharma, P.; Maselli, R.; Craviotto, V.; Spadaccini, M.; Repici, A. New artificial intelligence system: First validation study versus experienced endoscopists for colorectal polyp detection. Gut 2019, 69, 799–800. [Google Scholar] [CrossRef]
- Repici, A.; Badalamenti, M.; Maselli, R.; Correale, L.; Radaelli, F.; Rondonotti, E.; Ferrara, E.; Spadaccini, M.; Alkandari, A.; Fugazza, A.; et al. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology 2020, 159, 512–520.e7. [Google Scholar] [CrossRef]
- Karkanis, S.; Iakovidis, D.; Maroulis, D.; Karras, D.; Tzivras, M. Computer-aided tumor detection in endoscopic video using color wavelet features. IEEE Trans. Inf. Technol. Biomed. 2003, 7, 141–152. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Xiao, X.; Brown, J.R.G.; Berzin, T.M.; Tu, M.; Xiong, F.; Hu, X.; Liu, P.; Song, Y.; Zhang, D.; et al. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat. Biomed. Eng. 2018, 2, 741–748. [Google Scholar] [CrossRef]
- Lui, T.K.; Guo, C.-G.; Leung, W.K. Accuracy of artificial intelligence on histology prediction and detection of colorectal polyps: A systematic review and meta-analysis. Gastrointest. Endosc. 2020, 92, 11–22.e6. [Google Scholar] [CrossRef]
- Bisschops, R.; East, J.E.; Hassan, C.; Hazewinkel, Y.; Kamiński, M.F.; Neumann, H.; Pellisé, M.; Antonelli, G.; Balen, M.B.; Coron, E.; et al. Advanced imaging for detection and differentiation of colorectal neoplasia: European Society of Gastrointestinal Endoscopy (ESGE) Guideline—Update 2019. Endoscopy 2019, 51, 1155–1179. [Google Scholar] [CrossRef] [Green Version]
- Rex, D.K.; Kahi, C.; O’Brien, M.; Levin, T.; Pohl, H.; Rastogi, A.; Burgart, L.; Imperiale, T.; Ladabaum, U.; Cohen, J.; et al. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest. Endosc. 2011, 73, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Kominami, Y.; Yoshida, S.; Tanaka, S.; Sanomura, Y.; Hirakawa, T.; Raytchev, B.; Tamaki, T.; Koide, T.; Kaneda, K.; Chayama, K. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest. Endosc. 2016, 83, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Misawa, M.; Kudo, S.-E.; Mori, Y.; Nakamura, H.; Kataoka, S.; Maeda, Y.; Kudo, T.; Hayashi, T.; Wakamura, K.; Miyachi, H.; et al. Characterization of Colorectal Lesions Using a Computer-Aided Diagnostic System for Narrow-Band Imaging Endocytoscopy. Gastroenterology 2016, 150, 1531–1532.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, Y.; Kudo, S.-E.; Chiu, P.W.Y.; Singh, R.; Misawa, M.; Wakamura, K.; Kudo, T.; Hayashi, T.; Katagiri, A.; Miyachi, H.; et al. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: An international web-based study. Endoscopy 2016, 48, 1110–1118. [Google Scholar] [CrossRef]
- Takeda, K.; Kudo, S.-E.; Mori, Y.; Misawa, M.; Kudo, T.; Wakamura, K.; Katagiri, A.; Baba, T.; Hidaka, E.; Ishida, F.; et al. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy 2017, 49, 798–802. [Google Scholar] [CrossRef]
- Komeda, Y.; Handa, H.; Watanabe, T.; Nomura, T.; Kitahashi, M.; Sakurai, T.; Okamoto, A.; Minami, T.; Kono, M.; Arizumi, T.; et al. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology 2017, 93, 30–34. [Google Scholar] [CrossRef]
- Chen, P.-J.; Lin, M.-C.; Lai, M.-J.; Lin, J.-C.; Lu, H.H.-S.; Tseng, V.S. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology 2018, 154, 568–575. [Google Scholar] [CrossRef]
- Renner, J.; Phlipsen, H.; Haller, B.; Navarro-Avila, F.; Saint-Hill-Febles, Y.; Mateus, D.; Ponchon, T.; Poszler, A.; Abdelhafez, M.; Schmid, R.M.; et al. Optical classification of neoplastic colorectal polyps–A computer-assisted approach (the COACH study). Scand. J. Gastroenterol. 2018, 53, 1100–1106. [Google Scholar] [CrossRef]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Pérez, M.L.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019, 68, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Min, M.; Su, S.; He, W.; Bi, Y.; Ma, Z.; Liu, Y. Computer-aided diagnosis of colorectal polyps using linked color imaging colonoscopy to predict histology. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Montes, C.; Sánchez, F.J.; Bernal, J.; Córdova, H.; López-Cerón, M.; Cuatrecasas, M.; De Miguel, C.R.; García-Rodríguez, A.; Garcés-Durán, R.; Pellisé, M.; et al. Computer-aided prediction of polyp histology on white light colonoscopy using surface pattern analysis. Endoscopy 2018, 51, 261–265. [Google Scholar] [CrossRef]
- Lui, T.K.; Wong, K.K.; Mak, L.L.; Ko, M.K.; Tsao, S.K.; Leung, W.K. Endoscopic prediction of deeply submucosal invasive carcinoma with use of artificial intelligence. Endosc. Int. Open 2019, 7, E514–E520. [Google Scholar] [CrossRef] [Green Version]
- Horiuchi, H.; Tamai, N.; Kamba, S.; Inomata, H.; Ohya, T.R.; Sumiyama, K. Real-time computer-aided diagnosis of diminutive rectosigmoid polyps using an auto-fluorescence imaging system and novel color intensity analysis software. Scand. J. Gastroenterol. 2019, 54, 800–805. [Google Scholar] [CrossRef]
- Rodriguez-Diaz, E.; Baffy, G.; Lo, W.-K.; Mashimo, H.; Vidyarthi, G.; Mohapatra, S.S.; Singh, S.K. Real-time artificial intelligence–based histologic classification of colorectal polyps with augmented visualization. Gastrointest. Endosc. 2021, 93, 662–670. [Google Scholar] [CrossRef]
- Jin, E.H.; Lee, D.; Bae, J.H.; Kang, H.Y.; Kwak, M.-S.; Seo, J.Y.; Yang, J.I.; Yang, S.Y.; Lim, S.H.; Yim, J.Y.; et al. Improved Accuracy in Optical Diagnosis of Colorectal Polyps Using Convolutional Neural Networks with Visual Explanations. Gastroenterology 2020, 158, 2169–2179.e8. [Google Scholar] [CrossRef]
- Kudo, S.-E.; Misawa, M.; Mori, Y.; Hotta, K.; Ohtsuka, K.; Ikematsu, H.; Saito, Y.; Takeda, K.; Nakamura, H.; Ichimasa, K.; et al. Artificial Intelligence-assisted System Improves Endoscopic Identification of Colorectal Neoplasms. Clin. Gastroenterol. Hepatol. 2020, 18, 1874–1881.e2. [Google Scholar] [CrossRef]
- Mori, Y.; Kudo, S.-E.; East, J.E.; Rastogi, A.; Bretthauer, M.; Misawa, M.; Sekiguchi, M.; Matsuda, T.; Saito, Y.; Ikematsu, H.; et al. Cost savings in colonoscopy with artificial intelligence-aided polyp diagnosis: An add-on analysis of a clinical trial (with video). Gastrointest. Endosc. 2020, 92, 905–911.e1. [Google Scholar] [CrossRef]
- Hardy, N.P.; Mac Aonghusa, P.; Neary, P.M.; A Cahill, R. Intraprocedural Artificial Intelligence for Colorectal Cancer Detection and Characterisation in Endoscopy and Laparoscopy. Surg. Innov. 2021. [Google Scholar] [CrossRef] [PubMed]
- Barbeiro, S.; Libânio, D.; Castro, R.; Dinis-Ribeiro, M.; Pimentel-Nunes, P. Narrow-Band Imaging: Clinical Application in Gastrointestinal Endoscopy. GE Port. J. Gastroenterol. 2018, 26, 40–53. [Google Scholar] [CrossRef]
- Takemura, Y.; Yoshida, S.; Tanaka, S.; Kawase, R.; Onji, K.; Oka, S.; Tamaki, T.; Raytchev, B.; Kaneda, K.; Yoshihara, M.; et al. Computer-aided system for predicting the histology of colorectal tumors by using narrow-band imaging magnifying colonoscopy (with video). Gastrointest. Endosc. 2012, 75, 179–185. [Google Scholar] [CrossRef]
- Hirata, M.; Tanaka, S.; Oka, S.; Kaneko, I.; Yoshida, S.; Yoshihara, M.; Chayama, K. Evaluation of microvessels in colorectal tumors by narrow band imaging magnification. Gastrointest. Endosc. 2007, 66, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Maeyama, Y.; Mitsuyama, K.; Noda, T.; Nagata, S.; Nagata, T.; Yoshioka, S.; Yoshida, H.; Mukasa, M.; Sumie, H.; Kawano, H.; et al. Prediction of colorectal tumor grade and invasion depth through narrow-band imaging scoring. World J. Gastroenterol. 2018, 24, 4809–4820. [Google Scholar] [CrossRef] [PubMed]
- Tischendorf, J.J.W.; Groß, S.; Winograd, R.; Hecker, H.; Auer, R.; Behrens, A.; Trautwein, C.; Aach, T.; Stehle, T. Computer-aided classification of colorectal polyps based on vascular patterns: A pilot study. Endoscopy 2010, 42, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.; Trautwein, C.; Behrens, A.; Winograd, R.; Palm, S.; Lutz, H.H.; Schirin-Sokhan, R.; Hecker, H.; Aach, T.; Tischendorf, J.J. Computer-based classification of small colorectal polyps by using narrow-band imaging with optical magnification. Gastrointest. Endosc. 2011, 74, 1354–1359. [Google Scholar] [CrossRef]
- Tamaki, T.; Yoshimuta, J.; Kawakami, M.; Raytchev, B.; Kaneda, K.; Yoshida, S.; Takemura, Y.; Onji, K.; Miyaki, R.; Tanaka, S. Computer-aided colorectal tumor classification in NBI endoscopy using local features. Med. Image Anal. 2013, 17, 78–100. [Google Scholar] [CrossRef]
- Hirakawa, T.; Tamaki, T.; Raytchev, B.; Kaneda, K.; Koide, T.; Kominami, Y.; Yoshida, S.; Tanaka, S. SVM-MRF segmentation of colorectal NBI endoscopic images. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2014, 2014, 4739–4742. [Google Scholar]
- Häfner, M.; Tamaki, T.; Tanaka, S.; Uhl, A.; Wimmer, G.; Yoshida, S. Local fractal dimension based approaches for colonic polyp classification. Med. Image Anal. 2015, 26, 92–107. [Google Scholar] [CrossRef]
- Wimmer, G.; Tamaki, T.; Tischendorf, J.; Häfner, M.; Yoshida, S.; Tanaka, S.; Uhl, A. Directional wavelet based features for colonic polyp classification. Med. Image Anal. 2016, 31, 16–36. [Google Scholar] [CrossRef]
- Okamoto, T.; Koide, T.; Sugi, K.; Shimizu, T.; Hoang, A.-T.; Tamaki, T.; Raytchev, B.; Kaneda, K.; Kominami, Y.; Yoshida, S.; et al. Image segmentation of pyramid style identifier based on Support Vector Machine for colorectal endoscopic images. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2015, 2015, 2997–3000. [Google Scholar]
- Kudo, S.-E.; Mori, Y.; Wakamura, K.; Ikehara, N.; Ichimasa, K.; Wada, Y.; Kutsukawa, M.; Misawa, M.; Kudo, T.; Hayashi, T.; et al. Endocytoscopy Can Provide Additional Diagnostic Ability to Magnifying Chromoendoscopy for Colorectal Neoplasms: Endocytoscopy for Colorectal Neoplasms. J. Gastroenterol. Hepatol. 2014, 29, 83–90. [Google Scholar] [CrossRef]
- Häfner, M.; Gangl, A.; Kwitt, R.; Uhl, A.; Vécsei, A.; Wrba, F. Improving Pit-Pattern Classification of Endoscopy Images by a Combination of Experts. Med. Image Comput. Comput. Assist. Interv. 2009, 12 Pt 1, 247–254. [Google Scholar]
- Takemura, Y.; Yoshida, S.; Tanaka, S.; Onji, K.; Oka, S.; Tamaki, T.; Kaneda, K.; Yoshihara, M.; Chayama, K. Quantitative analysis and development of a computer-aided system for identification of regular pit patterns of colorectal lesions. Gastrointest. Endosc. 2010, 72, 1047–1051. [Google Scholar] [CrossRef] [Green Version]
- Neumann, H.; Fuchs, F.S.; Vieth, M.; Atreya, R.; Siebler, J.; Kiesslich, R.; Neurath, M.F. Review Article: In Vivo Imaging by Endocytoscopy: Review: Endocytoscopy. Aliment. Pharmacol. Ther. 2011, 33, 1183–1193. [Google Scholar] [CrossRef]
- Neumann, H.; Kudo, S.; Vieth, M.; Neurath, M.F. Real-time in vivo histologic examination using a probe-based endocytoscopy system for differentiating duodenal polyps. Endoscopy 2013, 45, E53–E54. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Kudo, S.-E.; Wakamura, K.; Misawa, M.; Ogawa, Y.; Kutsukawa, M.; Kudo, T.; Hayashi, T.; Miyachi, H.; Ishida, F.; et al. Novel computer-aided diagnostic system for colorectal lesions by using endocytoscopy (with videos). Gastrointest. Endosc. 2015, 81, 621–629. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Kudo, S.-E.; Mori, K. Potential of artificial intelligence-assisted colonoscopy using an endocytoscope (with video). Dig. Endosc. 2018, 30, 52–53. [Google Scholar] [CrossRef] [Green Version]
- Neumann, H.; Kiesslich, R.; Wallace, M.B.; Neurath, M.F. Confocal Laser Endomicroscopy: Technical Advances and Clinical Applications. Gastroenterology 2010, 139, 388–392.e2. [Google Scholar] [CrossRef]
- André, B.; Vercauteren, T.; Buchner, A.M.; Krishna, M.; Ayache, N.; Wallace, M.B. Software for automated classification of probe-based confocal laser endomicroscopy videos of colorectal polyps. World J. Gastroenterol. 2012, 18, 5560–5569. [Google Scholar] [CrossRef]
- Ştefănescu, D.; Streba, C.; Cârţână, E.T.; Săftoiu, A.; Gruionu, G.; Gruionu, L.G. Computer Aided Diagnosis for Confocal Laser Endomicroscopy in Advanced Colorectal Adenocarcinoma. PLoS ONE 2016, 11, e0154863. [Google Scholar] [CrossRef] [Green Version]
- Kuiper, T.; Alderlieste, Y.A.; Tytgat, K.M.A.J.; Vlug, M.S.; Nabuurs, J.A.; Bastiaansen, B.A.J.; Lowenberg, M.; Fockens, P.; Dekker, E. Automatic optical diagnosis of small colorectal lesions by laser-induced autofluorescence. Endoscopy 2014, 47, 56–62. [Google Scholar] [CrossRef]
- Rath, T.; Tontini, G.E.; Vieth, M.; Nägel, A.; Neurath, M.F.; Neumann, H. In vivo real-time assessment of colorectal polyp histology using an optical biopsy forceps system based on laser-induced fluorescence spectroscopy. Endoscopy 2016, 48, 557–562. [Google Scholar] [CrossRef]
- Aihara, H.; Saito, S.; Inomata, H.; Ide, D.; Tamai, N.; Ohya, T.R.; Kato, T.; Amitani, S.; Tajiri, H. Computer-aided diagnosis of neoplastic colorectal lesions using ‘real-time’ numerical color analysis during autofluorescence endoscopy. Eur. J. Gastroenterol. Hepatol. 2013, 25, 488–494. [Google Scholar] [CrossRef]
- Inomata, H.; Tamai, N.; Aihara, H.; Sumiyama, K.; Saito, S.; Kato, T.; Tajiri, H. Efficacy of a novel auto-fluorescence imaging system with computer-assisted color analysis for assessment of colorectal lesions. World J. Gastroenterol. 2013, 19, 7146–7153. [Google Scholar] [CrossRef]
- Hussain, A.; Malik, A.; Halim, M.U.; Ali, A.M. The use of robotics in surgery: A review. Int. J. Clin. Pract. 2014, 68, 1376–1382. [Google Scholar] [CrossRef]
- Albani, J.M. The role of robotics in surgery: A review. Mo. Med. 2007, 104, 166–172. [Google Scholar] [PubMed]
- Hirano, Y.; Kondo, H.; Yamaguchi, S. Robot-assisted surgery with Senhance robotic system for colon cancer: Our original single-incision plus 2-port procedure and a review of the literature. Tech. Coloproctol. 2021, 25, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Spanheimer, P.M.; Armstrong, J.G.; Fu, S.; Liao, J.; Regenbogen, S.E.; Byrn, J.C. Robotic proctectomy for rectal cancer: Analysis of 71 patients from a single institution. Int. J. Med. Robot. Comput. Assist. Surg. 2017, 13, e1841. [Google Scholar] [CrossRef]
- Zawadzki, M.; Krzystek-Korpacka, M.; Gamian, A.; Witkiewicz, W. Comparison of inflammatory responses following robotic and open colorectal surgery: A prospective study. Int. J. Color. Dis. 2017, 32, 399–407. [Google Scholar] [CrossRef]
- Kavalukas, S.L.; Ghuman, A.; Sharp, S.P.; Wexner, S.D. Robotic or laparoscopic surgery for rectal cancer—which is the best answer? A comprehensive review of non-oncological outcomes and learning curve. Mini-Invasive Surg. 2020, 2020. [Google Scholar] [CrossRef]
- Rodríguez, R.M.J.; Pavón, J.M.D.; Juan, F.D.L.P.D.; Sillero, E.P.; Dussort, J.M.H.C.; Padillo, J. Prospective Randomised Study: Robotic-Assisted Versus Conventional Laparoscopic Surgery in Colorectal Cancer Resection. Cir. Esp. 2011, 89, 432–438. [Google Scholar] [CrossRef]
- Lee, S.H.; Lim, S.; Kim, J.H.; Lee, K.Y. Robotic versus conventional laparoscopic surgery for rectal cancer: Systematic review and meta-analysis. Ann. Surg. Treat. Res. 2015, 89, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Oshiro, E.; Sánchez-Egido, I.; Moreno-Sierra, J.; Pérez, C.F.; Díaz, J.S.; Fernández-Represa, J.Á. Robotic Assistance May Reduce Conversion to Open in Rectal Carcinoma Laparoscopic Surgery: Systematic Review and Meta-Analysis: Meta-Analysis of Robotic vs Laparoscopic Rectal Cancer Surgery. Int. J. Med. Robot. 2012, 8, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Cho, M.S.; Baek, S.J.; Hur, H.; Min, B.S.; Baik, S.H.; Lee, K.Y.; Kim, N.K. Long-Term Oncologic Outcomes of Robotic Low Anterior Resection for Rectal Cancer: A Comparative Study with Laparoscopic Surgery. Ann. Surg. 2015, 261, 129–137. [Google Scholar] [CrossRef]
- Baek, S.J.; Kim, C.H.; Cho, M.S.; Bae, S.U.; Hur, H.; Min, B.S.; Baik, S.H.; Lee, K.Y.; Kim, N.K. Robotic surgery for rectal cancer can overcome difficulties associated with pelvic anatomy. Surg. Endosc. 2015, 29, 1419–1424. [Google Scholar] [CrossRef]
- Jayne, D.; Pigazzi, A.; Marshall, H.; Croft, J.; Corrigan, N.; Copeland, J.; Quirke, P.; West, N.; Rautio, T.; Thomassen, N.; et al. Effect of Robotic-Assisted vs Conventional Laparoscopic Surgery on Risk of Conversion to Open Laparotomy among Patients Undergoing Resection for Rectal Cancer: The ROLARR Randomized Clinical Trial. JAMA 2017, 318, 1569–1580. [Google Scholar] [CrossRef]
- Kim, H.J.; Choi, G.-S.; Park, J.S.; Park, S.Y.; Yang, C.S.; Lee, H.J. The Impact of Robotic Surgery on Quality of Life, Uri-nary and Sexual Function Following Total Mesorectal Excision for Rectal Cancer: A Propensity Score-Matched Analysis with Laparoscopic Surgery. Colorectal. Dis. 2018, 20, O103–O113. [Google Scholar] [CrossRef]
- Yang, S.-X.; Sun, Z.-Q.; Zhou, Q.-B.; Xu, J.-Z.; Chang, Y.; Xia, K.-K.; Wang, G.-X.; Li, Z.; Song, J.-M.; Zhang, Z.-Y.; et al. Security and Radical Assessment in Open, Laparoscopic, Robotic Colorectal Cancer Surgery: A Comparative Study. Technol. Cancer Res. Treat. 2018, 17. [Google Scholar] [CrossRef] [Green Version]
- Cruz, S.M.; Gomes, S.E.; Borralho, P.M.; Rodrigues, C.M.P.; Gaudêncio, S.P.; Pereira, F. In silico HCT116 human colon cancer cell-based models en route to the discovery of lead-like anticancer drugs. Biomolecules 2018, 8, 56. [Google Scholar] [CrossRef] [Green Version]
- Berishvili, V.P.; Voronkov, A.E.; Radchenko, E.V.; Palyulin, V.A. Machine Learning Classification Models to Improve the Docking-based Screening: A Case of PI3K-Tankyrase Inhibitors. Mol. Inform. 2018, 37, e1800030. [Google Scholar] [CrossRef]
- Torchilin, V.P. Passive and Active Drug Targeting: Drug Delivery to Tumors as an Example. Organotypic Models Drug Dev. 2009, 3–53. [Google Scholar] [CrossRef]
- Martel, S.; Mohammadi, M. Switching between Magnetotactic and Aerotactic Displacement Controls to Enhance the Efficacy of MC-1 Magneto-Aerotactic Bacteria as Cancer-Fighting Nanorobots. Micromachines 2016, 7, 97. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, R.; Mancini-Terracciano, C.; Voena, C.; Rengo, M.; Zerunian, M.; Ciardiello, A.; Grasso, S.; Mare’, V.; Paramatti, R.; Russomando, A.; et al. MR-based artificial intelligence model to assess response to therapy in locally advanced rectal cancer. Eur. J. Radiol. 2019, 118, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, Y.; Nie, K.; Sun, X.; Niu, T.; Yue, N.; Kwong, T.; Chang, P.; Chow, D.; Chen, J.-H.; et al. Machine learning for prediction of chemoradiation therapy response in rectal cancer using pre-treatment and mid-radiation multi-parametric MRI. Magn. Reson. Imaging 2019, 61, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.P.; Magee, D.; Cremolini, C.; Antoniotti, C.; Halbert, D.D.; Xiu, J.; Stafford, P.; Berry, D.A.; Oberley, M.J.; Shields, A.F.; et al. Clinical Validation of a Machine-learning–derived Signature Predictive of Outcomes from First-line Oxaliplatin-based Chemotherapy in Advanced Colorectal Cancer. Clin. Cancer Res. 2021, 27, 1174–1183. [Google Scholar] [CrossRef] [PubMed]
- Oyaga-Iriarte, E.; Insausti, A.; Sayar, O.; Aldaz, A. Prediction of irinotecan toxicity in metastatic colorectal cancer patients based on machine learning models with pharmacokinetic parameters. J. Pharmacol. Sci. 2019, 140, 20–25. [Google Scholar] [CrossRef]
- National Research Council (US). Committee on a Framework for Development a New Taxonomy of Disease. In Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Keshava, N.; Toh, T.S.; Yuan, H.; Yang, B.; Menden, M.P.; Wang, D. Defining subpopulations of differential drug response to reveal novel target populations. NPJ Syst. Biol. Appl. 2019, 5, 1–11. [Google Scholar] [CrossRef]
- Lee, J.; Kumar, S.; Lee, S.-Y.; Park, S.J.; Kim, M.-H. Development of Predictive Models for Identifying Potential S100A9 Inhibitors Based on Machine Learning Methods. Front. Chem. 2019, 7, 779. [Google Scholar] [CrossRef]
- Ding, D.; Han, S.; Zhang, H.; He, Y.; Li, Y. Predictive biomarkers of colorectal cancer. Comput. Biol. Chem. 2019, 83, 107106. [Google Scholar] [CrossRef]
- Pacheco, M.P.; Bintener, T.; Ternes, D.; Kulms, D.; Haan, S.; Letellier, E.; Sauter, T. Identifying and targeting cancer-specific metabolism with network-based drug target prediction. EBioMedicine 2019, 43, 98–106. [Google Scholar] [CrossRef] [Green Version]
- Nowak-Sliwinska, P.; Scapozza, L.; I Altaba, A.R. Drug repurposing in oncology: Compounds, pathways, phenotypes and computational approaches for colorectal cancer. Biochim. Biophys. Acta Bioenerg. 2019, 1871, 434–454. [Google Scholar] [CrossRef]
- Horta, A.B.; Salgado, C.; Fernandes, M.; Vieira, S.; Sousa, J.M.; Papoila, A.L.; Xavier, M. Clinical decision support tool for Co-management signalling. Int. J. Med. Inform. 2018, 113, 56–62. [Google Scholar] [CrossRef]
- Schmidt, C.M.D. Anderson Breaks With IBM Watson, Raising Questions About Artificial Intelligence in Oncology. J. Natl. Cancer Inst. 2017, 109, 5. [Google Scholar] [CrossRef] [Green Version]
- Miyano, S. Artificial Intelligence for Cancer Genomic Medicine: Understanding Cancer is Beyond Human Ability. Brain Nerve 2019, 71, 25–32. [Google Scholar]
- Potter, P. Hippocrates. In Diseases III; Hippocrates Volume VI; Harvard University Press: Cambridge, MA, USA, 1988. [Google Scholar]
- Abrahams, E. Right Drug-Right Patient-Right Time: Personalized Medicine Coalition. Clin. Transl. Sci. 2008, 1, 11–12. [Google Scholar] [CrossRef] [Green Version]
- East, J.E.; Vleugels, J.L.; Roelandt, P.; Bhandari, P.; Bisschops, R.; Dekker, E.; Hassan, C.; Horgan, G.; Kiesslich, R.; Longcroft-Wheaton, G.; et al. Advanced endoscopic imaging: European Society of Gastrointestinal Endoscopy (ESGE) Technology Review. Endoscopy 2016, 48, 1029–1045. [Google Scholar] [CrossRef] [Green Version]
- Imler, T.; Morea, J.; Kahi, C.; Xu, H.; Calley, C.; Imperiale, T. Multicenter Colonoscopy Quality Measurement Utilizing Natural Language Processing. Am. J. Gastroenterol. 2014, 109, S653. [Google Scholar] [CrossRef]
- Marzuki, M.F.M.; Yaacob, N.A.; Bin Yaacob, N.M.; Abu Hassan, M.R.; Ahmad, S.B.; Hairon, S.M.; Leclercq, W. Usable Mobile App for Community Education on Colorectal Cancer: Development Process and Usability Study. JMIR Hum. Factors 2019, 6, e12103. [Google Scholar] [CrossRef]
- Abadir, A.P.; Ali, M.F.; Karnes, W.; Samarasena, J.B. Artificial Intelligence in Gastrointestinal Endoscopy. Clin. Endosc. 2020, 53, 132–141. [Google Scholar] [CrossRef]
- Auger, S.D.; Jacobs, B.M.; Dobson, R.; Marshall, C.R.; Noyce, A.J. Big Data, Machine Learning and Artificial Intelligence: A Neurologist’s Guide. Pract. Neurol. 2020, 21, 4–11. [Google Scholar] [CrossRef]
- Chen, D.; Liu, S.; Kingsbury, P.; Sohn, S.; Storlie, C.B.; Habermann, E.B.; Naessens, J.M.; Larson, D.W.; Liu, H. Deep learning and alternative learning strategies for retrospective real-world clinical data. NPJ Digit. Med. 2019, 2, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Narla, A.; Kuprel, B.; Sarin, K.; Novoa, R.; Ko, J. Automated Classification of Skin Lesions: From Pixels to Practice. J. Investig. Dermatol. 2018, 138, 2108–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tom, E.; Keane, P.A.; Blazes, M.; Pasquale, L.R.; Chiang, M.F.; Lee, A.Y.; Lee, C.S.; Force, A.A.A.I.T. Protecting Data Privacy in the Age of AI-Enabled Ophthalmology. Transl. Vis. Sci. Technol. 2020, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Alagappan, M.; Brown, J.R.G.; Mori, Y.; Berzin, T.M. Artificial intelligence in gastrointestinal endoscopy: The future is almost here. World J. Gastrointest. Endosc. 2018, 10, 239–249. [Google Scholar] [CrossRef]
- Dash, S.; Shakyawar, S.K.; Sharma, M.; Kaushik, S. Big data in healthcare: Management, analysis and future prospects. J. Big Data 2019, 6, 54. [Google Scholar] [CrossRef] [Green Version]
- Supriya, M.; Deepa, A. Machine learning approach on healthcare big data: A review. Big Data Inf. Anal. 2020, 5, 58–75. [Google Scholar] [CrossRef]

| Author, Year | Country | Study Design | AI Algorithm | Type of Images | Outcomes |
|---|---|---|---|---|---|
| Fernández-Esparrach et al., 2016 [71] | Spain | Retrospective | WM-DOVA energy maps | 24 videos containing 31 colorectal polyps | Sensitivity: 70.4% Specificity: 72.4% |
| Geetha et al., 2016 [72] | India | Ex vivo | Hand crafted | Still images, 703 frames | Sensitivity: 95% Specificity: 97% |
| Yu et al., 2017 [73] | China | Ex vivo | CNN | Videos, ASU-Mayo 18 colonoscopy videos | Sensitivity: 71% PPV: 88% |
| Zhang et al., 2017 [74] | China | Ex vivo | CNN | Still images | Accuracy: 86% AUC: 1 |
| Billah et al., 2017 [75] | Bangladesh | Ex vivo | CNN | 14,000 still images | Sensitivity: 99% Specificity: 99% Accuracy: 99% |
| Misawa et al., 2018 [76] | Japan | Ex vivo | CNN | Videos | Per-frame sensitivity: 90% Specificity: 63.3% Accuracy: 76.5% Per-polyp sensitivity: 94% False positive rate: 60% |
| Urban et al., 2018 [77] | United States | Ex vivo | CNN | Videos | Sensitivity: 90% |
| Figueiredo et al., 2019 [78] | Portugal | Retrospective | SVM binary classifiers | 42 colonoscopy videos containing 1680 frames with polyps and 1360 frames without polyps | Sensitivity: 99.7% Specificity: 84.9% Accuracy: 91.1% |
| Klare et al., 2019 [79] | Germany | In vivo, prospective cohort | KoloPol software | Real-time colonoscopy | Per-polyp sensitivity: 75% ADR in CADe group vs colonoscopy group: 29% vs. 31% |
| Yamada et al., 2019 [80] | Japan | Ex vivo | CNN | Videos | Sensitivity: 97.3% Specificity: 99% AUC: 0.975 |
| Wang et al., 2019 [48] | China | Prospective, RCT | EndoScreener | Real-time colonoscopy | ADR in CADe group vs standard colonoscopy group: 29.1% vs. 20.3%, p < 0.001 |
| Liu et al., 2020 [81] | China | Prospective, RCT | Henan Tongyu | Real-time colonoscopy | ADR in CADe group vs control group: 39.2% vs. 24% |
| Su et al., 2020 [82] | China | Prospective, RCT | Deep CNNs | Real-time colonoscopy | ADR in CADe group vs control group: 28.9% vs. 16.5% |
| Ozawa et al., 2020 [83] | Japan | Ex vivo | CNN | 7077 images | Sensitivity: 92% Accuracy: 83% PPV: 86% |
| Gong et al., 2020 [84] | China | Prospective, RCT | ENDOANGEL | Real-time colonoscopy | ADR in CADe group vs. control group: 16% vs. 8% |
| Wang et al., 2020 [85] | China | Double-blind, RCT | EndoScreener | Real-time colonoscopy | ADR in CADe group (484 patients) vs control group (478 patients): 34.1% vs. 28% |
| Hassan et al., 2020 [86] | Italy | Retrospective | GI Genius | 338 videos | Per-lesion sensitivity: 99.7% |
| Repici et al., 2020 [87] | Italy | RCT | GI Genius | Real-time colonoscopy | ADR in CADe group vs. control group: 54.8% vs. 40.4% |
| Author, Year | Country | Study Type | Patients/ Polyps | Imaging Modality | AI Algorithm | Real-Time | Outcomes | Notes |
|---|---|---|---|---|---|---|---|---|
| Kominami et al., 2016 [93] | Japan | Prospective | 41/118 | Magnifying NBI | SVM | Yes | Sensitivity: 93% Specificity: 93.3% Accuracy: 93.2% PPV: 93% NPV: 93.3% | Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Misawa et al., 2016 [94] | Japan | Retrospective | NA/100 | Endocytoscopy with NBI | EndoBRAIN | No | Sensitivity: 84.5% Specificity: 97.6% Accuracy: 90% PPV: 98%, NPV: 82% | Histologic findings were used as the reference standard |
| Mori et al., 2016 [95] | Japan | Retrospective | 123/205 | Endocytoscopy | SVM | No | Sensitivity: 89% Specificity: 88% Accuracy: 89% PPV: 95% NPV: 76% | Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Takeda et al., 2017 [96] | Japan | Retrospective | 76/76 | Endocytoscopy | SVM | No | Sensitivity: 89.4% Specificity: 98.9% Accuracy: 94.1% PPV: 98.8% NPV: 90.1% | CADx system for differentiation between invasive CRC and adenomatous polyps. Histologic findings were used as the reference standard |
| Komeda et al., 2017 [97] | Japan | Retrospective | NA/NA | A combination of WLE, NBI and Chromoendoscopy | CNN | Yes | Accuracy: 75.1% | Histologic findings were used as the reference standard |
| Chen et al., 2018 [98] | Taiwan | Prospective | 193/284 | Magnifying NBI | CNN | No (real-time capability) | Sensitivity: 96.3% Specificity: 78.1% Accuracy: 90.1% PPV: 89.6% NPV: 91.5% | Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Mori et al., 2018 [49] | Japan | Prospective | 325/466 | Endocytoscopy with NBI and ΜΒ staining modes | SVM | Yes | Sensitivity: >90% Specificity: ~70% for identifying proximal diminutive adenomas. Accuracy: 98.1% NPV: 96.4% | Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Renner et al., 2018 [99] | Germany | Retrospective | NA/100 | WLE, NBI | Deep neural network | No | Sensitivity: 92.3% Specificity: 62.5% Accuracy: 78% PPV: 72.7% NPV: 88.2% | Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Byrne et al., 2019 [100] | Canada | Retrospective | NA/106 | NBI | CNN | Real-time capability | Sensitivity: 98% Specificity: 83% Accuracy: 94% PPV: 90% NPV: 97% | Diminutive polyps were involved. Histologic findings were used as the reference standard |
| Min et al., 2019 [101] | China | Prospective | 91/181 | LCI | Gaussian mixture model | No | Sensitivity: 83.3% Specificity: 70.1% Accuracy: 78.4% PPV: 82.6% NPV: 71.2% | Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Sánchez-Montes et al., 2019 [102] | Spain | Retrospective | NA/225 | WLE | SVMs | No | Sensitivity: 92.3% Specificity: 89.2% Accuracy: 91.1% PPV: 93.6% NPV: 87.1% | Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Lui et al., 2019 [103] | China | Retrospective | NA/76 | WLE, NBI | CNN | No | Sensitivity: 88.2% Specificity: 77.9% Accuracy: 85.5% | CADx system for invasive CRC diagnosis. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Horiuchi et al., 2019 [104] | Japan | Prospective | 95/429 | AFI | Color intensity analysis software | Yes | Sensitivity: 80% Specificity: 95.3% Accuracy: 91.5% PPV: 85.2% NPV: 93.4% | Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Ozawa et al., 2020 [83] | Japan | Retrospective | 174/309 | NBI | CNN | No (real-time capability) | Sensitivity: 97% PPV: 84% NPV: 88% | AI system for characterization and detection of colorectal polyps. Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Rodriguez-Diaz et al., 2020 [105] | United States | Prospective | 119/280 | Magnifying NBI | DL, a semantic segmentation model based on DeepLab V3+ framework with ResNet18-based feature extractor | Yes | Sensitivity: 96% Specificity: 84% NPV: 91%, HCR: 88% For diminutive colorectal polyps: Sensitivity: 95% Specificity: 88% NPV: 93%, HCR: 86% | Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Jin et al., 2020 [106] | South Korea | Prospective | NA/300 | NBI | CNN | No | Sensitivity: 83.3% Specificity: 91.7% Accuracy: 86.7% PPV: 93.8% NPV: 78.6% | Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
| Kudo et al., 2020 [107] | Japan | Retrospective | NA/2000 | Endocytoscopy with NBI and ΜΒ staining modes | EndoBRAIN | No | NBI Sensitivity: 96.9% Specificity: 94.3% Accuracy: 96% PPV: 96.9% NPV: 94.3% Stained images Sensitivity: 96.9% Specificity: 100% Accuracy: 98% PPV: 100% NPV: 94.6% | Diminutive polyps were involved. Endoscopists were used as controls. Histologic findings were used as the reference standard |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitsala, A.; Tsalikidis, C.; Pitiakoudis, M.; Simopoulos, C.; Tsaroucha, A.K. Artificial Intelligence in Colorectal Cancer Screening, Diagnosis and Treatment. A New Era. Curr. Oncol. 2021, 28, 1581-1607. https://doi.org/10.3390/curroncol28030149
Mitsala A, Tsalikidis C, Pitiakoudis M, Simopoulos C, Tsaroucha AK. Artificial Intelligence in Colorectal Cancer Screening, Diagnosis and Treatment. A New Era. Current Oncology. 2021; 28(3):1581-1607. https://doi.org/10.3390/curroncol28030149
Chicago/Turabian StyleMitsala, Athanasia, Christos Tsalikidis, Michail Pitiakoudis, Constantinos Simopoulos, and Alexandra K. Tsaroucha. 2021. "Artificial Intelligence in Colorectal Cancer Screening, Diagnosis and Treatment. A New Era" Current Oncology 28, no. 3: 1581-1607. https://doi.org/10.3390/curroncol28030149
APA StyleMitsala, A., Tsalikidis, C., Pitiakoudis, M., Simopoulos, C., & Tsaroucha, A. K. (2021). Artificial Intelligence in Colorectal Cancer Screening, Diagnosis and Treatment. A New Era. Current Oncology, 28(3), 1581-1607. https://doi.org/10.3390/curroncol28030149




