Clinicopathological Features of Non-Small Cell Lung Carcinoma with BRAF Mutation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Data
2.2. NGS Analysis
2.3. IHC
2.4. Statistical Analysis
3. Results
3.1. Case Series
3.2. Statistical Analysis
4. Discussion
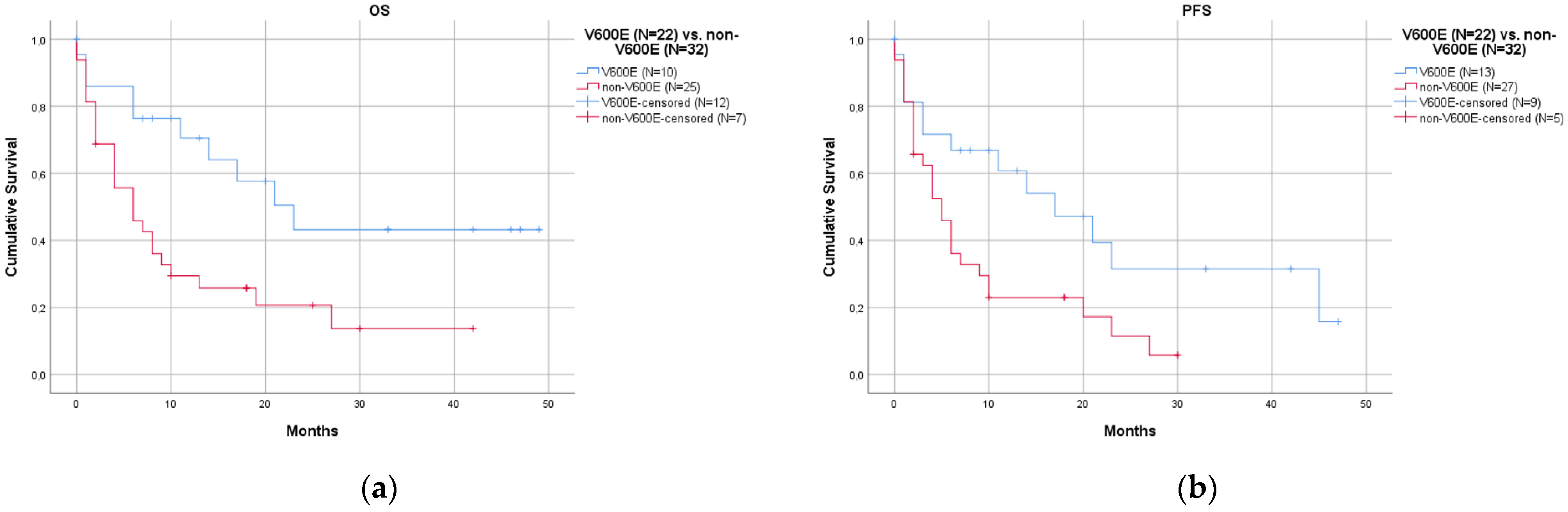
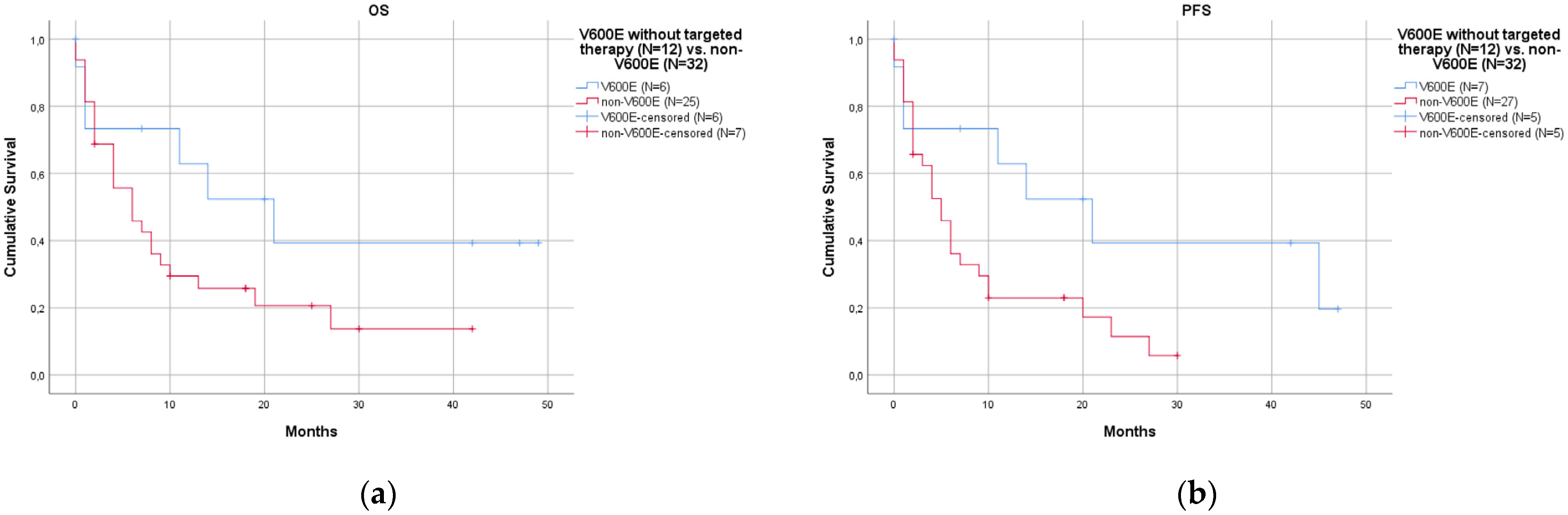
- Patients with the V600E mutation had a better prognosis than those with non-V600E mutations.
- Associated co-mutations were not rare (35.0%) and did not affect prognosis.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ibodeng, G.; Uche, I.N.; Mokua, R.; Galo, M.; Odigwe, B.; Galeas, J.N.; Dasgupta, S. A Snapshot of Lung Cancer: Where Are We Now?—A Narrative Review. Ann. Transl. Med. 2023, 11, 261. [Google Scholar] [CrossRef]
- Singh, N.; Temin, S.; Baker, S.; Blanchard, E.; Brahmer, J.R.; Celano, P.; Duma, N.; Ellis, P.M.; Elkins, I.B.; Haddad, R.Y.; et al. Therapy for Stage IV Non–Small-Cell Lung Cancer With Driver Alterations: ASCO Living Guideline. J. Clin. Oncol. 2022, 40, 3310–3322. [Google Scholar] [CrossRef] [PubMed]
- Proietti, I.; Skroza, N.; Michelini, S.; Mambrin, A.; Balduzzi, V.; Bernardini, N.; Marchesiello, A.; Tolino, E.; Volpe, S.; Maddalena, P.; et al. BRAF Inhibitors: Molecular Targeting and Immunomodulatory Actions. Cancers 2020, 12, 1823. [Google Scholar] [CrossRef] [PubMed]
- Crispo, F.; Notarangelo, T.; Pietrafesa, M.; Lettini, G.; Storto, G.; Sgambato, A.; Maddalena, F.; Landriscina, M. BRAF Inhibitors in Thyroid Cancer: Clinical Impact, Mechanisms of Resistance and Future Perspectives. Cancers 2019, 11, 1388. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO Consensus Guidelines for the Management of Patients with Metastatic Colorectal Cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef] [PubMed]
- Grassi, E.; Corbelli, J.; Papiani, G.; Barbera, M.A.; Gazzaneo, F.; Tamberi, S. Current Therapeutic Strategies in BRAF-Mutant Metastatic Colorectal Cancer. Front. Oncol. 2021, 11, 601722. [Google Scholar] [CrossRef]
- Malapelle, U.; Rossi, G.; Pisapia, P.; Barberis, M.; Buttitta, F.; Castiglione, F.; Cecere, F.L.; Grimaldi, A.M.; Iaccarino, A.; Marchetti, A.; et al. BRAF as a Positive Predictive Biomarker: Focus on Lung Cancer and Melanoma Patients. Crit. Rev. Oncol./Hematol. 2020, 156, 103118. [Google Scholar] [CrossRef]
- Guaitoli, G.; Zullo, L.; Tiseo, M.; Dankner, M.; Rose, A.A.; Facchinetti, F. Non-Small-Cell Lung Cancer: How to Manage BRAF-Mutated Disease. Drugs Context 2023, 12, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Puri, M.; Gawri, K.; Dawar, R. Therapeutic Strategies for BRAF Mutation in Non-Small Cell Lung Cancer: A Review. Front. Oncol. 2023, 13, 1141876. [Google Scholar] [CrossRef]
- Melosky, B.; Wheatley-Price, P.; Juergens, R.A.; Sacher, A.; Leighl, N.B.; Tsao, M.-S.; Cheema, P.; Snow, S.; Liu, G.; Card, P.B.; et al. The Rapidly Evolving Landscape of Novel Targeted Therapies in Advanced Non-Small Cell Lung Cancer. Lung Cancer 2021, 160, 136–151. [Google Scholar] [CrossRef]
- Owsley, J.; Stein, M.K.; Porter, J.; In, G.K.; Salem, M.; O’Day, S.; Elliott, A.; Poorman, K.; Gibney, G.; VanderWalde, A. Prevalence of Class I–III BRAF Mutations among 114,662 Cancer Patients in a Large Genomic Database. Exp. Biol. Med. 2021, 246, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Villaruz, L.C.; Socinski, M.A.; Abberbock, S.; Berry, L.D.; Johnson, B.E.; Kwiatkowski, D.J.; Iafrate, A.J.; Varella-Garcia, M.; Franklin, W.A.; Camidge, D.R.; et al. Clinicopathologic Features and Outcomes of Patients with Lung Adenocarcinomas Harboring BRAF Mutations in the Lung Cancer Mutation Consortium: BRAF Mutations in Lung Adenocarcinomas. Cancer 2015, 121, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.; Felicioni, L.; Malatesta, S.; Grazia Sciarrotta, M.; Guetti, L.; Chella, A.; Viola, P.; Pullara, C.; Mucilli, F.; Buttitta, F. Clinical Features and Outcome of Patients With Non–Small-Cell Lung Cancer Harboring BRAF Mutations. J. Clin. Oncol. 2011, 29, 3574–3579. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhang, L.-Q.; Huang, J.-F.; Liu, K.; Chuai, Z.-R.; Yang, Z.; Wang, Y.-X.; Shi, D.-C.; Liu, Q.; Huang, Q.; et al. BRAF Mutations in Patients with Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2014, 9, e101354. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M. A Narrative Review of BRAF Alterations in Human Tumors: Diagnostic and Predictive Implications. Precis. Cancer Med. 2020, 3, 26. [Google Scholar] [CrossRef]
- Nguyen-Ngoc, T.; Bouchaab, H.; Adjei, A.A.; Peters, S. BRAF Alterations as Therapeutic Targets in Non–Small-Cell Lung Cancer. J. Thorac. Oncol. 2015, 10, 1396–1403. [Google Scholar] [CrossRef]
- Dankner, M.; Wang, Y.; Fazelzad, R.; Johnson, B.; Nebhan, C.A.; Dagogo-Jack, I.; Myall, N.J.; Richtig, G.; Bracht, J.W.P.; Gerlinger, M.; et al. Clinical Activity of Mitogen-Activated Protein Kinase–Targeted Therapies in Patients With Non–V600 BRAF-Mutant Tumors. JCO Precis. Oncol. 2022, 6, e2200107. [Google Scholar] [CrossRef]
- Leonetti, A.; Facchinetti, F.; Rossi, G.; Minari, R.; Conti, A.; Friboulet, L.; Tiseo, M.; Planchard, D. BRAF in Non-Small Cell Lung Cancer (NSCLC): Pickaxing Another Brick in the Wall. Cancer Treat. Rev. 2018, 66, 82–94. [Google Scholar] [CrossRef]
- Mazieres, J.; Cropet, C.; Montané, L.; Barlesi, F.; Souquet, P.J.; Quantin, X.; Dubos-Arvis, C.; Otto, J.; Favier, L.; Avrillon, V.; et al. Vemurafenib in Non-Small-Cell Lung Cancer Patients with BRAFV600 and BRAFnonV600 Mutations. Ann. Oncol. 2020, 31, 289–294. [Google Scholar] [CrossRef]
- Odogwu, L.; Mathieu, L.; Blumenthal, G.; Larkins, E.; Goldberg, K.B.; Griffin, N.; Bijwaard, K.; Lee, E.Y.; Philip, R.; Jiang, X.; et al. FDA Approval Summary: Dabrafenib and Trametinib for the Treatment of Metastatic Non-Small Cell Lung Cancers Harboring BRAF V600E Mutations. Oncologist 2018, 23, 740–745. [Google Scholar] [CrossRef]
- Noeparast, A.; Teugels, E.; Giron, P.; Verschelden, G.; De Brakeleer, S.; Decoster, L.; De Grève, J. Non-V600 BRAF Mutations Recurrently Found in Lung Cancer Predict Sensitivity to the Combination of Trametinib and Dabrafenib. Oncotarget 2017, 8, 60094–60108. [Google Scholar] [CrossRef]
- Abuali, I.; Lee, C.-S.; Seetharamu, N. A Narrative Review of the Management of BRAF Non-V600E Mutated Metastatic Non-Small Cell Lung Cancer. Precis. Cancer Med. 2022, 5, 13. [Google Scholar] [CrossRef]
- Sakai, T.; Matsumoto, S.; Ueda, Y.; Shibata, Y.; Ikeda, T.; Nakamura, A.; Kodani, M.; Ohashi, K.; Furuya, N.; Izumi, H.; et al. Clinico-Genomic Features and Targetable Mutations in Non-Small Cell Lung Cancers Harboring BRAF Non-V600E Mutations: A Multi-Institutional Genomic Screening Study (LC-SCRUM-Asia). J. Thorac. Oncol. 2023, 18, 1538–1549. [Google Scholar] [CrossRef] [PubMed]
- Cardarella, S.; Ogino, A.; Nishino, M.; Butaney, M.; Shen, J.; Lydon, C.; Yeap, B.Y.; Sholl, L.M.; Johnson, B.E.; Jänne, P.A. Clinical, Pathologic, and Biologic Features Associated with BRAF Mutations in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2013, 19, 4532–4540. [Google Scholar] [CrossRef]
- Sheikine, Y.; Pavlick, D.; Klempner, S.J.; Trabucco, S.E.; Chung, J.H.; Rosenzweig, M.; Wang, K.; Velcheti, V.; Frampton, G.M.; Peled, N.; et al. BRAF in Lung Cancers: Analysis of Patient Cases Reveals Recurrent BRAF Mutations, Fusions, Kinase Duplications, and Concurrent Alterations. JCO Precis. Oncol. 2018, 2, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Ambrosini-Spaltro, A.; Dubini, A.; Pieri, F.; Ravaglia, C.; Delmonte, A.; Poletti, V. PD-L1 Expression in NSCLC: Role of Cell Blocks and Concordance between Samples. Diagn. Cytopathol. 2021, 49, 303–310. [Google Scholar] [CrossRef]
- Wong, D.W.-S.; Leung, E.L.-H.; So, K.K.-T.; Tam, I.Y.-S.; Sihoe, A.D.-L.; Cheng, L.-C.; Ho, K.-K.; Au, J.S.-K.; Chung, L.-P.; Pik Wong, M.; et al. The EML4-ALK Fusion Gene Is Involved in Various Histologic Types of Lung Cancers from Nonsmokers with Wild-Type EGFR and KRAS. Cancer 2009, 115, 1723–1733. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Nambirajan, A.; Borczuk, A.; Chen, G.; Minami, Y.; Moreira, A.L.; Motoi, N.; Papotti, M.; Rekhtman, N.; Russell, P.A.; et al. Immunocytochemistry for Predictive Biomarker Testing in Lung Cancer Cytology. Cancer Cytopathol. 2019, 127, 325–339. [Google Scholar] [CrossRef]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J.; et al. Pembrolizumab versus Docetaxel for Previously Treated, PD-L1-Positive, Advanced Non-Small-Cell Lung Cancer (KEYNOTE-010): A Randomised Controlled Trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Perrone, F.; Mazzaschi, G.; Minari, R.; Verzè, M.; Azzoni, C.; Bottarelli, L.; Nizzoli, R.; Pluchino, M.; Altimari, A.; Gruppioni, E.; et al. Multicenter Observational Study on Metastatic Non-Small Cell Lung Cancer Harboring BRAF Mutations: Focus on Clinical Characteristics and Treatment Outcome of V600E and Non-V600E Subgroups. Cancers 2022, 14, 2019. [Google Scholar] [CrossRef]
- O’Leary, C.G.; Andelkovic, V.; Ladwa, R.; Pavlakis, N.; Zhou, C.; Hirsch, F.; Richard, D.; O’Byrne, K. Targeting BRAF Mutations in Non-Small Cell Lung Cancer. Transl. Lung Cancer Res. 2019, 8, 1119–1124. [Google Scholar] [CrossRef]
- Jiang, L.; Yu, H.; Ness, S.; Mao, P.; Guo, F.; Tang, J.; Guo, Y. Comprehensive Analysis of Co-Mutations Identifies Cooperating Mechanisms of Tumorigenesis. Cancers 2022, 14, 415. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Heymach, J.V. Co-Occurring Genomic Alterations in Non-Small-Cell Lung Cancer Biology and Therapy. Nat. Rev. Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, J.; Xu, Y.; Cai, S.; Li, T.; Wang, G.; Li, C.; Zhao, L.; Hu, Y. Co-Occurring Genomic Alterations and Immunotherapy Efficacy in NSCLC. NPJ Precis. Oncol. 2022, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Canale, M.; Andrikou, K.; Priano, I.; Cravero, P.; Pasini, L.; Urbini, M.; Delmonte, A.; Crinò, L.; Bronte, G.; Ulivi, P. The Role of TP53 Mutations in EGFR-Mutated Non-Small-Cell Lung Cancer: Clinical Significance and Implications for Therapy. Cancers 2022, 14, 1143. [Google Scholar] [CrossRef]
- Stockhammer, P.; Grant, M.; Wurtz, A.; Foggetti, G.; Expósito, F.; Gu, J.; Zhao, H.; Choi, J.; Chung, S.; Li, F.; et al. Co-Occurring Alterations in Multiple Tumor Suppressor Genes Are Associated with Worse Outcomes in Patients with EGFR-Mutant Lung Cancer. J. Thorac. Oncol. 2023. [Google Scholar] [CrossRef]
- Dudnik, E.; Peled, N.; Nechushtan, H.; Wollner, M.; Onn, A.; Agbarya, A.; Moskovitz, M.; Keren, S.; Popovits-Hadari, N.; Urban, D.; et al. BRAF Mutant Lung Cancer: Programmed Death Ligand 1 Expression, Tumor Mutational Burden, Microsatellite Instability Status, and Response to Immune Check-Point Inhibitors. J. Thorac. Oncol. 2018, 13, 1128–1137. [Google Scholar] [CrossRef]
- Gibson, A.J.W.; Pabani, A.; Dean, M.L.; Martos, G.; Cheung, W.Y.; Navani, V. Real-World Treatment Patterns and Effectiveness of Targeted and Immune Checkpoint Inhibitor-Based Systemic Therapy in BRAF Mutation-Positive NSCLC. JTO Clin. Res. Rep. 2023, 4, 100460. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, L.; Zhao, C.; Zhou, F.; Jiang, T.; Guo, H.; Shi, J.; Chen, P.; Tang, Z.; Mao, S.; et al. Efficacy of Immune Checkpoint Inhibitors in Advanced Non-Small Cell Lung Cancer Harboring BRAF Mutations. Transl. Lung Cancer Res. 2023, 12, 219–229. [Google Scholar] [CrossRef]
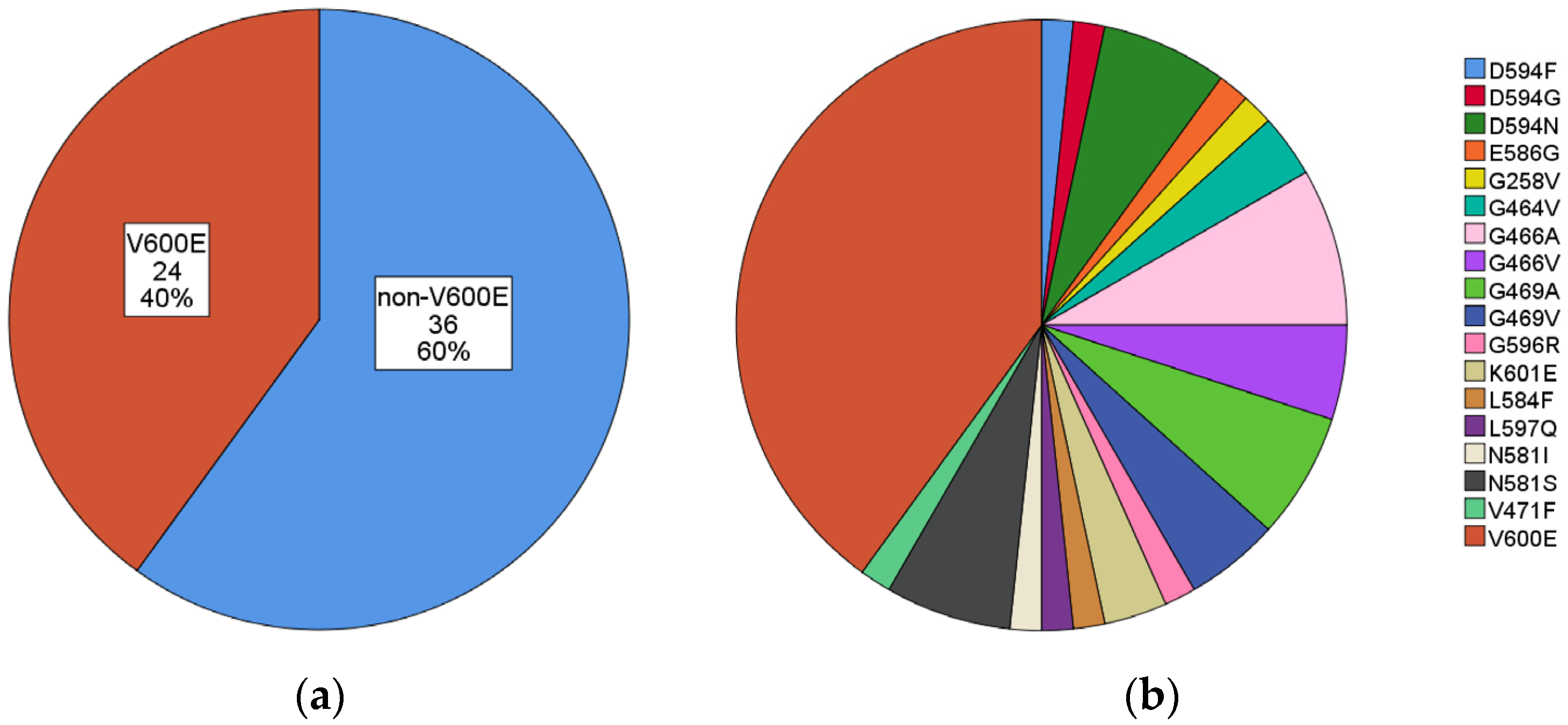

| Mutation | N. (%) | Mutation | N. (%) |
|---|---|---|---|
| V600E | 24 (40.0) | V600E | 24 (40.0) |
| non-V600E | 36 (60.0) | G466A | 5 (8.3) |
| D594N | 4 (6.7) | ||
| G469A | 4 (6.7) | ||
| N581S | 4 (6.7) | ||
| G466V | 3 (5.0) | ||
| G469V | 3 (5.0) | ||
| G464V | 2 (3.3) | ||
| K601E | 2 (3.3) | ||
| D594F | 1 (1.7) | ||
| D594G | 1 (1.7) | ||
| E586G | 1 (1.7) | ||
| G258V | 1 (1.7) | ||
| G596R | 1 (1.7) | ||
| L584F | 1 (1.7) | ||
| L597Q | 1 (1.7) | ||
| N581I | 1 (1.7) | ||
| V471F | 1 (1.7) | ||
| Total | 60 |
| Gene | N. (%) | N. (%) | |
|---|---|---|---|
| KRAS | 4 (19.0) | ||
| KRAS, EGFR | 1 (4.8) | ||
| KRAS, MET | 1 (4.8) | ||
| KRAS, PAM2K1 | 1 (4.8) | ||
| KRAS, PIK3CA | 1 (4.8) | ||
| KRAS (all) | 8 (38.1) | ||
| PIK3CA | 4 (19.0) | ||
| PIK3CA (all) | 5 (23.8) | ||
| IDH1 | 2 (9.5) | ||
| FGFR3 | 2 (9.5) | ||
| EGFR | 1 (4.8) | ||
| EGFR (all) | 2 (9.5) | ||
| ESR1 | 1 (4.8) | ||
| GNAQ | 1 (4.8) | ||
| CDH4 | 1 (4.8) | ||
| AKT1, MTOR | 1 (4.8) | ||
| Total | 21 |
| Overall | V600E | Non-V600E | p-Value | with Co-Mutations | without Co-Mutations | p-Value | ||
|---|---|---|---|---|---|---|---|---|
| N. | 60 | 24/60 (40.0%) | 36/60 (60.0%) | 21/60 (35.0%) | 39/60 (65.0%) | |||
| Age (mean ± SD) | 71.2 ± 9.9 | 71.3 ± 11.1 | 71.1 ± 9.1 | 0.92 | 70.0 ± 10.8 | 71.8 ± 9.5 | 0.74 | |
| Sex | male | 37/60 (61.7%) | 12/24 (50.0%) | 25/36 (69.4%) | 0.13 | 15/21 (71.4%) | 22/39 (56.4%) | 0.26 |
| female | 23/60 (38.3%) | 12/24 (50.0%) | 11/36 (30.6%) | 6/21 (28.6%) | 17/39 (43.6%) | |||
| Histotype | adenocarcinoma | 55/60 (91.7%) | 24/24 (100.0%) | 31/36 (86.1%) | 0.6 | 18/21 (85.7%) | 37/39 (94.9%) | 0.21 |
| squamous cell carcinoma | 1/60 (1.7%) | 0/24 | 1/36 (2.8%) | 0/21 | 1/39 (2.6%) | |||
| NSCLC, NOS and other histotypes | 4 (6.7%) | 0/24 | 4/36 (11.1%) | 3/21 (14.3%) | 1/39 (2.6%) | |||
| Material | cytological | 19/60 (31.7%) | 8/24 (33.3%) | 11/36 (30.6%) | 0.76 | 5/21 (23.8%) | 14/39 (35.9%) | 0.97 |
| bioptic | 32/60 (53.3%) | 11/24 (45.8%) | 21/36 (58.3%) | 15/21 (71.4%) | 17/39 (43.6%) | |||
| surgical | 9/60 (15.0%) | 5/24 (20.8%) | 4/36 (11.1%) | 1/21 (4.8%) | 8/39 (20.5%) | |||
| Smoking status (data available in 35 cases) | non-smokers | 2/35 (5.7%) | 2/14 (14.3%) | 0/21 | 0.86 | 1/13 (7.7%) | 1/22 (4.5%) | 0.07 |
| current smokers | 11/35 (31.4%) | 3/14 (21.4%) | 8/21 (38.1%) | 1/13 (7.7%) | 10/22 (45.5%) | |||
| ex-smokers | 22/35 (62.9%) | 9/14 (64.3%) | 13/21 (61.9%) | 11/13 (84.6%) | 11/22 (50.0%) | |||
| non-smokers | 2/35 (5.7%) | 2/14 (14.3%) | 0/21 | 0.08 | 1/13 (7.7%) | 1/22 (4.5%) | 0.7 | |
| smokers (current and ex-) | 33/35 (94.3%) | 12/14 (85.7%) | 21/21 (100.0%) | 12/13 (92.3%) | 21/22 (95.5%) | |||
| PD-L1 (total: 53) | TPS < 1% | 7/53 (13.2%) | 3/21 (14.3%) | 4/32 (12.5%) | 0.81 | 4/19 (21.1%) | 3/34 (8.8%) | 0.79 |
| TPS 1–49% | 25/53 (47.2%) | 9/21 (42.9%) | 16/32 (50.0%) | 7/19 (36.8%) | 18/34 (52.9%) | |||
| TPS ≥ 50% | 21/53 (39.6%) | 9/21 (42.9%) | 12/32 (37.5%) | 8/19 (42.1%) | 13/34 (38.2%) | |||
| ALK (total: 42) | negative | 42 (100%) | 17 | 25 | 16 | 26 | ||
| positive | 0 | |||||||
| ROS1 (total: 42) | negative | 42 (100%) | 17 | 25 | 16 | 26 | ||
| positive | 0 | |||||||
| Co-mutations | 21/60 (35.0%) | 6/24 (25.0%) | 15/36 (41.7%) | 0.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ambrosini-Spaltro, A.; Rengucci, C.; Capelli, L.; Chiadini, E.; Calistri, D.; Bennati, C.; Cravero, P.; Limarzi, F.; Nosseir, S.; Panzacchi, R.; et al. Clinicopathological Features of Non-Small Cell Lung Carcinoma with BRAF Mutation. Curr. Oncol. 2023, 30, 10019-10032. https://doi.org/10.3390/curroncol30110728
Ambrosini-Spaltro A, Rengucci C, Capelli L, Chiadini E, Calistri D, Bennati C, Cravero P, Limarzi F, Nosseir S, Panzacchi R, et al. Clinicopathological Features of Non-Small Cell Lung Carcinoma with BRAF Mutation. Current Oncology. 2023; 30(11):10019-10032. https://doi.org/10.3390/curroncol30110728
Chicago/Turabian StyleAmbrosini-Spaltro, Andrea, Claudia Rengucci, Laura Capelli, Elisa Chiadini, Daniele Calistri, Chiara Bennati, Paola Cravero, Francesco Limarzi, Sofia Nosseir, Riccardo Panzacchi, and et al. 2023. "Clinicopathological Features of Non-Small Cell Lung Carcinoma with BRAF Mutation" Current Oncology 30, no. 11: 10019-10032. https://doi.org/10.3390/curroncol30110728
APA StyleAmbrosini-Spaltro, A., Rengucci, C., Capelli, L., Chiadini, E., Calistri, D., Bennati, C., Cravero, P., Limarzi, F., Nosseir, S., Panzacchi, R., Valli, M., Ulivi, P., & Rossi, G. (2023). Clinicopathological Features of Non-Small Cell Lung Carcinoma with BRAF Mutation. Current Oncology, 30(11), 10019-10032. https://doi.org/10.3390/curroncol30110728