Application of CRISPR/Cas9 Technology in Cancer Treatment: A Future Direction
Abstract
:1. Introduction
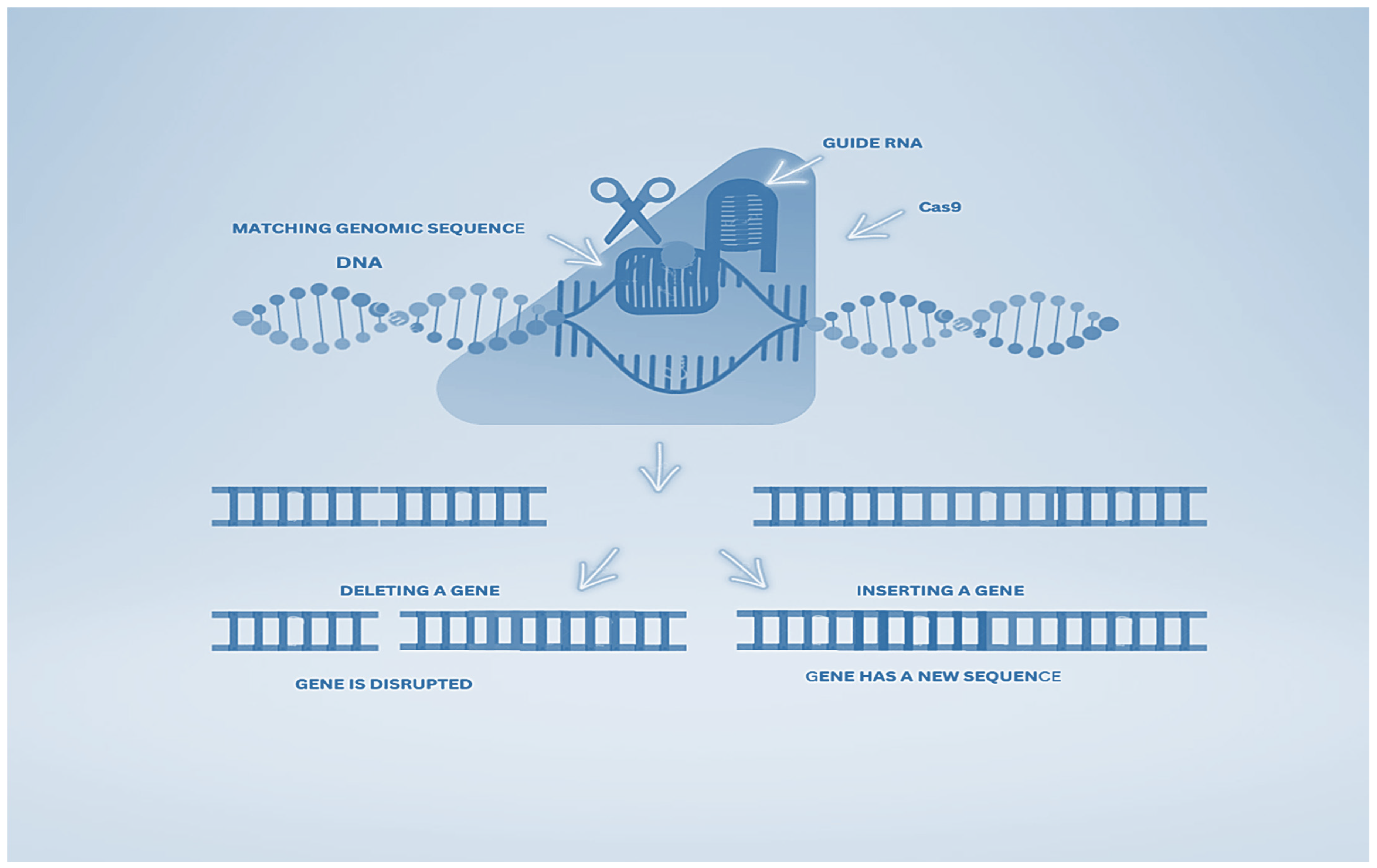
1.1. Behind CRISPR-Cas9
1.2. CRISPR/Cas9 Apparatus
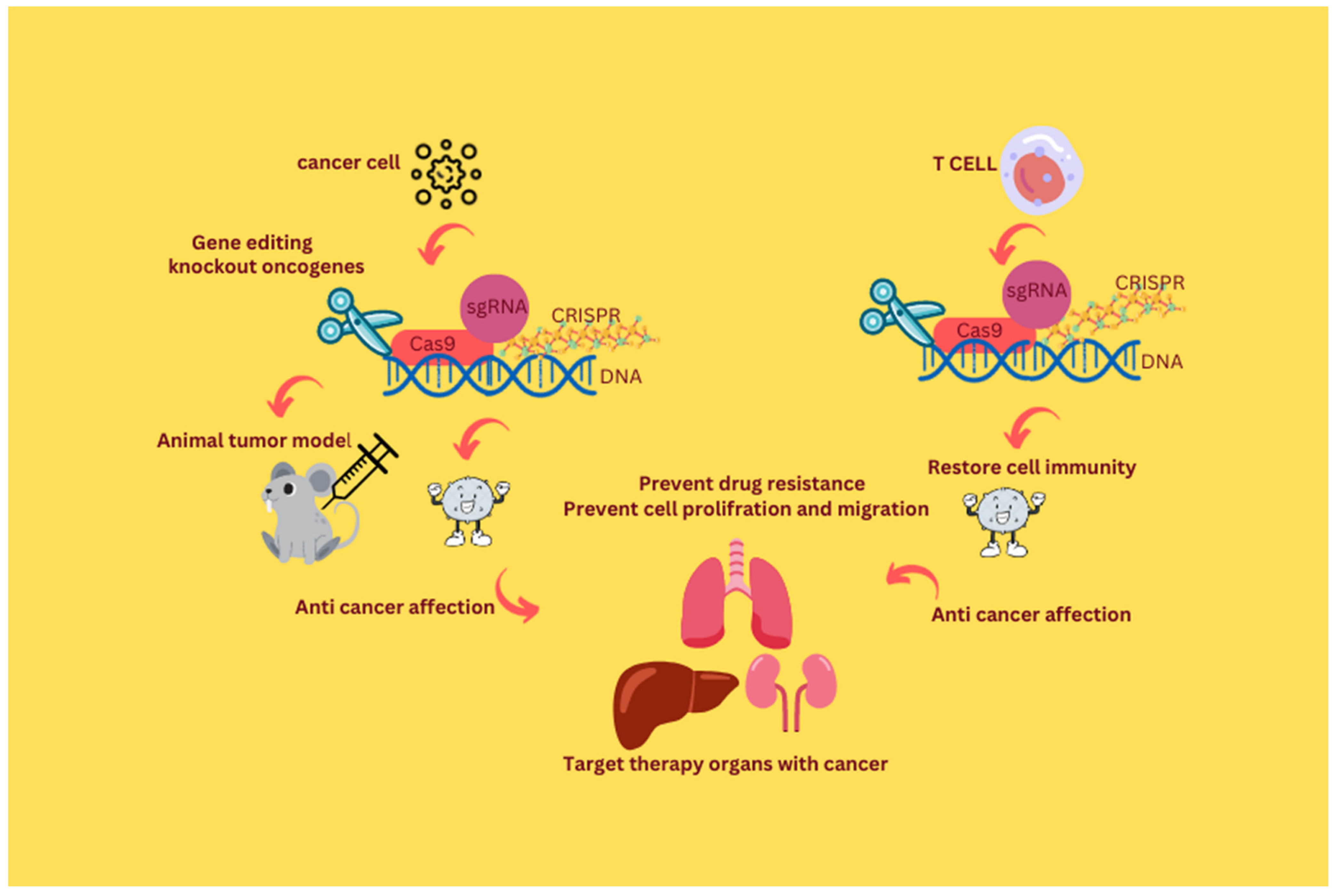
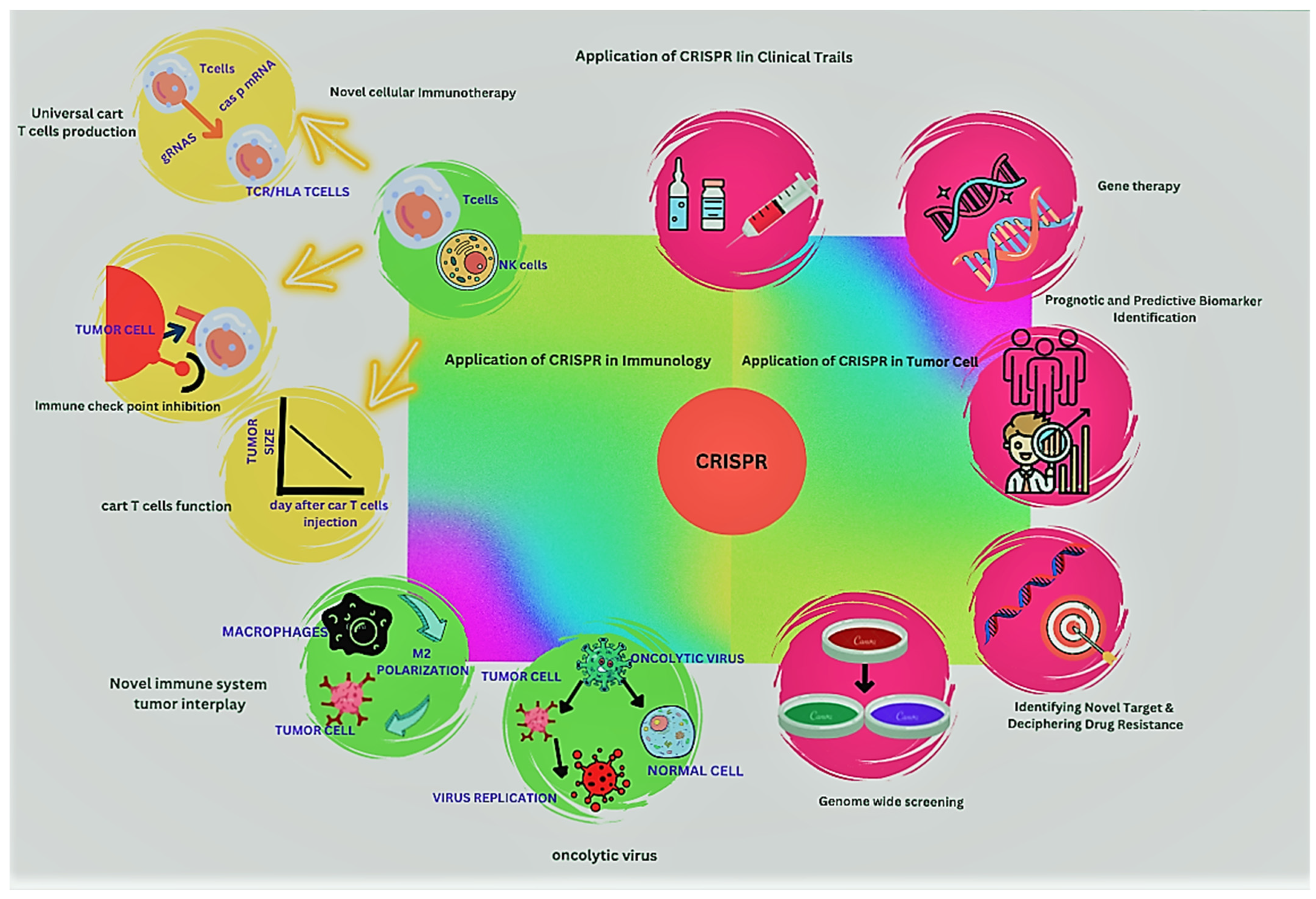
2. The Era of CRISPR/Cas9 in Therapeutic Oncology
2.1. CRISPR/Cas9 in Brain Cancer
2.2. CRISPR/Cas9 in Hepatocellular Carcinoma
2.3. CRISPR/Cas9 in Colorectal Cancer
2.4. CRISPR/Cas9 in Renal Cell Carcinoma
2.5. Application of CRISPR/Cas9 in Patient-Derived Organoids
3. CRISPR/Cas9 in Oncolytic Viruses
4. Editing the Cancer Epigenome
5. Clinical Trials of CRISPR/Cas9
6. CRISPR/Cas9 in Cancer Immunotherapy
7. CRISPR/Cas9 in the Elimination or Inactivation of Carcinogenic Viral Infections
8. Limitations of the CRISPR-Cas9 System
9. Conclusions and Future Directions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, X.; Kriz, A.J.; Sharp, P.A. Target specificity of the CRISPR-Cas9 system. Quant. Biol. 2014, 2, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Wong, N.; Liu, W.; Wang, X. WU-CRISPR: Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015, 16, 218. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, G.; Andersen, T.; Zhou, P.; Pu, W.T. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PLoS ONE 2014, 9, e105779. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef]
- Wood, A.J.; Lo, T.-W.; Zeitler, B.; Pickle, C.S.; Ralston, E.J.; Lee, A.H.; Amora, R.; Miller, J.C.; Leung, E.; Meng, X. Targeted genome editing across species using ZFNs and TALENs. Science 2011, 333, 307. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end joining pathway. Annu. Rev. Biochem. 2010, 79, 181. [Google Scholar] [CrossRef] [PubMed]
- Taleei, R.; Nikjoo, H. The non-homologous end-joining (NHEJ) pathway for the repair of DNA double-strand breaks: I. A mathematical model. Radiat. Res. 2013, 179, 530–539. [Google Scholar] [CrossRef]
- Zhu, K. In Vivo Gene Editing of Muscles and Muscle Stem Cells Using AAV-CRISPR. Doctoral Dissertation, Graduate School of Arts & Sciences, Harvard University, Cambridge, MA, USA, 2018. [Google Scholar]
- Gupta, R.M.; Musunuru, K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Investig. 2014, 124, 4154–4161. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D.; Beumer, K.J. Genome engineering with TALENs and ZFNs: Repair pathways and donor design. Methods 2014, 69, 137–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaFountaine, J.S.; Fathe, K.; Smyth, H.D. Delivery and therapeutic applications of gene editing technologies ZFNs, TALENs, and CRISPR/Cas9. Int. J. Pharm. 2015, 494, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kweon, J.; Kim, J.-S. TALENs and ZFNs are associated with different mutation signatures. Nat. Methods 2013, 10, 185. [Google Scholar] [CrossRef]
- Chen, S.; Oikonomou, G.; Chiu, C.N.; Niles, B.J.; Liu, J.; Lee, D.A.; Antoshechkin, I.; Prober, D.A. A large-scale in vivo analysis reveals that TALENs are significantly more mutagenic than ZFNs generated using context-dependent assembly. Nucleic Acids Res. 2013, 41, 2769–2778. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, H.; Yamamoto, T. Genome editing using zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). In Targeted Genome Editing Using Site-Specific Nucleases; Springer: Tokyo, Japan, 2015; pp. 3–24. [Google Scholar]
- Wani, A.K.; Akhtar, N.; Singh, R.; Prakash, A.; Raza, S.H.A.; Cavalu, S.; Chopra, C.; Madkour, M.; Elolimy, A.; Hashem, N.M. Genome centric engineering using ZFNs, TALENs and CRISPR-Cas9 systems for trait improvement and disease control in Animals. Vet. Res. Commun. 2022, 47, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Church, G.M. Genome Editing and Engineering: From TALENs, ZFNs and CRISPRs to Molecular Surgery; Cambridge University Press: Cambridge, UK, 2018. [Google Scholar]
- Gaj, T.; Gersbach, C.A.; Barbas III, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Zhang, H.-X.; Zhang, Y.; Yin, H. Genome editing with mRNA encoding ZFN, TALEN, and Cas9. Mol. Ther. 2019, 27, 735–746. [Google Scholar] [CrossRef]
- Kim, H.; Kim, M.; Im, S.-K.; Fang, S.J.L.a.r. Mouse Cre-LoxP system: General principles to determine tissue-specific roles of target genes. Lab. Anim. Res. 2018, 34, 147–159. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Pawlik, K.M.; Napierala, J.S.; Napierala, M. A CRISPR-Cas9, Cre-lox, and Flp-FRT cascade strategy for the precise and efficient integration of exogenous DNA into cellular genomes. CRISPR J. 2020, 3, 470–486. [Google Scholar] [CrossRef] [PubMed]
- Odintsova, I.; Pisareva, L.; Khryapenkov, A. Worldwide cancer epidemiology. Sib. J. Oncol. 2016, 1, 95–101. [Google Scholar]
- La Vecchia, C.; Negri, E.; Carioli, G. Progress in cancer epidemiology: Avoided deaths in Europe over the last three decades. Eur. J. Cancer Prev. 2022, 31, 388–392. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell. Signal. 2007, 128, 683–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zugazagoitia, J.; Guedes, C.; Ponce, S.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. Current challenges in cancer treatment. Clin. Ther. 2016, 38, 1551–1566. [Google Scholar] [CrossRef]
- Yuan, Y.; Jiang, Y.-C.; Sun, C.-K.; Chen, Q.-M. Role of the tumor microenvironment in tumor progression and the clinical applications. Oncol. Rep. 2016, 35, 2499–2515. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Baig, A.A.; Hussain, M.; Saeed, M.U.; Bilal, M.; Ahmed, N.; Chopra, H.; Hassan, M.; Rachamalla, M.; Putnala, S.K. Narrative on Hydrogen Therapy and its Clinical Applications: Safety and Efficacy. Curr. Pharm. Des. 2022, 28, 2519–2537. [Google Scholar] [PubMed]
- Al-Mhanna, S.B.; Ghazali, W.S.W.; Mohamed, M.; Rabaan, A.A.; Santali, E.Y.; Alestad, J.H.; Santali, E.Y.; Arshad, S.; Ahmed, N.; Afolabi, H.A. Effectiveness of physical activity on immunity markers and quality of life in cancer patient: A systematic review. PeerJ 2022, 10, e13664. [Google Scholar] [CrossRef]
- Bukhari, B.; Naveed, M.; Makhdoom, S.I.; Jabeen, K.; Asif, M.F.; Batool, H.; Ahmed, N.; Chan, Y.Y. A comparison between organic and inorganic nanoparticles: Prime nanoparticles for tumor curation. Nano 2021, 16, 2130011. [Google Scholar] [CrossRef]
- Xia, A.-L.; He, Q.-F.; Wang, J.-C.; Zhu, J.; Sha, Y.-Q.; Sun, B.; Lu, X.-J. Applications and advances of CRISPR-Cas9 in cancer immunotherapy. J. Med. Genet. 2019, 56, 4–9. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, Y. CRISPR/Cas9 genome editing: Fueling the revolution in cancer immunotherapy. Curr. Res. Transl. Med. 2018, 66, 39–42. [Google Scholar] [CrossRef]
- Wu, H.-y.; Cao, C.-y. The application of CRISPR-Cas9 genome editing tool in cancer immunotherapy. Brief. Funct. Genom. 2019, 18, 129–132. [Google Scholar] [CrossRef]
- Chen, M.; Mao, A.; Xu, M.; Weng, Q.; Mao, J.; Ji, J. CRISPR-Cas9 for cancer therapy: Opportunities and challenges. Cancer Lett. 2019, 447, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Krupovic, M.; Forterre, P. History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. J. Bacteriol. 2018, 200, e00580-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharathkumar, N.; Sunil, A.; Meera, P.; Aksah, S.; Kannan, M.; Saravanan, K.M.; Anand, T. CRISPR/Cas-Based modifications for therapeutic applications: A review. Mol. Biotechnol. 2022, 64, 355–372. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Luo, M.; Hayes, R.P.; Kim, J.; Ng, S.; Ding, F.; Liao, M.; Ke, A. Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR-Cas system. Cell 2017, 170, 48–60.e11. [Google Scholar] [CrossRef] [PubMed]
- Shams, F.; Bayat, H.; Mohammadian, O.; Mahboudi, S.; Vahidnezhad, H.; Soosanabadi, M.; Rahimpour, A. Advance trends in targeting homology-directed repair for accurate gene editing: An inclusive review of small molecules and modified CRISPR-Cas9 systems. BioImpacts 2022, 12, 371. [Google Scholar] [CrossRef] [PubMed]
- Kulshrestha, S.; Chauhan, S.; Singh, G. With CRISPR/CAS9, Fighting against the Dark Empire of Diseases. Guident 2022, 15, 29–32. [Google Scholar]
- Xu, C.L.; Tsang, S.H. The history of CRISPR: From discovery to the present. In CRISPR Genome Surgery in Stem Cells and Disease Tissues; Elsevier: Amsterdam, The Netherlands, 2022; pp. 1–6. [Google Scholar]
- Khan, Z.; Ali, Z.; Khan, A.A.; Sattar, T.; Zeshan, A.; Saboor, T.; Binyamin, B. History and Classification of CRISPR/Cas System. In The CRISPR/Cas Tool Kit for Genome Editing; Springer: Berlin/Heidelberg, Germany, 2022; pp. 29–52. [Google Scholar]
- Van der Oost, J.; Jore, M.M.; Westra, E.R.; Lundgren, M.; Brouns, S.J. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009, 34, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Vigouroux, A.; Bikard, D. CRISPR tools to control gene expression in bacteria. Microbiol. Mol. Biol. Rev. 2020, 84, e00077-19. [Google Scholar] [CrossRef] [PubMed]
- Burmistrz, M.; Krakowski, K.; Krawczyk-Balska, A. RNA-targeting CRISPR–Cas systems and their applications. Int. J. Mol. Sci. 2020, 21, 1122. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.R.; Cocozaki, A.; Li, H.; Terns, R.M.; Terns, M.P. Target RNA capture and cleavage by the Cmr type III-B CRISPR–Cas effector complex. Genes Dev. 2014, 28, 2432–2443. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Anders, C.; Jinek, M. In vitro enzymology of Cas9. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 546, pp. 1–20. [Google Scholar]
- Kampmann, M. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem. Biol. 2018, 13, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Baisya, D.; Ramesh, A.; Schwartz, C.; Lonardi, S.; Wheeldon, I. Genome-wide functional screens enable the prediction of high activity CRISPR-Cas9 and-Cas12a guides in Yarrowia lipolytica. Nat. Commun. 2022, 13, 922. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhou, K.; Ma, L.; Gressel, S.; Doudna, J.A. A Cas9–guide RNA complex preorganized for target DNA recognition. Science 2015, 348, 1477–1481. [Google Scholar] [CrossRef]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Kimberland, M.L.; Hou, W.; Alfonso-Pecchio, A.; Wilson, S.; Rao, Y.; Zhang, S.; Lu, Q. Strategies for controlling CRISPR/Cas9 off-target effects and biological variations in mammalian genome editing experiments. J. Biotechnol. 2018, 284, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Monsur, M.B.; Shao, G.; Lv, Y.; Ahmad, S.; Wei, X.; Hu, P.; Tang, S. Base editing: The ever expanding clustered regularly interspaced short palindromic repeats (CRISPR) tool kit for precise genome editing in plants. Genes Dev. 2020, 11, 466. [Google Scholar] [CrossRef]
- Chen, S.; Yao, Y.; Zhang, Y.; Fan, G. CRISPR system: Discovery, development and off-target detection. Cell. Signal. 2020, 70, 109577. [Google Scholar] [CrossRef]
- Pavlinov, I.; Farkhondeh, A.; Yang, S.; Xu, M.; Cheng, Y.-S.; Beers, J.; Zou, J.; Liu, C.; Might, M.; Rodems, S. Generation of two gene corrected human isogenic iPSC lines (NCATS-CL6104 and NCATS-CL6105) from a patient line (NCATS-CL6103) carrying a homozygous p. R401X mutation in the NGLY1 gene using CRISPR/Cas9. Stem Cell Res. 2021, 56, 102554. [Google Scholar] [CrossRef]
- Zon, G.; Biotechnologies, T. Glen Report 32–24: Very Fast CRISPR (vfCRISPR) “On Demand”. Available online: https://www.glenresearch.com/reports/gr32-24 (accessed on 14 October 2022).
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010, 11, 181–190. [Google Scholar] [CrossRef]
- Mojica, F.J.; Rodriguez-Valera, F. The discovery of CRISPR in archaea and bacteria. FEBS J. 2016, 283, 3162–3169. [Google Scholar] [CrossRef]
- Wang, R.; Preamplume, G.; Terns, M.P.; Terns, R.M.; Li, H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: Recognition and cleavage. Structure 2011, 19, 257–264. [Google Scholar] [CrossRef] [Green Version]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- MacPherson, C.R.; Scherf, A. Flexible guide-RNA design for CRISPR applications using Protospacer Workbench. Nat. Biotechnol. 2015, 33, 805–806. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef]
- Leenay, R.T.; Maksimchuk, K.R.; Slotkowski, R.A.; Agrawal, R.N.; Gomaa, A.A.; Briner, A.E.; Barrangou, R.; Beisel, C.L. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol. Cell 2016, 62, 137–147. [Google Scholar] [CrossRef]
- Hasham, K.; Ahmed, N.; Zeshan, B. Circulating microRNAs in oncogenic viral infections: Potential diagnostic biomarkers. SN Appl. Sci. 2020, 2, 1–13. [Google Scholar] [CrossRef]
- Facchino, S.; Abdouh, M.; Bernier, G. Brain cancer stem cells: Current status on glioblastoma multiforme. Cancers 2011, 3, 1777–1797. [Google Scholar] [CrossRef]
- Zuckermann, M.; Kawauchi, D.; Gronych, J. “CRISPR” validation of recessive brain cancer genes in vivo. Oncotarget 2015, 6, 17865. [Google Scholar] [CrossRef]
- Lee, E.X.; Lam, D.H.; Wu, C.; Yang, J.; Tham, C.K.; Ng, W.H.; Wang, S. Glioma gene therapy using induced pluripotent stem cell derived neural stem cells. Mol. Pharm. 2011, 8, 1515–1524. [Google Scholar] [CrossRef]
- Vaubel, R.A.; Tian, S.; Remonde, D.; Schroeder, M.A.; Mladek, A.C.; Kitange, G.J.; Caron, A.; Kollmeyer, T.M.; Grove, R.; Peng, S.J.C.C.R. Genomic and Phenotypic Characterization of a Broad Panel of Patient-Derived Xenografts Reflects the Diversity of Glioblastoma Characterization of Glioblastoma Patient-Derived Xenografts. Clin. Cancer Res. 2020, 26, 1094–1104. [Google Scholar] [CrossRef] [PubMed]
- Vo, B.T.; Kwon, J.A.; Li, C.; Finkelstein, D.; Xu, B.; Orr, B.A.; Sherr, C.J.; Roussel, M.F. Mouse medulloblastoma driven by CRISPR activation of cellular Myc. Sci. Rep. 2018, 8, 8733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratan, Z.A.; Son, Y.-J.; Haidere, M.F.; Uddin, B.M.M.; Yusuf, M.A.; Zaman, S.B.; Kim, J.-H.; Banu, L.A.; Cho, J. CRISPR-Cas9: A promising genetic engineering approach in cancer research. Ther. Adv. Med. Oncol. 2018, 10, 1758834018755089. [Google Scholar] [CrossRef] [PubMed]
- Zuckermann, M.; Hovestadt, V.; Knobbe-Thomsen, C.B.; Zapatka, M.; Northcott, P.A.; Schramm, K.; Belic, J.; Jones, D.T.; Tschida, B.; Moriarity, B. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat. Commun. 2015, 6, 7391. [Google Scholar] [CrossRef]
- Mou, H.; Kennedy, Z.; Anderson, D.G.; Yin, H.; Xue, W. Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med. 2015, 7, 53. [Google Scholar] [CrossRef]
- Yin, H.; Sun, L.; Pu, Y.; Yu, J.; Feng, W.; Dong, C.; Zhou, B.; Du, D.; Zhang, Y.; Chen, Y. Ultrasound-controlled crispr/cas9 system augments sonodynamic therapy of hepatocellular carcinoma. ACS Cent. Sci. 2021, 7, 2049–2062. [Google Scholar] [CrossRef]
- Qi, Y.; Liu, Y.; Yu, B.; Hu, Y.; Zhang, N.; Zheng, Y.; Yang, M.; Xu, F.J. A lactose-derived CRISPR/Cas9 delivery system for efficient genome editing in vivo to treat orthotopic hepatocellular carcinoma. Adv. Sci. 2020, 7, 2001424. [Google Scholar] [CrossRef]
- Liu, Y.; Qi, X.; Zeng, Z.; Wang, L.; Wang, J.; Zhang, T.; Xu, Q.; Shen, C.; Zhou, G.; Yang, S. CRISPR/Cas9-mediated p53 and Pten dual mutation accelerates hepatocarcinogenesis in adult hepatitis B virus transgenic mice. Sci. Rep. 2017, 7, 2796. [Google Scholar] [CrossRef]
- Xu, T.; Li, L.; Liu, Y.-c.; Cao, W.; Chen, J.-s.; Hu, S.; Liu, Y.; Li, L.-y.; Zhou, H.; Meng, X.-M. CRISPR/Cas9-related technologies in liver diseases: From feasibility to future diversity. Int. J. Biol. Sci. 2020, 16, 2283. [Google Scholar] [CrossRef]
- Lv, W.; Li, T.; Wang, S.; Wang, H.; Li, X.; Zhang, S.; Wang, L.; Xu, Y.; Wei, W. The Application of the CRISPR/Cas9 System in the Treatment of Hepatitis B Liver Cancer. Technol. Cancer Res. Treat. 2021, 20, 15330338211045206. [Google Scholar] [CrossRef]
- Pankowicz, F.P.; Jarrett, K.E.; Lagor, W.R.; Bissig, K.-D.J.G. CRISPR/Cas9: At the cutting edge of hepatology. Gut Pathog. 2017, 66, 1329–1340. [Google Scholar] [CrossRef]
- Matano, M.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Mohd Salim, N.H.; Mussa, A.; Ahmed, N.; Ahmad, S.; Yean Yean, C.; Hassan, R.; Uskoković, V.; Mohamud, R.; Che Jalil, N.A. The Immunosuppressive Effect of TNFR2 Expression in the Colorectal Cancer Microenvironment. Biomedicines 2023, 11, 173. [Google Scholar] [CrossRef]
- Munro, M.J.; Tan, S.T.; Gray, C. Applications for Colon Organoid Models in Cancer Research. Organoids 2023, 2, 37–49. [Google Scholar] [CrossRef]
- Takeda, H.; Kataoka, S.; Nakayama, M.; Ali, M.A.; Oshima, H.; Yamamoto, D.; Park, J.-W.; Takegami, Y.; An, T.; Jenkins, N. CRISPR-Cas9–mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes. Proc. Natl. Acad. Sci. USA 2019, 116, 15635–15644. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 17009. [Google Scholar] [CrossRef]
- Yoshino, H.; Yonemori, M.; Miyamoto, K.; Tatarano, S.; Kofuji, S.; Nohata, N.; Nakagawa, M.; Enokida, H. microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis through revival of TWIST1 in renal cell carcinoma. Oncotarget 2017, 8, 20881. [Google Scholar] [CrossRef]
- Yoshino, H.; Yonezawa, T.; Yonemori, M.; Miyamoto, K.; Sakaguchi, T.; Sugita, S.; Osako, Y.; Tatarano, S.; Nakagawa, M.; Enokida, H. Downregulation of microRNA-1274a induces cell apoptosis through regulation of BMPR1B in clear cell renal cell carcinoma. Oncol. Rep. 2018, 39, 173–181. [Google Scholar] [CrossRef]
- Zhu, S.-j.; Wang, X.; Hu, S.-l.; Fang, Y.; Guan, B.-x.; Li, J.; Li, G.; Xu, J.-y. Clinical Significance and Biological Function of miR-1274a in Non-small Cell Lung Cancer. Mol. Biotechnol. 2022, 64, 9–16. [Google Scholar] [CrossRef]
- Yue, L.; Lin, H.; Yuan, S.; Wu, L.; Chen, G.; Wang, J.; Feng, J. miR-1251-5p Overexpression Inhibits Proliferation, Migration, and Immune Escape in Clear Cell Renal Cell Carcinoma by Targeting NPTX2. J. Oncol. 2022, 2022, 3058588. [Google Scholar] [CrossRef]
- Fenner, A. RCC classification using miRNA signatures. Nat. Rev. Urol. 2011, 8, 120. [Google Scholar] [CrossRef]
- Sanli, O.; Dobruch, J.; Knowles, M.A.; Burger, M.; Alemozaffar, M.; Nielsen, M.E.; Lotan, Y. Bladder cancer. Nat. Rev. Dis. Prim. 2017, 3, 17022. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Tian, T.; Li, W.; Xu, H.; Zhan, C.; Wu, X.; Wang, C.; Wu, X.; Wu, W.; Zheng, S. Long non-coding RNA in bladder cancer. Clin. Chim. Acta 2020, 503, 113–121. [Google Scholar] [CrossRef]
- Terracciano, D.; Ferro, M.; Terreri, S.; Lucarelli, G.; D’Elia, C.; Musi, G.; de Cobelli, O.; Mirone, V.; Cimmino, A. Urinary long noncoding RNAs in nonmuscle-invasive bladder cancer: New architects in cancer prognostic biomarkers. Transl. Res. 2017, 184, 108–117. [Google Scholar] [CrossRef]
- Quan, J.; Pan, X.; Zhao, L.; Li, Z.; Dai, K.; Yan, F.; Liu, S.; Ma, H.; Lai, Y. LncRNA as a diagnostic and prognostic biomarker in bladder cancer: A systematic review and meta-analysis. OncoTargets Ther. 2018, 11, 6415. [Google Scholar] [CrossRef]
- Duan, W.; Du, L.; Jiang, X.; Wang, R.; Yan, S.; Xie, Y.; Yan, K.; Wang, Q.; Wang, L.; Zhang, X. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget 2016, 7, 78850. [Google Scholar] [CrossRef]
- Aberle, M.R.; Burkhart, R.A.; Tiriac, H.; Olde Damink, S.; Dejong, C.H.; Tuveson, D.A.; van Dam, R.M. Patient-derived organoid models help define personalized management of gastrointestinal cancer. J. Br. Surg. 2018, 105, e48–e60. [Google Scholar] [CrossRef]
- Wang, J.; Feng, X.; Li, Z.; Chen, Y.; Huang, W. Patient-derived organoids as a model for tumor research. Prog. Mol. Biol. Transl. Sci. 2022, 189, 259–326. [Google Scholar]
- Bartfeld, S.; Clevers, H. Stem cell-derived organoids and their application for medical research and patient treatment. J. Mol. Med. 2017, 95, 729–738. [Google Scholar] [CrossRef]
- Zhou, Z.; Cong, L.; Cong, X. Patient-derived organoids in precision medicine: Drug screening, organoid-on-a-chip and living organoid biobank. Front. Oncol. 2021, 11, 5625. [Google Scholar] [CrossRef]
- Drost, J.; Clevers, H. Translational applications of adult stem cell-derived organoids. Development 2017, 144, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Clevers, H.; Sato, T. Modeling human digestive diseases with CRISPR-Cas9–modified organoids. Gastroenterology 2019, 156, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Nagle, P.W.; Plukker, J.T.M.; Muijs, C.T.; van Luijk, P.; Coppes, R.P. Patient-derived tumor organoids for prediction of cancer treatment response. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2018; pp. 258–264. [Google Scholar]
- Ramakrishna, G.; Babu, P.E.; Singh, R.; Trehanpati, N. Application of CRISPR-Cas9 based gene editing to study the pathogenesis of colon and liver cancer using organoids. Hepatol. Int. 2021, 15, 1309–1317. [Google Scholar] [CrossRef]
- Yang, H.; Sun, L.; Liu, M.; Mao, Y. Patient-derived organoids: A promising model for personalized cancer treatment. Gastroenterol. Rep. 2018, 6, 243–245. [Google Scholar] [CrossRef]
- Okamoto, T.; Natsume, Y.; Yamanaka, H.; Fukuda, M.; Yao, R. A protocol for efficient CRISPR-Cas9-mediated knock-in in colorectal cancer patient-derived organoids. STAR Protoc. 2021, 2, 100780. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Huang, J.; Tong, A.; Yang, H. Oncolytic viruses for cancer therapy: Barriers and recent advances. Mol. Ther.-Oncolytics 2019, 15, 234–247. [Google Scholar] [CrossRef]
- Tysome, J.R.; Li, X.; Wang, S.; Wang, P.; Gao, D.; Du, P.; Chen, D.; Gangeswaran, R.; Chard, L.S.; Yuan, M. A Novel Therapeutic Regimen to Eradicate Established Solid Tumors with an Effective Induction of Tumor-Specific ImmunitySequential Use of Two Oncolytic Viruses for Cancer Treatment. Clin. Cancer Res. 2012, 18, 6679–6689. [Google Scholar] [CrossRef]
- Chulpanova, D.S.; Solovyeva, V.V.; Kitaeva, K.V.; Dunham, S.P.; Khaiboullina, S.F.; Rizvanov, A.A. Recombinant viruses for cancer therapy. Biomedicines 2018, 6, 94. [Google Scholar] [CrossRef]
- Macedo, N.; Miller, D.M.; Haq, R.; Kaufman, H.L. Clinical landscape of oncolytic virus research in 2020. J. Immunother. Cancer 2020, 8, e001486. [Google Scholar] [CrossRef]
- Yuan, M.; Webb, E.; Lemoine, N.R.; Wang, Y.J.V. CRISPR-Cas9 as a powerful tool for efficient creation of oncolytic viruses. Viruses 2016, 8, 72. [Google Scholar] [CrossRef]
- Yi, L.; Li, J. CRISPR-Cas9 therapeutics in cancer: Promising strategies and present challenges. Biochim. Biophys. Acta-Rev. Cancer 2016, 1866, 197–207. [Google Scholar] [CrossRef]
- Lin, C.; Li, H.; Hao, M.; Xiong, D.; Luo, Y.; Huang, C.; Yuan, Q.; Zhang, J.; Xia, N. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing of HSV-1 virus in human cells. Sci. Rep. 2016, 6, 34531. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Gao, Y.; Dominguez, A.A.; Qi, L.S. CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 2021, 23, 11–22. [Google Scholar] [CrossRef]
- Hilton, I.B.; D’ippolito, A.M.; Vockley, C.M.; Thakore, P.I.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Thakore, P.I.; D’ippolito, A.M.; Song, L.; Safi, A.; Shivakumar, N.K.; Kabadi, A.M.; Reddy, T.E.; Crawford, G.E.; Gersbach, C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 2015, 12, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Sarvari, P.; Sarvari, P.; Ramírez-Díaz, I.; Mahjoubi, F.; Rubio, K. Advances of Epigenetic Biomarkers and Epigenome Editing for Early Diagnosis in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 9521. [Google Scholar] [CrossRef]
- Pei, W.-D.; Zhang, Y.; Yin, T.-L.; Yu, Y. Epigenome editing by CRISPR/Cas9 in clinical settings: Possibilities and challenges. Brief. Funct. Genom. 2020, 19, 215–228. [Google Scholar] [CrossRef]
- Tothova, Z.; Krill-Burger, J.M.; Popova, K.D.; Landers, C.C.; Sievers, Q.L.; Yudovich, D.; Belizaire, R.; Aster, J.C.; Morgan, E.A.; Tsherniak, A. Multiplex CRISPR/Cas9-based genome editing in human hematopoietic stem cells models clonal hematopoiesis and myeloid neoplasia. Cell Stem Cell 2017, 21, 547–555.e548. [Google Scholar] [CrossRef]
- Sachdeva, M.; Sachdeva, N.; Pal, M.; Gupta, N.; Khan, I.; Majumdar, M.; Tiwari, A. CRISPR/Cas9: Molecular tool for gene therapy to target genome and epigenome in the treatment of lung cancer. Cancer Gene Ther. 2015, 22, 509–517. [Google Scholar] [CrossRef]
- Pabst, G.; Foßelteder, J.; Schlacher, A.; Auinger, L.; Martinez-Krams, D.; Ediriwickrema, A.; Kashofer, K.; Beham-Schmid, C.; Greinix, H.T.; Woelfler, A. Modeling the Development of SRSF2 Mutated Myeloid Malignancies By CRISPR/Cas9 Mediated Genome Engineering of Primary Human Hematopoietic Stem and Progenitor Cells. Blood 2021, 138, 2160. [Google Scholar] [CrossRef]
- Valletta, S.; Dolatshad, H.; Bartenstein, M.; Yip, B.H.; Bello, E.; Gordon, S.; Yu, Y.; Shaw, J.; Roy, S.; Scifo, L. ASXL1 mutation correction by CRISPR/Cas9 restores gene function in leukemia cells and increases survival in mouse xenografts. Oncotarget 2015, 6, 44061. [Google Scholar] [CrossRef] [PubMed]
- Castaño, J.; Herrero, A.B.; Bursen, A.; González, F.; Marschalek, R.; Gutiérrez, N.C.; Menendez, P. Expression of MLL-AF4 or AF4-MLL fusions does not impact the efficiency of DNA damage repair. Oncotarget 2016, 7, 30440. [Google Scholar] [CrossRef]
- Barnabas, G.D.; Lee, J.S.; Shami, T.; Harel, M.; Beck, L.; Selitrennik, M.; Jerby-Arnon, L.; Erez, N.; Ruppin, E.; Geiger, T. Serine Biosynthesis Is a Metabolic Vulnerability in IDH2-Driven Breast Cancer ProgressionSerine Biosynthesis as Metabolic Vulnerability of IDH2. Cancer Res. 2021, 81, 1443–1456. [Google Scholar] [CrossRef]
- Ivy, K.S.; Cote, C.H.; Ferrell Jr, P.B. IDH2 Mutations Induce Altered STAT Signaling and Cytokine Responses Which Are Restored by Enasidenib. Blood 2018, 132, 1468. [Google Scholar] [CrossRef]
- Ritter, M.U.; Secker, B.; Nasri, M.; Klimiankou, M.; Dannenmann, B.; Amend, D.; Haaf, J.; Mir, P.; Bernhard, R.; Steiert, I. Efficient Correction of HAX1 Mutations in Primary HSPCs of Severe Congenital Neutropenia Patients Using CRISPR/CAS9 GENE-Editing. Blood 2020, 136, 22. [Google Scholar] [CrossRef]
- Reimer, J.; Knöß, S.; Labuhn, M.; Charpentier, E.M.; Göhring, G.; Schlegelberger, B.; Klusmann, J.-H.; Heckl, D. CRISPR-Cas9-induced t (11; 19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica 2017, 102, 1558. [Google Scholar] [CrossRef]
- Matsumoto, A.; Yoshida, T.; Shima, T.; Yamasaki, K.; Tadagaki, K.; Kondo, N.; Kuwahara, Y.; Zhang, D.-E.; Okuda, T. C11orf21, a novel RUNX1 target gene, is down-regulated by RUNX1-ETO. BBA Adv. 2022, 2, 100047. [Google Scholar] [CrossRef]
- Almosailleakh, M.; Schwaller, J. Murine models of acute myeloid leukaemia. Int. J. Mol. Sci. 2019, 20, 453. [Google Scholar] [CrossRef] [PubMed]
- Christen, F.; Hablesreiter, R.; Hoyer, K.; Hennch, C.; Maluck-Böttcher, A.; Segler, A.; Madadi, A.; Frick, M.; Bullinger, L.; Briest, F. Modeling clonal hematopoiesis in umbilical cord blood cells by CRISPR/Cas9. Leukemia 2022, 36, 1102–1110. [Google Scholar] [CrossRef]
- Liu, T.; Shen, J.K.; Li, Z.; Choy, E.; Hornicek, F.J.; Duan, Z. Development and potential applications of CRISPR-Cas9 genome editing technology in sarcoma. Cancer Lett. 2016, 373, 109–118. [Google Scholar] [CrossRef]
- Sweeney, C.L.; Pavel-Dinu, M.; Choi, U.; Brault, J.; Liu, T.; Koontz, S.; Li, L.; Theobald, N.; Lee, J.; Bello, E.A. Correction of X-CGD patient HSPCs by targeted CYBB cDNA insertion using CRISPR/Cas9 with 53BP1 inhibition for enhanced homology-directed repair. Gene Ther. 2021, 28, 373–390. [Google Scholar] [CrossRef]
- Frøkjær-Jensen, C. Exciting prospects for precise engineering of Caenorhabditis elegans genomes with CRISPR/Cas9. Genetics 2013, 195, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Arribere, J.A.; Bell, R.T.; Fu, B.X.; Artiles, K.L.; Hartman, P.S.; Fire, A.Z. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 2014, 198, 837–846. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Xin, H.; Roy, B.; Dai, J.; Miao, Y.; Gao, G. Heritable genome editing with CRISPR/Cas9 in the silkworm, Bombyx mori. PLoS ONE 2014, 9, e101210. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Dong, F.; Yu, X.; Huang, L.; Jiang, Y.; Hu, Z.; Chen, P.; Lu, C.; Pan, M. Excision of nucleopolyhedrovirus form transgenic silkworm using the CRISPR/Cas9 system. Front. Microbiol. 2018, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Huang, L.; Dong, F.; Hu, Z.; Qin, Q.; Long, J.; Cao, M.; Chen, P.; Lu, C.; Pan, M.-H. Establishment of a baculovirus-inducible CRISPR/Cas9 system for antiviral research in transgenic silkworms. Appl. Microbiol. Biotechnol. 2018, 102, 9255–9265. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.Z.; Ciccaglione, K.M.; Tournier, V.; Zaratiegui, M. Implementation of the CRISPR-Cas9 system in fission yeast. Nat. Commun. 2014, 5, 5344. [Google Scholar] [CrossRef]
- Bassett, A.R.; Liu, J.-L. CRISPR/Cas9 and genome editing in Drosophila. J. Genet. Genom. 2014, 41, 7–19. [Google Scholar] [CrossRef]
- Oishi, I.; Yoshii, K.; Miyahara, D.; Kagami, H.; Tagami, T. Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep. 2016, 6, 23980. [Google Scholar] [CrossRef]
- Chojnacka-Puchta, L.; Sawicka, D. CRISPR/Cas9 gene editing in a chicken model: Current approaches and applications. J. Appl. Genet. 2020, 61, 221–229. [Google Scholar] [CrossRef]
- Ming, Z.; Vining, B.; Bagheri-Fam, S.; Harley, V. SOX9 in organogenesis: Shared and unique transcriptional functions. Cell. Mol. Life Sci. 2022, 79, 522. [Google Scholar] [CrossRef] [PubMed]
- Fear, V.S.; Forbes, C.A.; Anderson, D.; Rauschert, S.; Syn, G.; Shaw, N.; Jamieson, S.; Ward, M.; Baynam, G.; Lassmann, T. CRISPR single base editing, neuronal disease modelling and functional genomics for genetic variant analysis: Pipeline validation using Kleefstra syndrome EHMT1 haploinsufficiency. Stem Cell Res. Ther. 2022, 13, 69. [Google Scholar] [CrossRef]
- Kurata, M.; Yamamoto, K.; Moriarity, B.S.; Kitagawa, M.; Largaespada, D.A. CRISPR/Cas9 library screening for drug target discovery. J. Hum. Genet. 2018, 63, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Du, Z.; Zhou, F.-M.; Dong, P.; Pfeffer, L.M. Applications of CRSIPR/Cas9 in cancer research. Cancer Med. Anti Cancer Drugs 2016, 1, 1000103. [Google Scholar] [CrossRef]
- Wang, J.; Song, X.; Guo, C.; Wang, Y.; Yin, Y.J.C.s. Establishment of MAGEC 2-knockout cells and functional investigation of MAGEC 2 in tumor cells. Cancer Sci. 2016, 107, 1888–1897. [Google Scholar] [CrossRef]
- Hang, C.Y.; Moriya, S.; Ogawa, S.; Parhar, I.S. Deep brain photoreceptor (val-opsin) gene knockout using CRISPR/Cas affects chorion formation and embryonic hatching in the zebrafish. PLoS ONE 2016, 11, e0165535. [Google Scholar] [CrossRef]
- Niu, Y.; Shen, B.; Cui, Y.; Chen, Y.; Wang, J.; Wang, L.; Kang, Y.; Zhao, X.; Si, W.; Li, W. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef]
- Endo, M.; Mikami, M.; Toki, S. Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 2015, 56, 41–47. [Google Scholar] [CrossRef]
- Zeng, X.; Luo, Y.; Vu, N.T.Q.; Shen, S.; Xia, K.; Zhang, M. CRISPR/Cas9-mediated mutation of OsSWEET14 in rice cv. Zhonghua11 confers resistance to Xanthomonas oryzae pv. oryzae without yield penalty. BMC Plant Biol. 2020, 20, 313. [Google Scholar]
- Ma, L.; Zhang, D.; Miao, Q.; Yang, J.; Xuan, Y.; Hu, Y. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 2017, 58, 863–873. [Google Scholar] [CrossRef]
- Yan, L.; Wei, S.; Wu, Y.; Hu, R.; Li, H.; Yang, W.; Xie, Q. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 2015, 8, 1820–1823. [Google Scholar] [CrossRef]
- Ryder, P.; McHale, M.; Fort, A.; Spillane, C. Generation of stable nulliplex autopolyploid lines of Arabidopsis thaliana using CRISPR/Cas9 genome editing. Plant Cell Rep. 2017, 36, 1005–1008. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Gui, W.; Ya-Nan, Z.; Xiang, H.; Wen, W. Advances and perspectives in the application of CRISPR/Cas9 in insects. Zool. Res. 2016, 37, 136. [Google Scholar]
- Gratz, S.J.; Rubinstein, C.D.; Harrison, M.M.; Wildonger, J.; O’Connor-Giles, K.M. CRISPR-Cas9 genome editing in Drosophila. Curr. Protoc. Mol. Biol. 2015, 111, 31–32. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Kumar, R.; Das, A.; Won, S.Y.; Shukla, P. CRISPR-Cas9 system: A genome-editing tool with endless possibilities. J. Biotechnol. 2020, 319, 36–53. [Google Scholar] [CrossRef]
- Wang, L.; Yang, L.; Guo, Y.; Du, W.; Yin, Y.; Zhang, T.; Lu, H. Enhancing targeted genomic DNA editing in chicken cells using the CRISPR/Cas9 system. PLoS ONE 2017, 12, e0169768. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-E.; Park, C.-H.; Powell, A.; Martin, J.; Donovan, D.M.; Telugu, B.P. Targeted gene knockin in porcine somatic cells using CRISPR/Cas ribonucleoproteins. Int. J. Mol. Sci. 2016, 17, 810. [Google Scholar] [CrossRef] [PubMed]
- Lackner, D.H.; Carré, A.; Guzzardo, P.M.; Banning, C.; Mangena, R.; Henley, T.; Oberndorfer, S.; Gapp, B.V.; Nijman, S.; Brummelkamp, T.R. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat. Commun. 2015, 6, 10237. [Google Scholar] [CrossRef]
- Ahmad, G.; Amiji, M. Use of CRISPR/Cas9 gene-editing tools for developing models in drug discovery. Drug Discov. Today 2018, 23, 519–533. [Google Scholar] [CrossRef]
- Ghosh, S.; Tibbit, C.; Liu, J.-L. Effective knockdown of Drosophila long non-coding RNAs by CRISPR interference. Nucleic Acids Res. 2016, 44, e84. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Zhao, J.; Yang, C.; Luo, W.; Xiong, T.; Li, Y.; Fang, X.; Gao, G.; Singh, C.O.; Madsen, L. CRISPR/Cascade 9-mediated genome editing-challenges and opportunities. Front. Genet. 2018, 9, 240. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Zhang, X. SOCS2 affects the proliferation, migration, and invasion of nasopharyngeal carcinoma cells via regulating EphA1. Neoplasma 2020, 67, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Mon, H.; Xu, J.; Lee, J.M.; Kusakabe, T. CRISPR/Cas9-mediated knockout of factors in non-homologous end joining pathway enhances gene targeting in silkworm cells. Sci. Rep. 2015, 5, 18103. [Google Scholar] [CrossRef]
- Ledford, H. CRISPR fixes disease gene in viable human embryos. Nature 2017, 548, 13–14. [Google Scholar] [CrossRef] [Green Version]
- Baumann, K. Biotechnology: CRISPR-Cas becoming more human. Nat. Rev. Drug Discov. 2017, 16, 601–602. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Zhang, X.; Chai, Y.; Zhu, Z.; Yi, P.; Feng, G.; Li, W.; Ou, G. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell 2014, 30, 625–636. [Google Scholar] [CrossRef]
- Costa, M.A.; de Araújo, E.F.; van den Berg, C.; Bastos, C.A. A tecnologia CRISPR/Cas9 aplicada ao modelo biológico Drosophila melanogaster/CRISPR/Cas9 technology applied to the Drosophila melanogaster biological model. Braz. J. Dev. 2022, 8, 27610–27642. [Google Scholar] [CrossRef]
- Hryhorowicz, M.; Grześkowiak, B.; Mazurkiewicz, N.; Śledziński, P.; Lipiński, D.; Słomski, R. Improved delivery of CRISPR/Cas9 system using magnetic nanoparticles into porcine fibroblast. Mol. Biotechnol. 2019, 61, 173–180. [Google Scholar] [CrossRef]
- Li, W.; Ou, G. The application of somatic CRISPR-Cas9 to conditional genome editing in Caenorhabditis elegans. Genesis 2016, 54, 170–181. [Google Scholar] [CrossRef]
- Jiang, C.; Lin, X.; Zhao, Z. Applications of CRISPR/Cas9 technology in the treatment of lung cancer. Trends Mol. Med. 2019, 25, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Xue, J.; Deng, T.; Zhou, X.; Yu, K.; Deng, L.; Huang, M.; Yi, X.; Liang, M.; Wang, Y. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020, 26, 732–740. [Google Scholar] [CrossRef]
- White, M.K.; Khalili, K. CRISPR/Cas9 and cancer targets: Future possibilities and present challenges. Oncotarget 2016, 7, 12305. [Google Scholar] [CrossRef] [PubMed]
- Kick, L.; Kirchner, M.; Schneider, S. CRISPR-Cas9: From a bacterial immune system to genome-edited human cells in clinical trials. Bioengineered 2017, 8, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhou, Y.; Zhang, M.; Ge, W.; Li, Y.; Yang, L.; Wei, G.; Han, L.; Wang, H.; Yu, S. CRISPR/Cas9-Engineered Universal CD19/CD22 Dual-Targeted CAR-T Cell Therapy for Relapsed/Refractory B-cell Acute Lymphoblastic LeukemiaCRISPR/Cas9-Engineered Universal CAR-T Therapy for ALL. Clin. Cancer Res. 2021, 27, 2764–2772. [Google Scholar] [CrossRef] [PubMed]
- Tipanee, J.; Samara-Kuko, E.; Gevaert, T.; Chuah, M.K.; VandenDriessche, T. Universal allogeneic CAR T cells engineered with Sleeping Beauty transposons and CRISPR-CAS9 for cancer immunotherapy. Mol. Ther. 2022, 30, 3155–3175. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Sharma, A.R.; Bhattacharya, M.; Lee, S.-S.; Chakraborty, C. CRISPR-Cas9: A preclinical and clinical perspective for the treatment of human diseases. Mol. Ther. 2021, 29, 571–586. [Google Scholar] [CrossRef] [PubMed]
- Mollanoori, H.; Teimourian, S. Therapeutic applications of CRISPR/Cas9 system in gene therapy. Biotechnol. Lett. 2018, 40, 907–914. [Google Scholar] [CrossRef]
- Karapurkar, J.K.; Antao, A.M.; Kim, K.-S.; Ramakrishna, S. CRISPR-Cas9 based genome editing for defective gene correction in humans and other mammals. Prog. Mol. Biol. Transl. Sci. 2021, 181, 185–229. [Google Scholar]
- Liu, D.; Zhao, X.; Tang, A.; Xu, X.; Liu, S.; Zha, L.; Ma, W.; Zheng, J.; Shi, M. CRISPR screen in mechanism and target discovery for cancer immunotherapy. Biochim. Biophys. Acta-Rev. Cancer 2020, 1874, 188378. [Google Scholar] [CrossRef]
- Mollanoori, H.; Shahraki, H.; Rahmati, Y.; Teimourian, S. CRISPR/Cas9 and CAR-T cell, collaboration of two revolutionary technologies in cancer immunotherapy, an instruction for successful cancer treatment. Hum. Immunol. 2018, 79, 876–882. [Google Scholar] [CrossRef]
- Razeghian, E.; Nasution, M.K.; Rahman, H.S.; Gardanova, Z.R.; Abdelbasset, W.K.; Aravindhan, S.; Bokov, D.O.; Suksatan, W.; Nakhaei, P.; Shariatzadeh, S. A deep insight into CRISPR/Cas9 application in CAR-T cell-based tumor immunotherapies. Stem Cell Res. Ther. 2021, 12, 428. [Google Scholar] [CrossRef]
- Li, C.; Mei, H.; Hu, Y. Applications and explorations of CRISPR/Cas9 in CAR T-cell therapy. Brief. Funct. Genom. 2020, 19, 175–182. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, Y.; Kang, Z.; Li, S. Inhibiting PDE7A enhances the protective effects of neural stem cells on neurodegeneration and memory deficits in sevoflurane-exposed mice. Eneuro 2021, 8, ENEURO.0071-21.2021. [Google Scholar] [CrossRef]
- Mandal, P.K.; Ferreira, L.M.; Collins, R.; Meissner, T.B.; Boutwell, C.L.; Friesen, M.; Vrbanac, V.; Garrison, B.S.; Stortchevoi, A.; Bryder, D. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 2014, 15, 643–652. [Google Scholar] [CrossRef]
- Georgiadis, C.; Preece, R.; Nickolay, L.; Etuk, A.; Petrova, A.; Ladon, D.; Danyi, A.; Humphryes-Kirilov, N.; Ajetunmobi, A.; Kim, D. Long terminal repeat CRISPR-CAR-coupled “universal” T cells mediate potent anti-leukemic effects. Mol. Ther.-Nucleic Acids 2018, 26, 1215–1227. [Google Scholar] [CrossRef] [Green Version]
- Ottaviano, G.; Georgiadis, C.; Syed, F.; Gkazi, S.A.; Zhan, H.; Etuk, A.; Chu, J.; Pinner, D.; Inglott, S.; Gilmour, K. TT52CAR19: Phase 1 Trial of CRISPR/Cas9 Edited Allogeneic CAR19 T Cells for Paediatric Relapsed/Refractory B-ALL. Blood 2021, 138, 4838. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, J.; Manoharan, E.N.E.A.; Alvin, B.Y.; Milone, M.C. CRISPR/Cas9-mediated Gene Knockout Followed by Negative Selection Leads to a Complete TCR Depletion in orthoCAR19 T Cells. Bio-Protocol 2022, 12, e4485. [Google Scholar] [CrossRef]
- Azangou-Khyavy, M.; Ghasemi, M.; Khanali, J.; Boroomand-Saboor, M.; Jamalkhah, M.; Soleimani, M.; Kiani, J. CRISPR/Cas: From tumor gene editing to T cell-based immunotherapy of cancer. Front. Immunol. 2020, 11, 2062. [Google Scholar] [CrossRef]
- Ghaffari, S.; Khalili, N.; Rezaei, N. CRISPR/Cas9 revitalizes adoptive T-cell therapy for cancer immunotherapy. J. Exp. Clin. Cancer Res. 2021, 40, 269. [Google Scholar] [CrossRef]
- He, Y.; Wu, Y.; Wang, Y.; Wang, X.; Xing, S.; Li, H.; Guo, S.; Yu, X.; Dai, S.; Zhang, G. Applying CRISPR/Cas13 to construct exosomal PD-L1 ultrasensitive biosensors for dynamic monitoring of tumor progression in immunotherapy. Adv. Ther. 2020, 3, 2000093. [Google Scholar] [CrossRef]
- Kozani, P.S.; Shokrgozar, M.A.; Evazalipour, M.; Roudkenar, M.H. CRISPR/Cas9-medaited knockout of endogenous T-cell receptor in Jurkat cells and generation of NY-ESO-1-specific T cells: An in vitro study. Int. Immunopharmacol. 2022, 110, 109055. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zi, Z.; Jin, Y.; Li, G.; Shao, K.; Cai, Q.; Ma, X.; Wei, F. CRISPR/Cas9-mediated PD-1 disruption enhances human mesothelin-targeted CAR T cell effector functions. Cancer Immunol. Immunother. 2019, 68, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Hu, B.; Shao, J.; Shen, B.; Du, J.; Du, Y.; Zhou, J.; Yu, L.; Zhang, L.; Chen, F. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 2016, 6, 20072. [Google Scholar] [CrossRef]
- Wang, S.-W.; Gao, C.; Zheng, Y.-M.; Yi, L.; Lu, J.-C.; Huang, X.-Y.; Cai, J.-B.; Zhang, P.-F.; Cui, Y.-H.; Ke, A.-W. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol. Cancer 2022, 21, 57. [Google Scholar] [CrossRef] [PubMed]
- Deets, K.A.; Doyle, R.N.; Rauch, I.; Vance, R.E. Inflammasome activation leads to cDC1-independent cross-priming of CD8 T cells by epithelial cell-derived antigen. eLife 2021, 10, e72082. [Google Scholar] [CrossRef]
- Buquicchio, F.A.; Satpathy, A.T. Interrogating immune cells and cancer with CRISPR-Cas9. Trends Immunol. 2021, 42, 432–446. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, W.; Xing, L.; Lin, L.; Wen, K.; Tai, Y.-T.; Hideshima, T.; Cang, Y.; Anderson, K.C. Genome-Wide CRISPR-Cas9 Screening Reveals a Role for TRAF2 in Resistance to IMiDs in Multiple Myeloma. Blood 2018, 132, 1917. [Google Scholar] [CrossRef]
- Wei, J.; Long, L.; Zheng, W.; Dhungana, Y.; Lim, S.A.; Guy, C.; Wang, Y.; Wang, Y.-D.; Qian, C.; Xu, B. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 2019, 576, 471–476. [Google Scholar] [CrossRef]
- Li, W. Development and Application of CRISPR-Mediated Genetic Screening in Oncology. In Proceedings of the 2020 7th International Conference on Biomedical and Bioinformatics Engineering, Kyoto, Japan, 6–9 November 2020; pp. 130–135. [Google Scholar]
- Han, P.; Dai, Q.; Fan, L.; Lin, H.; Zhang, X.; Li, F.; Yang, X. Genome-wide CRISPR screening identifies JAK1 deficiency as a mechanism of T-cell resistance. Front. Immunol. 2019, 10, 251. [Google Scholar] [CrossRef]
- Li, F.; Huang, Q.; Luster, T.A.; Hu, H.; Zhang, H.; Ng, W.-L.; Khodadadi-Jamayran, A.; Wang, W.; Chen, T.; Deng, J. In Vivo Epigenetic CRISPR Screen Identifies Asf1a as an Immunotherapeutic Target in Kras-Mutant Lung AdenocarcinomaAsf1a Regulates Sensitivity to Immune-Checkpoint Therapy. Cancer Discov. 2020, 10, 270–287. [Google Scholar] [CrossRef] [PubMed]
- Suresh, S.; Chen, B.; Zhu, J.; Golden, R.J.; Lu, C.; Evers, B.M.; Novaresi, N.; Smith, B.; Zhan, X.; Schmid, V. eIF5B drives integrated stress response-dependent translation of PD-L1 in lung cancer. Nat. Cancer 2020, 1, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Fan, S.; Wen, C.; Du, X. CRISPR/Cas9 for cancer treatment: Technology, clinical applications and challenges. Brief. Funct. Genom. 2020, 19, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Inturi, R.; Jemth, P. CRISPR/Cas9-based inactivation of human papillomavirus oncogenes E6 or E7 induces senescence in cervical cancer cells. Virology 2021, 562, 92–102. [Google Scholar] [CrossRef]
- Wu, X.; Ma, W.; Mei, C.; Chen, X.; Yao, Y.; Liu, Y.; Qin, X.; Yuan, Y. Description of CRISPR/Cas9 development and its prospect in hepatocellular carcinoma treatment. J. Exp. Clin. Cancer Res. 2020, 39, 97. [Google Scholar] [CrossRef]
- Yuen, K.-S.; Chan, C.-P.; Kok, K.-H.; Jin, D.-Y. Mutagenesis and genome engineering of Epstein–Barr virus in cultured human cells by CRISPR/Cas9. In In Vitro Mutagenesis; Springer: Berlin/Heidelberg, Germany, 2017; pp. 23–31. [Google Scholar]
- Mohammadzadeh, I.; Qujeq, D.; Yousefi, T.; Ferns, G.A.; Maniati, M.; Vaghari-Tabari, M. CRISPR/Cas9 gene editing: A new therapeutic approach in the treatment of infection and autoimmunity. IUBMB Life 2020, 72, 1603–1621. [Google Scholar] [CrossRef]
- Wei, J.; Yan, J.; Su, S.; Shao, J.; Zhao, Y.; Xu, Q.; Yang, Y.; Zou, Z.; Huang, X.; Liu, B.J.A.o.O. A phase I/II Trial of CRISPR-Cas9-mediated PD-1 knockout Epstein-Barr virus cytotoxic lymphocytes (EBV-CTLs) for advanced stage EBV associated malignancies-Trial in progress. Ann. Oncol. 2018, 29, v36. [Google Scholar] [CrossRef]
- Wollebo, H.S.; Bellizzi, A.; Kaminski, R.; Hu, W.; White, M.K.; Khalili, K.J.P.o. CRISPR/Cas9 system as an agent for eliminating polyomavirus JC infection. PLoS ONE 2015, 10, e0136046. [Google Scholar] [CrossRef]
- Zhen, S.; Li, X. Application of CRISPR-Cas9 for long noncoding RNA genes in cancer research. Hum. Gene Ther. 2019, 30, 3–9. [Google Scholar] [CrossRef]
- Akram, F.; Ul Haq, I.; Ahmed, Z.; Khan, H.; Ali, M.S. CRISPR-Cas9, a promising therapeutic tool for cancer therapy: A review. Protein Pept. Lett. 2020, 27, 931–944. [Google Scholar]
- Foss, D.V.; Hochstrasser, M.L.; Wilson, R.C.J.T. Clinical applications of CRISPR-based genome editing and diagnostics. Transfusion 2019, 59, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Chow, R.D.; Chen, S. Cancer CRISPR screens in vivo. Trends Cancer 2018, 4, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Ansari, I.; Chaturvedi, A.; Chitkara, D.; Singh, S. CRISPR/Cas mediated epigenome editing for cancer therapy. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2022; pp. 570–583. [Google Scholar]
- Eoh, J.; Gu, L. Biomaterials as vectors for the delivery of CRISPR–Cas9. Biomater. Sci. 2019, 7, 1240–1261. [Google Scholar] [CrossRef]
- Nymark, M.; Sharma, A.K.; Sparstad, T.; Bones, A.M.; Winge, P. A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 2016, 6, 24951. [Google Scholar] [CrossRef]
- Ebina, H.; Misawa, N.; Kanemura, Y.; Koyanagi, Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013, 3, 2510. [Google Scholar] [CrossRef]
- Dai, W.-J.; Zhu, L.-Y.; Yan, Z.-Y.; Xu, Y.; Wang, Q.-L.; Lu, X.-J. CRISPR-Cas9 for in vivo gene therapy: Promise and hurdles. Mol. Ther.-Nucleic Acids 2016, 5, e349. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.-T.; Zhou, N.; Huang, J.; Koirala, P.; Xu, M.; Fung, R.; Wu, F.; Mo, Y.-Y. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 2015, 43, e17. [Google Scholar] [CrossRef] [Green Version]
- Chuang, C.-K.; Chen, C.-H.; Huang, C.-L.; Su, Y.-H.; Peng, S.-H.; Lin, T.-Y.; Tai, H.-C.; Yang, T.-S.; Tu, C.-F. Generation of GGTA1 mutant pigs by direct pronuclear microinjection of CRISPR/Cas9 plasmid vectors. Anim. Biotechnol. 2017, 28, 174–181. [Google Scholar] [CrossRef]
- Senís, E.; Fatouros, C.; Große, S.; Wiedtke, E.; Niopek, D.; Mueller, A.K.; Börner, K.; Grimm, D. CRISPR/Cas9-mediated genome engineering: An adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 2014, 9, 1402–1412. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, F.; Gao, G. CRISPR-based therapeutic genome editing: Strategies and in vivo delivery by AAV vectors. Cell 2020, 181, 136–150. [Google Scholar] [CrossRef]
- Li, A.; Tanner, M.R.; Lee, C.M.; Hurley, A.E.; De Giorgi, M.; Jarrett, K.E.; Davis, T.H.; Doerfler, A.M.; Bao, G.; Beeton, C. AAV-CRISPR gene editing is negated by pre-existing immunity to Cas9. Mol. Ther. 2020, 28, 1432–1441. [Google Scholar] [CrossRef] [PubMed]
| Properties | CRISPR | ZNFs | TALENs | FLP-FRT | CRE-LOXP | Bibliography |
|---|---|---|---|---|---|---|
| DNA binding moiety | RNA | Protein | Protein | Flippase recombination target | Site-specific recombinases | [16,17,18,19,20,21] |
| Ease of targeting multiple targets | High | Low | Low | High | High | |
| Complexity of design | Simple | Very complex | Complex | Simple | Simple | |
| Nuclease | Cas | FokI | FokI | Recombinase | Recombinase | |
| Off-target effects | Variable | Moderate | - | Specific | Specific | |
| Toxicity | Low | Variable to high | Low | Low | Variable | |
| Target recognition size | 22 nucleotides | 18–36 nucleotides | 30–40 nucleotides | 20–35 nucleotides | 38 nucleotides |
| Years | Findings | Bibliography |
|---|---|---|
| 1987 | Discovery of the CRISPR clustered repeats | [38] |
| 2000 | Acceptance of the widespread presence of CRISPR families in prokaryotes | [39] |
| 2002 | The Cas gene was discovered and given the name “CRISPR.” | [34] |
| 2005 | Adaptive immunity function was proposed, and foreign origins of spacers were identified using PAM | [40] |
| 2007 | First experimental proof that CRISPR conferred adaptive immunity | [41] |
| 2008 | CRISPR acts upon DNA target | [42,43] |
| Discovered the function of crRNA | ||
| 2009 | Cleavage of RNA by Type III B Cmr CRISPR complex | [44] |
| 2010 | Cleavage of target DNA via DSBs through Cas9 was guided by spacer sequences | [45] |
| 2011 | Discovery of tracrRNA in conjunction with Cas9 that formed a duplex structure with crRNA | [45] |
| 2012 | Characterization of Cas9’s DNA targeting in vitro | [46] |
| 2013 | Mammalian cell genome editing for the first time | [47] |
| Discovery of dCas9, CRISPRi, and CRISPRa | ||
| 2014 | Crystal structure of Cas9 in guide RNA and target DNA, genome-wide functional screening with Cas9, and crystal structure of apo-cas9 | [48,49,50] |
| 2015 | CRISPR/Cas9 was used to edit human embryos but with prominent off-target effects, CRISPR/Cas9 was used to develop virus-resistant tomato plants, and discovery of Cas 12a (Cpf1) | [51,52] |
| 2016 | The invention of base editor (BE) | [53] |
| Discovery of Cas13a (C2c2) | ||
| 2019 | The invention of nCATS by CRISPR/Cas9 | [54] |
| 2020 | Discovery of the vfCRISPR | [55] |
| Disease | Target Cells | Gene/s | Aim/Repair Pathway | Format/Delivery | Reference |
|---|---|---|---|---|---|
| Myeloid malignancies | LSK | (TET2, RUNX1), (SMC3, TET2), (NF1, EZH2, and DNMT3A) | Knock out/NHEJ | Two-vector system/Lentivirus | [116] |
| Myeloid malignancies | RN2 with constitutive Cas9 expression | 192 chromatin regulatory domains | One-vector system/Lentivirus | [117] | |
| MDS | K562 | SRSF2 | Point mutation/HDR | CRISPR vector and ssODN/Electroporation | [118] |
| MDS, CMML, AML | KBM5 | ASXL1 | Mutation correction/HDR | [119] | |
| MLL | HEK293 | MLL and AF4 | Chromosomal rearrangements/HDR | CRISPR vector and template plasmid/Lipofection | [120] |
| AML | K562 | IDH2 | Knock in/HDR | CRISPR vector and template plasmid/Nucleofection | [121] |
| AML | Primary AML blasts | IDH2 | Mutation correction/HDR | Two-vector system/Lentivirus | [122] |
| SCN | iPSC | HAX1 | CRISPR vector and ssODN/Lipofectamine | [123] | |
| Pediatric AML | Human HPSC | MLL and ENL | Chromosomal rearrangements/NHEJ | One-vector system/Lentivirus | [124] |
| AML | Human HPSC | RUNX1 and ETO | One-vector system/Electroporation | [125] | |
| AML and MDS | Human HPSC | (TET2, U2AF1), (DNMT3A, RUNX1), (ASXL1, TP53), (EZH2, STAG2), (SMC3, TP53, and SRSF2) | Knock out/NHEJ | One-vector system/Lentivirus | [126] |
| MDS | U937 | ASXL1 | Two-vector system/Electroporation | [119] | |
| CHIP | Human HPSC | DNMT3A and TET2 | One-vector system/Lentivirus | [127] | |
| CHIP | LSK | FLT3, DNMT3A, SMC3, EZH2, RUNX1, and NF1 | RNP/Electroporation | [128] | |
| XCGD | PLB | CYBB | Mutation correction/NHEJ | One-vector system/Lentivirus | [129] |
| Different Approaches | Organisms | Genes | References |
|---|---|---|---|
| Gene knockout | Invertebrates | [130,131] | |
| Caenorhabditis elegans | (unc-1, csr-1, dpy-3, and mes-6) | ||
| Silkworm | (BmKMO and BmTH), (BmBLOS2 and Bm-ok), and (Bmtan and BmWnt1) | [132,133,134] | |
| Yeast | (ADE-2) | [135] | |
| Drosophila | Yellow, white, and AGO1 | [136] | |
| Vertebrates | [137,138] | ||
| Chicken | Stra8 and Myostatin | ||
| Human | (MED12 and DMRT1), (OCIAD1 and DMRT3), (NF1 and NF2), (CUL3 and H69), (TADA2B and TADA1), and (MAGEC2 and S100A4) | [139,140,141,142,143] | |
| Mouse | Rp9 | ||
| Zebrafish | cyp19a1a, valopa, and valopb | [144] | |
| Monkey | Ppar-γ and Rag1 | [145] | |
| Plants | [146,147,148,149,150] | ||
| Rice and Arabidopsis Tobacco Sorghum | (IAA2 and CDK), (PDS3 and OsSWEET11), and (TTG1 and OsSWEET14) | ||
| Gene knock-in | Invertebrates | [151] | |
| Silkworm | Bmku70 | ||
| Drosophila | Yellow locus, white locus, and nanos | [136,152] | |
| Caenorhabditis elegans | unc-119 | [130] | |
| Plants | [153] | ||
| Tobacco | No | ||
| Arabidopsis | PDS3 and AtFLS2 | ||
| Rice | WDV | [154] | |
| Vertebrates | [155] | ||
| Mouse | Rosa26, KRAS, p53, and LKB1 | ||
| Chicken | yRad52 | [154] | |
| Pig | COL1A | [156] | |
| Human | DACT1, IFIT1, and EGR1 | [157] | |
| Zebrafish | Fus, Zebrafish th, and tardbp | [158] | |
| Gene knockdown and silencing approaches | Invertebrates | [159] | |
| Caenorhabditis elegant | TRHR-1 | ||
| Drosophila | roX1 and roX2 | ||
| Silkworm | No | ||
| Vertebrates | [160,161] | ||
| Mouse Chicken Pig Zebrafish Human | No EPHA1, mmp21, and Nr1 No | ||
| Gene correction | Invertebrates | [162] | |
| Silkworm | No | ||
| Drosophila | |||
| Caenorhabditis elegans | |||
| Vertebrates | [163,164] | ||
| Zebrafish | No | ||
| Chicken | |||
| Human | MYBPC3 | ||
| Pig | No | ||
| Mouse | Hemophilia B and Pde6b | ||
| Conditional approaches | Invertebrates | [165] | |
| Caenorhabditis elegans | dpy-5, unc-76, and lon-2 | ||
| Silkworm | No | ||
| Drosophila | wg, bam, cid, nos, ms(3)k81, and wg | [166] | |
| Vertebrates | [165] | ||
| Zebrafish | tyr, insra; insrb, and ascl1a | ||
| Human | puroR and Ctnnb1 | ||
| Chicken | No | [138] | |
| Pig | PFFs | [167] | |
| Mouse | Kras, Mecp2, Lkb1, Ispd, and p53 | [158,168] | |
| Objective Trails | Cell Markers | Library Markers | CRISPR/Cas9 Delivery Methods | Immune Selective Pressure | Significant Targets | References |
|---|---|---|---|---|---|---|
| Antigen processing and presentation pathway (IFN-y-pathway) | (Melanoma cell lines) | (411,123 sgRNAs targeting >50 genes) | (Lentiviral vector) | (NY-ESO-1-specific TCR T cells) | APLNR | [190] |
| 9872 sgRNAs targeting 2368 genes | (PD-1 blockade) | PTPN2 | [191] | |||
| (T-cell activation regulators) | (Jurkat T cells) | Total of 250,000 sgRNAs targeting all distinct Refseq-annotated (hg19) protein-coding genes. | - | FAM49B | [192] | |
| (T-cell stimulation regulators) | (Primary human CD8+ T cells) | (19,114 genes targeted by 77,441 sgRNAs) | (Lentiviral infection with Cas9 protein electroporation through single-guide RNA-sgRNA) | - | (RASA2, SCS1), (CBLB, TCEB2) | [193] |
| Chromatin regulators | B16F10 melanoma cells | >100 genes | (Lentiviral vector) | (Pmei-1 T cells, OT-I T cells) | (PBAF, PBRM1, ARID2) | [194] |
| Tumor infiltration and degranulation regulators | (Human CD8 T cells and mouse) | (128,209 specific genes) | - | (DHX-37) | [195] | |
| (IFNg-independent signaling pathway) | (IFNGR1-deficient melanoma cells) | (GeCKO library) | (MART-1 T cells) | (TRAF-2) | [196] | |
| (Metabolic regulators of T cell) | (OT-1 T cells) | (3017 genes linked with metabolism) | - | (Regnase-1) | [197] | |
| (Targets of cell membrane) | (CD8 T cells of mouse) | (1658 genes encoding membrane protein of mouse) | (Sleeping Beauty transposon system and AVV vector) | - | (Lag 3, Mgat5), (PDIA3, Emp-1) | [198] |
| Antigen processing and presentation pathway (IFN-y-pathway) | Melanoma cells (B16-F10) | (Brief genome-wide sgRNA library) | (Lentiviral vector) | (Mouse NK cells) | (Jak-1) | [199] |
| Epigenetic regulators | (KrasG12D/Trp53−/− lung cancer cells) | (524 genes encoding epigenetic regulation) | (Anti-PD-1 antibody) | (Asf1a) | [200] | |
| (Regulators of PD-L1) | (Adenocarcinoma cell line H358 cells of human lung) | (GeCKO version 2 library of human) | - | (eIF5B) | [201] | |
| (Gene regulatory programs in Foxp-3 expression) | (Primary mouse Tregs) | (Brie library) | (Retroviral vector) | - | (Rnf20, Usp22) | [195] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabaan, A.A.; AlSaihati, H.; Bukhamsin, R.; Bakhrebah, M.A.; Nassar, M.S.; Alsaleh, A.A.; Alhashem, Y.N.; Bukhamseen, A.Y.; Al-Ruhimy, K.; Alotaibi, M.; et al. Application of CRISPR/Cas9 Technology in Cancer Treatment: A Future Direction. Curr. Oncol. 2023, 30, 1954-1976. https://doi.org/10.3390/curroncol30020152
Rabaan AA, AlSaihati H, Bukhamsin R, Bakhrebah MA, Nassar MS, Alsaleh AA, Alhashem YN, Bukhamseen AY, Al-Ruhimy K, Alotaibi M, et al. Application of CRISPR/Cas9 Technology in Cancer Treatment: A Future Direction. Current Oncology. 2023; 30(2):1954-1976. https://doi.org/10.3390/curroncol30020152
Chicago/Turabian StyleRabaan, Ali A., Hajir AlSaihati, Rehab Bukhamsin, Muhammed A. Bakhrebah, Majed S. Nassar, Abdulmonem A. Alsaleh, Yousef N. Alhashem, Ammar Y. Bukhamseen, Khalil Al-Ruhimy, Mohammed Alotaibi, and et al. 2023. "Application of CRISPR/Cas9 Technology in Cancer Treatment: A Future Direction" Current Oncology 30, no. 2: 1954-1976. https://doi.org/10.3390/curroncol30020152
APA StyleRabaan, A. A., AlSaihati, H., Bukhamsin, R., Bakhrebah, M. A., Nassar, M. S., Alsaleh, A. A., Alhashem, Y. N., Bukhamseen, A. Y., Al-Ruhimy, K., Alotaibi, M., Alsubki, R. A., Alahmed, H. E., Al-Abdulhadi, S., Alhashem, F. A., Alqatari, A. A., Alsayyah, A., Farahat, R. A., Abdulal, R. H., Al-Ahmed, A. H., ... Mohapatra, R. K. (2023). Application of CRISPR/Cas9 Technology in Cancer Treatment: A Future Direction. Current Oncology, 30(2), 1954-1976. https://doi.org/10.3390/curroncol30020152









