Radiomic and Artificial Intelligence Analysis with Textural Metrics Extracted by Contrast-Enhanced Mammography and Dynamic Contrast Magnetic Resonance Imaging to Detect Breast Malignant Lesions
Abstract
:1. Introduction
2. Methods
2.1. Patient Selection
2.2. Imaging Protocol
2.3. Image Processing
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Definition of Textural Features
Appendix A.1. First-Order Gray-Level Statistics
- Mean, the mean gray level of .
- Mode, the most frequent element(s) of array .
- Median, the sample median of or the 50th percentile of .
- Standard deviation (STD)
- Mean Absolute Deviation (MAD), the mean of the absolute deviation of all voxel intensities around the mean intensity value.
- Range, the range of intensity values of .where is the maximum intensity value of and is the minimum intensity value of .
- Interquartile range (IQR), the interquartile range is defined as the 75th minus the 25th percentile of .
- Kurtosis:where is the mean of
- Variance, Variance is the square of the standard deviation:where is the mean of .
- Skewness:where is the mean of .
Appendix A.2. Gray Level Co-Occurrence Matrix (GLCM)
- -
- be the normalized (i.e., ) co-occurrence matrix, generalized for any and α
- -
- ,
- -
- ,
- -
- be the mean of , where ,
- -
- be the mean of , where ,
- -
- be the standard deviation of , where ,
- -
- be the standard deviation of , where .
- Energy
- Contrast
- Entropy
- Homogeneity
- Correlation
- Sum Average
- Dissimilarity
- Autocorrelation
Appendix A.3. Gray Level Run-Length Matrix (GLRLM)
- -
- be the th entry in the given run-length matrix p, generalized for any direction θ,
- -
- be the number of discrete intensity values in the image,
- -
- be the maximum run length,
- -
- be the total numbers of runs, where ,
- -
- be the sum distribution of the number of runs with run length , where ,
- -
- be the sum distribution of the number of runs with run length , where ,
- -
- be the number of voxels in the image, where ,
- -
- be the mean run length, where ,
- -
- be the mean gray level, where .
- Short-Run Emphasis (SRE)
- Long-Run Emphasis (LRE)
- Gray Level Nonuniformity (GLN)
- Run-Length Nonuniformity (RLN)
- Run Percentage (RP)
- Low Gray Level Run Emphasis (LGRE)
- High Gray Level Run Emphasis (HGRE)
- Short-Run Low Gray Level Emphasis (SRLGE)
- Short-Run High Gray Level Emphasis (SRHGE)
- Long-Run Low Gray Level Emphasis (LRLGE)
- Long-Run High Gray Level Emphasis (LRHGE)
- Gray Level Variance (GLV)
- Run-Length Variance (RLV)
Appendix A.4. Gray Level Size Zone Matrix (GLSZM)
- -
- be the th entry in the given GLSZM ,
- -
- be the number of discrete intensity values in the image,
- -
- be the size of the largest, homogeneous region in the volume of interest,
- -
- be the total number of homogeneous regions (zones), where ,
- -
- be the sum distribution of the number of zones with size , where ,
- -
- be the sum distribution of the number of zones with gray level , where ,
- -
- be the number of voxels in the image, where ,
- -
- be the mean zone size, where ,
- -
- be the mean gray level, where .
- Small Zone Emphasis (SZE)
- Large Zone Emphasis (LZE)
- Gray Level Nonuniformity (GLN)
- Zone Size Nonuniformity (ZSN)
- Zone Percentage (ZP)
- Low Gray Level Zone Emphasis (LGZE)
- High Gray Level Zone Emphasis (HGZE)
- Small Zone Low Gray Level Emphasis (SZLGE)
- Small Zone High Gray Level Emphasis (SZHGE)
- Large Zone Low Gray Level Emphasis (LZLGE)
- Large Zone High Gray Level Emphasis (LZHGE)
- Gray Level Variance (GLV)
- Zone Size Variance (ZSV)
Appendix A.5. Neighborhood Gray Tone Difference Matrix (NGTDM)
- -
- be the number of voxels with gray level ,
- -
- be the total number of voxels,
- -
- be generalized for any distance ,
- -
- be the maximum discrete intensity level in the image,
- -
- be the probability of gray level ,
- -
- be the total number of gray levels present in the image.
- Coarseness:where is a small number to prevent coarseness from becoming infinite.
- Contrast
- Busyness
- Complexity
- Strengthwhere is a small number to prevent strength from becoming infinite.
References
- Patel, B.K.; Lobbes, M.; Lewin, J. Contrast Enhanced Spectral Mammography: A Review. Semin. Ultrasound CT MRI 2018, 39, 70–79. [Google Scholar] [CrossRef]
- Heywang-Köbrunner, S.; Viehweg, P.; Heinig, A.; Küchler, C. Contrast-enhanced MRI of the breast: Accuracy, value, controversies, solutions. Eur. J. Radiol. 1997, 24, 94–108. [Google Scholar] [CrossRef]
- Viehweg, P.; Paprosch, I.; Strassinopoulou, M.; Heywang-Köbrunner, S.H. Contrast-enhanced magnetic resonance imaging of the breast: Interpretation guidelines. Top. Magn. Reson. Imaging 1998, 9, 17–43. [Google Scholar] [CrossRef] [PubMed]
- Dromain, C.; Balleyguier, C.; Muller, S.; Mathieu, M.-C.; Rochard, F.; Opolon, P.; Sigal, R. Evaluation of Tumor Angiogenesis of Breast Carcinoma Using Contrast-Enhanced Digital Mammography. Am. J. Roentgenol. 2006, 187, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Baltzer, P.A.; Bickel, H.; Spick, C.; Wengert, G.; Woitek, R.; Kapetas, P.; Clauser, P.; Helbich, T.H.; Pinker, K. Potential of Noncontrast Magnetic Resonance Imaging with Diffusion-Weighted Imaging in Characterization of Breast Lesions. Investig. Radiol. 2018, 53, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Pinker, K.; Moy, L.; Sutton, E.J.; Mann, R.; Weber, M.; Thakur, S.; Jochelson, M.S.; Bago-Horvath, Z.; Morris, E.A.; Baltzer, P.A.; et al. Diffusion-Weighted Imaging with Apparent Diffusion Coefficient Mapping for Breast Cancer Detection as a Stand-Alone Parameter: Comparison with Dynamic Contrast-Enhanced and Multiparametric Magnetic Resonance Imaging. Investig. Radiol. 2018, 53, 587–595. [Google Scholar] [CrossRef]
- Liu, Z.; Feng, B.; Li, C.; Chen, Y.; Chen, Q.; Li, X.; Guan, J.; Chen, X.; Cui, E.; Li, R.; et al. Preoperative prediction of lymphovascular invasion in invasive breast cancer with dynamic contrast-enhanced-MRI-based radiomics. J. Magn. Reson. Imaging 2019, 50, 847–857. [Google Scholar] [CrossRef]
- Li, L.; Roth, R.; Germaine, P.; Ren, S.; Lee, M.; Hunter, K.; Tinney, E.; Liao, L. Contrast-enhanced spectral mammography (CESM) versus breast magnetic resonance imaging (MRI): A retrospective comparison in 66 breast lesions. Diagn. Interv. Imaging 2017, 98, 113–123. [Google Scholar] [CrossRef]
- Fallenberg, E.M.; Dromain, C.; Diekmann, F.; Engelken, F.; Krohn, M.; Singh, J.M.; Ingold-Heppner, B.; Winzer, K.J.; Bick, U.; Renz, D.M. Contrast-enhanced spectral mammography versus MRI: Initial results in the detection of breast cancer and assessment of tumour size. Eur. Radiol. 2014, 24, 256–264. [Google Scholar] [CrossRef]
- Lewin, J.M.; Isaacs, P.K.; Vance, V.; Larke, F.J. Dual-Energy Contrast-enhanced Digital Subtraction Mammography: Feasibility. Radiology 2003, 229, 261–268. [Google Scholar] [CrossRef]
- Jochelson, M.S.; Dershaw, D.D.; Sung, J.; Heerdt, A.; Thornton, C.; Moskowitz, C.S.; Ferrara, J.; Morris, E. Bilateral Contrast-enhanced Dual-Energy Digital Mammography: Feasibility and Comparison with Conventional Digital Mammography and MR Imaging in Women with Known Breast Carcinoma. Radiology 2013, 266, 743–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tagliafico, A.S.; Bignotti, B.; Rossi, F.; Signori, A.; Sormani, M.P.; Valdora, F.; Calabrese, M.; Houssami, N. Diagnostic performance of contrast-enhanced spectral mammography: Systematic review and meta-analysis. Breast 2016, 28, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Liney, G.P.; Sreenivas, M.; Gibbs, P.; Bsc, R.G.-A.; Turnbull, L.W. Breast lesion analysis of shape technique: Semiautomated vs. manual morphological description. J. Magn. Reson. Imaging 2006, 23, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Fusco, R.; Sansone, M.; Filice, S.; Carone, G.; Amato, D.M.; Sansone, C.; Petrillo, A. Pattern Recognition Approaches for Breast Cancer DCE-MRI Classification: A Systematic Review. J. Med. Biol. Eng. 2016, 36, 449–459. [Google Scholar] [CrossRef] [Green Version]
- Nie, K.; Chen, J.-H.; Yu, H.J.; Chu, Y.; Nalcioglu, O.; Su, M.-Y. Quantitative Analysis of Lesion Morphology and Texture Features for Diagnostic Prediction in Breast MRI. Acad. Radiol. 2008, 15, 1513–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.-T.; Shan, Q.-Y.; Chen, S.-L.; Li, B.; Feng, S.-T.; Xu, E.-J.; Li, X.; Long, J.-Y.; Xie, X.-Y.; Lu, M.-D.; et al. CT-based radiomics for preoperative prediction of early recurrent hepatocellular carcinoma: Technical reproducibility of acquisition and scanners. Radiol. Med. 2020, 125, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Bignotti, B.; Bianchi, L.; Picasso, R.; Martinoli, C.; Tagliafico, A.S. Radiomics of peripheral nerves MRI in mild carpal and cubital tunnel syndrome. Radiol. Med. 2019, 125, 197–203. [Google Scholar] [CrossRef]
- Santone, A.; Brunese, M.C.; Donnarumma, F.; Guerriero, P.; Mercaldo, F.; Reginelli, A.; Miele, V.; Giovagnoni, A.; Brunese, L. Radiomic features for prostate cancer grade detection through formal verification. Radiol. Med. 2021, 126, 688–697. [Google Scholar] [CrossRef]
- Fusco, R.; Sansone, M.; Filice, S.; Granata, V.; Catalano, O.; Amato, D.M.; Di Bonito, M.; D’Aiuto, M.; Capasso, I.; Rinaldo, M.; et al. Integration of DCE-MRI and DW-MRI Quantitative Parameters for Breast Lesion Classification. BioMed Res. Int. 2015, 2015, 237863. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Y.; Zhang, K.; Liu, Y.; Cui, J.; Tao, J.; Wang, Y.; Wang, S. Invasive ductal breast cancer: Preoperative predict Ki-67 index based on radiomics of ADC maps. Radiol. Med. 2020, 125, 109–116. [Google Scholar] [CrossRef]
- Chianca, V.; Albano, D.; Messina, C.; Vincenzo, G.; Rizzo, S.; Del Grande, F.; Sconfienza, L.M. An update in musculoskeletal tumors: From quantitative imaging to radiomics. Radiol. Med. 2021, 126, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, M.; Ninatti, G.; Cozzi, L.; Voulaz, E.; Gennaro, N.; Barajon, I.; Ricci, F.; Carlo-Stella, C.; Zucali, P.; Sollini, M.; et al. Computed tomography (CT)-derived radiomic features differentiate prevascular mediastinum masses as thymic neoplasms versus lymphomas. Radiol. Med. 2020, 125, 951–960. [Google Scholar] [CrossRef]
- Karmazanovsky, G.; Gruzdev, I.; Tikhonova, V.; Kondratyev, E.; Revishvili, A. Computed tomography-based radiomics approach in pancreatic tumors characterization. Radiol. Med. 2021, 126, 1388–1395. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Pirovano, M.; Ciocca, M.; Gibelli, D.; Floridi, C.; Oliva, G. Radiomic analysis of the optic nerve at the first episode of acute optic neuritis: An indicator of optic nerve pathology and a predictor of visual recovery? Radiol. Med. 2021, 126, 698–706. [Google Scholar] [CrossRef]
- Benedetti, G.; Mori, M.; Panzeri, M.M.; Barbera, M.; Palumbo, D.; Sini, C.; Muffatti, F.; Andreasi, V.; Steidler, S.; Doglioni, C.; et al. CT-derived radiomic features to discriminate histologic characteristics of pancreatic neuroendocrine tumors. Radiol. Med. 2021, 126, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Nazari, M.; Shiri, I.; Hajianfar, G.; Oveisi, N.; Abdollahi, H.; Deevband, M.R.; Oveisi, M.; Zaidi, H. Noninvasive Fuhrman grading of clear cell renal cell carcinoma using computed tomography radiomic features and machine learning. Radiol. Med. 2020, 125, 754–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crivelli, P.; Ledda, R.E.; Parascandolo, N.; Fara, A.; Soro, D.; Conti, M. A New Challenge for Radiologists: Radiomics in Breast Cancer. BioMed Res. Int. 2018, 2018, 6120703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maglogiannis, I.; Zafiropoulos, E.; Anagnostopoulos, I. An intelligent system for automated breast cancer diagnosis and prognosis using SVM based classifiers. Appl. Intell. 2007, 30, 24–36. [Google Scholar] [CrossRef]
- Zheng, Y.; Englander, S.; Baloch, S.; Zacharaki, E.I.; Fan, Y.; Schnall, M.D.; Shen, D. STEP: Spatiotemporal enhancement pattern for MR-based breast tumor diagnosis. Med. Phys. 2009, 36, 3192–3204. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Sinha, S.; Ms, F.A.L.-Q.; Debruhl, N.D.; Sayre, J.; Farria, D.; Gorczyca, D.P.; Bassett, L.W. Multifeature analysis of Gd-enhanced MR images of breast lesions. J. Magn. Reson. Imaging 1997, 7, 1016–1026. [Google Scholar] [CrossRef]
- Vomweg, T.W.; Buscema, P.M.; Kauczor, H.U.; Teifke, A.; Intraligi, M.; Terzi, S.; Heussel, C.P.; Achenbach, T.; Rieker, O.; Mayer, D.; et al. Improved artificial neural networks in prediction of malignancy of lesions in contrast-enhanced MR-mammography. Med. Phys. 2003, 30, 2350–2359. [Google Scholar] [CrossRef] [PubMed]
- Sathya, D.J.; Geetha, K. Mass classification in breast DCE-MR images using an artificial neural network trained via a bee colony optimization algorithm. ScienceAsia 2013, 39, 294. [Google Scholar] [CrossRef] [Green Version]
- Sathya, J.; Geetha, K. Experimental Investigation of Classification Algorithms for Predicting Lesion Type on Breast DCE-MR Images. Int. J. Comput. Appl. 2013, 82, 1–8. [Google Scholar] [CrossRef]
- Fusco, R.; Sansone, M.; Petrillo, A.; Sansone, C. A Multiple Classifier System for Classification of Breast Lesions Using Dynamic and Morphological Features in DCE-MRI. In Joint IAPR International Workshops on Statistical Techniques in Pattern Recognition (SPR) and Structural and Syntactic Pattern Recognition (SSPR); Springer: Berlin/Heidelberg, Germany, 2012; pp. 684–692. [Google Scholar] [CrossRef] [Green Version]
- Degenhard, A.; Tanner, C.; Hayes, C.; Hawkes, D.J.; O Leach, M. The UK MRI Breast Screening Study Comparison between radiological and artificial neural network diagnosis in clinical screening. Physiol. Meas. 2002, 23, 727–739. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Fusco, R.; Sansone, M.; Sansone, C.; Petrillo, A. Segmentation and classification of breast lesions using dynamic and textural features in Dynamic Contrast Enhanced-Magnetic Resonance Imaging. In Proceedings of the 25th IEEE International Symposium on Computer-Based Medical Systems (CBMS), Rome, Italy, 20–22 June 2012; pp. 1–4. [Google Scholar]
- Abdolmaleki, P.; Buadu, L.D.; Naderimansh, H. Feature extraction and classification of breast cancer on dynamic magnetic resonance imaging using artificial neural network. Cancer Lett. 2001, 171, 183–191. [Google Scholar] [CrossRef]
- Agner, S.C.; Soman, S.; Libfeld, E.; McDonald, M.; Thomas, K.; Englander, S.; Rosen, M.A.; Chin, D.; Nosher, J.; Madabhushi, A. Textural Kinetics: A Novel Dynamic Contrast-Enhanced (DCE)-MRI Feature for Breast Lesion Classification. J. Digit. Imaging 2010, 24, 446–463. [Google Scholar] [CrossRef] [Green Version]
- Levman, J.; Leung, T.; Causer, P.; Plewes, D.; Martel, A.L. Classification of dynamic contrast-enhanced magnetic resonance breast lesions by support vector machines. IEEE Trans. Med. Imaging 2008, 27, 688–696. [Google Scholar] [CrossRef] [Green Version]
- Levman, J.; Martel, A.L. Computer-aided diagnosis of breast cancer from magnetic resonance imaging examinations by custom radial basis function vector machine. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 30 August–4 September 2010; Volume 2010, pp. 5577–5580. [Google Scholar]
- Fusco, R.; Piccirillo, A.; Sansone, M.; Granata, V.; Rubulotta, M.; Petrosino, T.; Barretta, M.; Vallone, P.; Di Giacomo, R.; Esposito, E.; et al. Radiomics and Artificial Intelligence Analysis with Textural Metrics Extracted by Contrast-Enhanced Mammography in the Breast Lesions Classification. Diagnostics 2021, 11, 815. [Google Scholar] [CrossRef]
- Fusco, R.; Piccirillo, A.; Sansone, M.; Granata, V.; Vallone, P.; Barretta, M.L.; Petrosino, T.; Siani, C.; Di Giacomo, R.; Di Bonito, M.; et al. Radiomic and Artificial Intelligence Analysis with Textural Metrics, Morphological and Dynamic Perfusion Features Extracted by Dynamic Contrast-Enhanced Magnetic Resonance Imaging in the Classification of Breast Lesions. Appl. Sci. 2021, 11, 1880. [Google Scholar] [CrossRef]
- Vallières, M.; Freeman, C.R.; Skamene, S.R.; El Naqa, I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys. Med. Biol. 2015, 60, 5471–5496. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- R-Tools Technology Inc. Available online: https://www.r-tt.com/ (accessed on 15 October 2020).
- Tibshirani, R. The lasso Method for Variable Selection in the Cox Model. Statist. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B Statist. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Bruce, P.; Bruce, A. Practical Statistics for Data Scientists; O’Reilly Media, Inc.: Newton, MA, USA, 2017. [Google Scholar]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: New York, NY, USA, 2013; Volume 112, p. 18. [Google Scholar]
- Wang, Q.; Luo, Z.; Huang, J.; Feng, Y.; Liu, Z. A Novel Ensemble Method for Imbalanced Data Learning: Bagging of Extrapolation-SMOTE SVM. Comput. Intell. Neurosci. 2017, 2017, 1827016. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Angelov, P.P.; Soares, E.A. A self-adaptive synthetic over-sampling technique for imbalanced classification. Int. J. Intell. Syst. 2020, 35, 923–943. [Google Scholar] [CrossRef]
- Chen, Z.; Lin, T.; Xia, X.; Xu, H.; Ding, S. A synthetic neighborhood generation-based ensemble learning for the imbalanced data classification. Appl. Intell. 2017, 48, 2441–2457. [Google Scholar] [CrossRef]
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–6 June 2008; pp. 1322–1328. [Google Scholar]
- Bernardi, D.; Belli, P.; Benelli, E.; Brancato, B.; Bucchi, L.; Calabrese, M.; Carbonaro, L.A.; Caumo, F.; Cavallo-Marincola, B.; Clauser, P.; et al. Digital breast tomosynthesis (DBT): Recommendations from the Italian College of Breast Radiologists (ICBR) by the Italian Society of Medical Radiology (SIRM) and the Italian Group for Mammography Screening (GISMa). Radiol. Med. 2017, 122, 723–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bucchi, L.; Belli, P.; Benelli, E.; Bernardi, D.; Brancato, B.; Calabrese, M.; Carbonaro, L.A.; Caumo, F.; Cavallo-Marincola, B.; Clauser, P.; et al. Recommendations for breast imaging follow-up of women with a previous history of breast cancer: Position paper from the Italian Group for Mammography Screening (GISMa) and the Italian College of Breast Radiologists (ICBR) by SIRM. Radiol. Med. 2016, 121, 891–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Losurdo, L.; Fanizzi, A.; Basile, T.M.A.; Bellotti, R.; Bottigli, U.; Dentamaro, R.; Didonna, V.; Lorusso, V.; Massafra, R.; Tamborra, P.; et al. Radiomics Analysis on Contrast-Enhanced Spectral Mammography Images for Breast Cancer Diagnosis: A Pilot Study. Entropy 2019, 21, 1110. [Google Scholar] [CrossRef] [Green Version]
- Fanizzi, A.; Losurdo, L.; Basile, T.M.A.; Bellotti, R.; Bottigli, U.; Delogu, P.; Diacono, D.; Didonna, V.; Fausto, A.; Lombardi, A.; et al. Fully Automated Support System for Diagnosis of Breast Cancer in Contrast-Enhanced Spectral Mammography Images. J. Clin. Med. 2019, 8, 891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Forgia, D.; Fanizzi, A.; Campobasso, F.; Bellotti, R.; Didonna, V.; Lorusso, V.; Moschetta, M.; Massafra, R.; Tamborra, P.; Tangaro, S.; et al. Radiomic Analysis in Contrast-Enhanced Spectral Mammography for Predicting Breast Cancer Histological Outcome. Diagnostics 2020, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Marino, M.A.; Leithner, D.; Sung, J.; Avendano, D.; Morris, E.A.; Pinker, K.; Jochelson, M.S. Radiomics for Tumor Characterization in Breast Cancer Patients: A Feasibility Study Comparing Contrast-Enhanced Mammography and Magnetic Resonance Imaging. Diagnostics 2020, 10, 492. [Google Scholar] [CrossRef]
- Jiang, T.; Song, J.; Wang, X.; Niu, S.; Zhao, N.; Dong, Y.; Wang, X.; Luo, Y.; Jiang, X. Intratumoral and Peritumoral Analysis of Mammography, Tomosynthesis, and Multiparametric MRI for Predicting Ki-67 Level in Breast Cancer: A Radiomics-Based Study. Mol. Imaging Biol. 2021, 1–10. [Google Scholar] [CrossRef]
- Zhao, Y.-F.; Chen, Z.; Zhang, Y.; Zhou, J.; Chen, J.-H.; Lee, K.E.; Combs, F.J.; Parajuli, R.; Mehta, R.S.; Wang, M.; et al. Diagnosis of Breast Cancer Using Radiomics Models Built Based on Dynamic Contrast Enhanced MRI Combined with Mammography. Front. Oncol. 2021, 11, 774248. [Google Scholar] [CrossRef]
- Niu, S.; Wang, X.; Zhao, N.; Liu, G.; Kan, Y.; Dong, Y.; Cui, E.-N.; Luo, Y.; Yu, T.; Jiang, X. Radiomic Evaluations of the Diagnostic Performance of DM, DBT, DCE MRI, DWI, and Their Combination for the Diagnosisof Breast Cancer. Front. Oncol. 2021, 11, 725922. [Google Scholar] [CrossRef]


| Benign (31 Lesions) | Number | Percentage Value (%) |
| Fibrosis | 6 | 19.35 |
| Ductal hyperplasia | 8 | 25.81 |
| Fibroadenoma | 9 | 29.03 |
| Dysplasia | 4 | 12.90 |
| Adenosis | 4 | 12.90 |
| Malignant (48 Lesions) | Number | Percentage Value (%) |
| Infiltrating lobular carcinoma | 7 | 14.58 |
| Infiltrating ductal carcinoma | 30 | 62.50 |
| Ductal carcinoma in situ | 9 | 18.75 |
| Tubular Carcinoma | 2 | 4.17 |
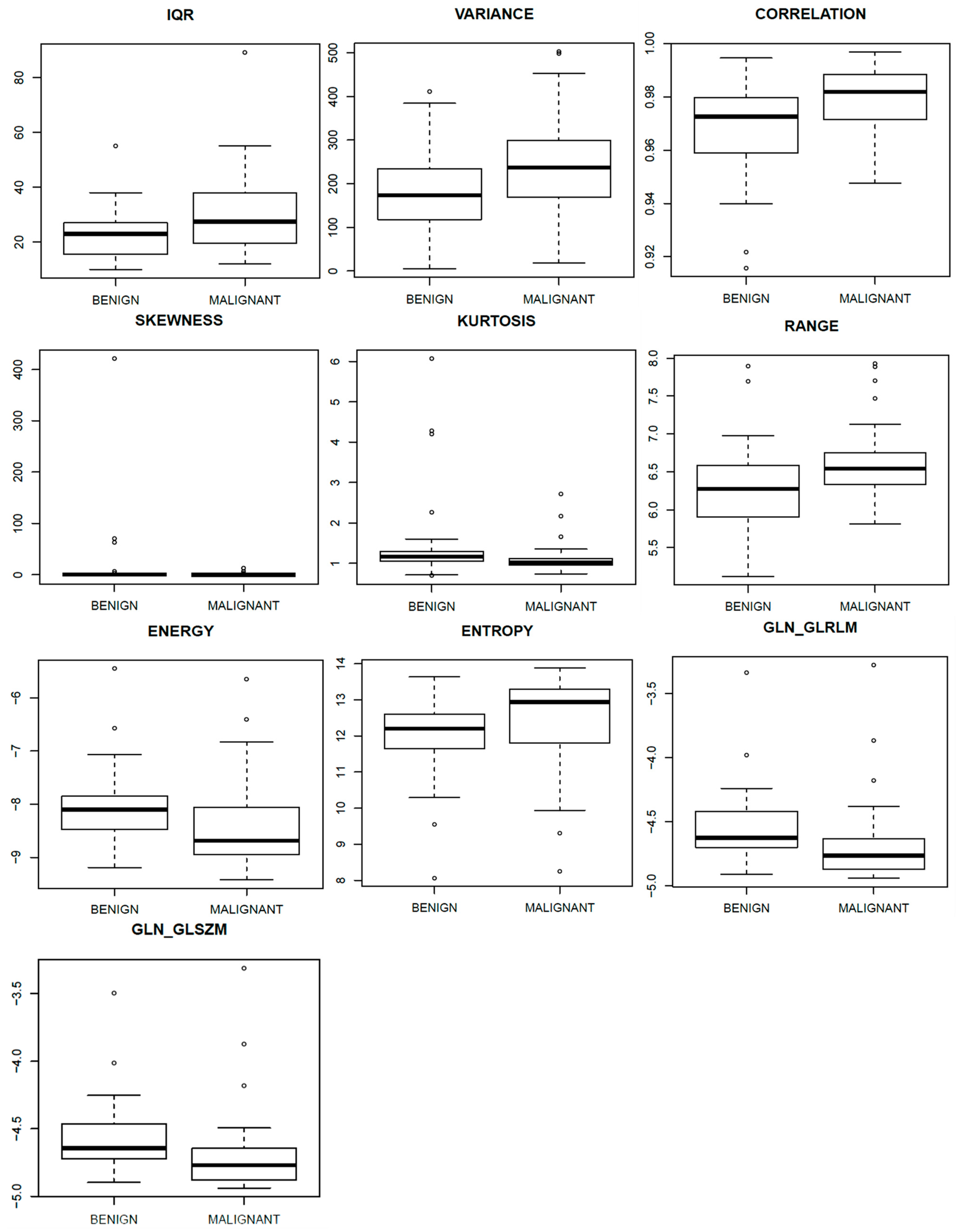
| Mammography Projection | Textural Parameters | AUC Values | p-Value |
|---|---|---|---|
| CC-view | IQR | 0.67 | 0.01 |
| Variance | 0.68 | 0.01 | |
| Correlation | 0.69 | 0.000 | |
| MLO view | Kurtosis | 0.71 | 0.000 |
| Skewness | 0.71 | 0.000 | |
| Magnetic Resonance Images | Textural Parameters | AUC Values | p-Value |
| Range | 0.72 | 0.001 | |
| Energy | 0.72 | 0.001 | |
| Entropy | 0.70 | 0.003 | |
| GLN_GLRLM | 0.72 | 0.001 | |
| GLN_GLSZM | 0.70 | 0.002 |
| Classifier | Cross-Validation | ACC | SENS | SPEC | PPV | NPV | AUC |
|---|---|---|---|---|---|---|---|
| CEM–CCview | |||||||
| Performance of classifiers trained with balanced data (with ADASYN function) and a subset of 34 textural features (AUC ≥ 0.60). | |||||||
| TREE | 10-fold CV | 0.74 | 0.74 | 0.78 | 0.76 | 0.74 | 0.73 |
| Performance of classifiers trained with balanced data (with SASYNO function) and a subset of 4 robust textural features (by LASSO and λminMSE). | |||||||
| LDA | 10-fold CV | 0.71 | 0.71 | 0.71 | 0.71 | 0.71 | 0.76 |
| LOOCV | 0.71 | 0.71 | 0.71 | 0.71 | 0.71 | 0.75 | |
| SVM | 10-fold CV | 0.71 | 0.71 | 0.71 | 0.71 | 0.71 | 0.77 |
| Performance of classifiers trained with balanced data (with SASYNO function) and a subset of 3 robust textural features (by LASSO and λ1SE). | |||||||
| LDA | 10-fold CV | 0.71 | 0.71 | 0.71 | 0.71 | 0.71 | 0.76 |
| LOOCV | 0.71 | 0.71 | 0.71 | 0.71 | 0.71 | 0.75 | |
| NNET | 10-fold CV | 0.70 | 0.71 | 0.69 | 0.69 | 0.70 | 0.74 |
| LOOCV | 0.70 | 0.73 | 0.67 | 0.69 | 0.71 | 0.74 | |
| SVM | 10-fold CV | 0.71 | 0.71 | 0.71 | 0.71 | 0.71 | 0.75 |
| LOOCV | 0.72 | 0.73 | 0.71 | 0.71 | 0.72 | 0.76 | |
| CEM–early MLO view | |||||||
| Performance of classifiers trained with balanced data (with ADASYN function) and all 48 textural features | |||||||
| LDA | 10-fold CV | 0.76 | 0.65 | 0.87 | 0.82 | 0.74 | 0.73 |
| LOOCV | 0.75 | 0.60 | 0.87 | 0.81 | 0.72 | 0.71 | |
| Performance of classifiers trained with balanced data (with ADASYN function) and a subset of 7 robust textural features (by LASSO and λminMSE). | |||||||
| LDA | 10-fold CV | 0.66 | 0.54 | 0.75 | 0.65 | 0.65 | 0.72 |
| LOOCV | 0.66 | 0.56 | 0.75 | 0.66 | 0.66 | 0.7 | |
| Performance of classifiers trained with balanced data (with ADASYN function) and a subset of 14 robust textural features (by LASSO and λ1SE). | |||||||
| LDA | 10-fold CV | 0.62 | 0.52 | 0.69 | 0.60 | 0.62 | 0.71 |
| LOOCV | 0.66 | 0.56 | 0.75 | 0.66 | 0.66 | 0.7 | |
| CEM–late MLO view | |||||||
| Performance of classifiers trained with balanced data (with ADASYN function) and all 48 textural features | |||||||
| LDA | 10-fold CV | 0.78 | 0.71 | 0.84 | 0.79 | 0.77 | 0.78 |
| LOOCV | 0.78 | 0.69 | 0.86 | 0.80 | 0.76 | 0.77 | |
| Performance of classifiers trained with balanced data (with ADASYN function) and a subset of 17 robust textural features (by LASSO and λminMSE). | |||||||
| LDA | 10-fold CV | 0.75 | 0.71 | 0.77 | 0.72 | 0.75 | 0.8 |
| LOOCV | 0.73 | 0.71 | 0.75 | 0.71 | 0.75 | 0.8 | |
| NNET | 10-fold CV | 0.72 | 0.65 | 0.77 | 0.70 | 0.72 | 0.78 |
| LOOCV | 0.72 | 0.69 | 0.75 | 0.70 | 0.74 | 0.72 | |
| Performance of classifiers trained with balanced data (with ADASYN function) and a subset of 14 robust textural features (by LASSO and λ1SE). | |||||||
| LDA | 10-fold CV | 0.71 | 0.69 | 0.71 | 0.67 | 0.73 | 0.78 |
| LOOCV | 0.70 | 0.69 | 0.71 | 0.67 | 0.73 | 0.78 | |
| NNET | 10-fold CV | 0.71 | 0.67 | 0.75 | 0.70 | 0.72 | 0.74 |
| LOOCV | 0.74 | 0.69 | 0.79 | 0.73 | 0.75 | 0.74 | |
| CEM–CC + early MLO + late view | |||||||
| Performance of classifiers trained with balanced data (with ADASYN function) and a subset of 15 robust textural features (by LASSO and λminMSE). | |||||||
| LDA | 10-fold CV | 0.75 | 0.69 | 0.81 | 0.77 | 0.75 | 0.82 |
| LOOCV | 0.76 | 0.71 | 0.81 | 0.77 | 0.76 | 0.81 | |
| NNET | 10-fold CV | 0.77 | 0.75 | 0.80 | 0.77 | 0.78 | 0.79 |
| LOOCV | 0.79 | 0.75 | 0.81 | 0.78 | 0.79 | 0.81 | |
| SVM | 10-fold CV | 0.72 | 0.73 | 0.70 | 0.69 | 0.75 | 0.79 |
| LOOCV | 0.76 | 0.73 | 0.80 | 0.76 | 0.77 | 0.81 | |
| Performance of classifiers trained with balanced data (with ADASYN function) and a subset of 8 robust textural features (by LASSO and λ1SE). | |||||||
| NNET | 10-fold CV | 0.72 | 0.73 | 0.72 | 0.70 | 0.75 | 0.78 |
| DCE-MRI | |||||||
| Performance of classifiers trained with balanced data (with ADASYN function) and all 48 textural features | |||||||
| LDA | 10-fold CV | 0.73 | 0.69 | 0.77 | 0.73 | 0.73 | 0.71 |
| LOOCV | 0.70 | 0.65 | 0.75 | 0.70 | 0.70 | 0.7 | |
| Performance of classifiers trained with balanced data (with SASYNO function) and a subset of 15 robust textural features (by LASSO and λminMSE). | |||||||
| SVM | 10-fold CV | 0.74 | 0.73 | 0.75 | 0.74 | 0.73 | 0.72 |
| LOOCV | 0.70 | 0.69 | 0.71 | 0.70 | 0.69 | 0.71 | |
| Classifier | Cross-Validation | ACC | SENS | SPEC | PPV | NPV | AUC |
|---|---|---|---|---|---|---|---|
| Performance for classifiers trained with balanced data (with ADASYN function) and a subset of 18 robust textural features (by LASSO and λminMSE). | |||||||
| LDA | 10-fold CV | 0.84 | 0.73 | 0.92 | 0.90 | 0.79 | 0.88 |
| LOOCV | 0.80 | 0.71 | 0.88 | 0.85 | 0.77 | 0.87 | |
| SVM | 10-fold CV | 0.84 | 0.81 | 0.87 | 0.85 | 0.83 | 0.86 |
| LOOCV | 0.83 | 0.79 | 0.87 | 0.84 | 0.82 | 0.86 | |
| Performance for classifiers trained with balanced data (with SASYNO function) and a subset of 3 robust textural features (by LASSO and λminMSE). | |||||||
| LDA | 10-fold CV | 0.79 | 0.79 | 0.79 | 0.79 | 0.79 | 0.88 |
| LOOCV | 0.79 | 0.79 | 0.79 | 0.79 | 0.79 | 0.89 | |
| SVM | 10-fold CV | 0.80 | 0.79 | 0.79 | 0.79 | 0.79 | 0.86 |
| LOOCV | 0.79 | 0.77 | 0.81 | 0.80 | 0.78 | 0.87 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fusco, R.; Di Bernardo, E.; Piccirillo, A.; Rubulotta, M.R.; Petrosino, T.; Barretta, M.L.; Mattace Raso, M.; Vallone, P.; Raiano, C.; Di Giacomo, R.; et al. Radiomic and Artificial Intelligence Analysis with Textural Metrics Extracted by Contrast-Enhanced Mammography and Dynamic Contrast Magnetic Resonance Imaging to Detect Breast Malignant Lesions. Curr. Oncol. 2022, 29, 1947-1966. https://doi.org/10.3390/curroncol29030159
Fusco R, Di Bernardo E, Piccirillo A, Rubulotta MR, Petrosino T, Barretta ML, Mattace Raso M, Vallone P, Raiano C, Di Giacomo R, et al. Radiomic and Artificial Intelligence Analysis with Textural Metrics Extracted by Contrast-Enhanced Mammography and Dynamic Contrast Magnetic Resonance Imaging to Detect Breast Malignant Lesions. Current Oncology. 2022; 29(3):1947-1966. https://doi.org/10.3390/curroncol29030159
Chicago/Turabian StyleFusco, Roberta, Elio Di Bernardo, Adele Piccirillo, Maria Rosaria Rubulotta, Teresa Petrosino, Maria Luisa Barretta, Mauro Mattace Raso, Paolo Vallone, Concetta Raiano, Raimondo Di Giacomo, and et al. 2022. "Radiomic and Artificial Intelligence Analysis with Textural Metrics Extracted by Contrast-Enhanced Mammography and Dynamic Contrast Magnetic Resonance Imaging to Detect Breast Malignant Lesions" Current Oncology 29, no. 3: 1947-1966. https://doi.org/10.3390/curroncol29030159
APA StyleFusco, R., Di Bernardo, E., Piccirillo, A., Rubulotta, M. R., Petrosino, T., Barretta, M. L., Mattace Raso, M., Vallone, P., Raiano, C., Di Giacomo, R., Siani, C., Avino, F., Scognamiglio, G., Di Bonito, M., Granata, V., & Petrillo, A. (2022). Radiomic and Artificial Intelligence Analysis with Textural Metrics Extracted by Contrast-Enhanced Mammography and Dynamic Contrast Magnetic Resonance Imaging to Detect Breast Malignant Lesions. Current Oncology, 29(3), 1947-1966. https://doi.org/10.3390/curroncol29030159





