Cost Savings of Expedited Care with Upfront Next-Generation Sequencing Testing versus Single-Gene Testing among Patients with Metastatic Non-Small Cell Lung Cancer Based on Current Canadian Practices
Abstract
:1. Introduction
2. Materials and Methods
2.1. Model Framework
2.2. Model Structure
2.2.1. NGS
2.2.2. Exclusionary
2.2.3. Sequential
2.2.4. Non-Comprehensive Sequential
2.2.5. Rapid Panel
2.3. Model Assumptions
2.4. Model Inputs
2.4.1. Population Inputs
2.4.2. Clinical Inputs
2.4.3. Cost Inputs
2.5. Model Outputs
2.6. Impact of Increasing the Proportion of Patients Tested with NGS
2.7. Sensitivity Analysis
3. Results
3.1. Total Cost for Patients Undergoing Testing with NGS versus Single-Gene Testing Strategies
3.2. Impact of Increasing the Proportion of Patients Tested with NGS
3.3. Sensitivity Analysis
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Canadian Cancer Statistics Advisory Committee in collaboration with the Canadian Cancer Society. Canadian Cancer Statistics; Statistics Canada and the Public Health Agency of Canada: Toronto, ON, Canada, 2021. [Google Scholar]
- Canadian Cancer Statistics Advisory Committee. Canadian Cancer Statistics: A 2020 Special Report on Lung Cancer; Canadian Cancer Society: Toronto, ON, Canada, 2020. [Google Scholar]
- Lung Cancer Canada. An Overview of Lung Cancer. Available online: https://www.lungcancercanada.ca/Lung-Cancer.aspx (accessed on 29 September 2022).
- Barlesi, F.; Mazieres, J.; Merlio, J.P.; Debieuvre, D.; Mosser, J.; Lena, H.; Ouafik, L.; Besse, B.; Rouquette, I.; Westeel, V.; et al. Routine molecular profiling of cancer: Results of a one-year nationwide program of the French CooperativeThoracic Intergroup (IFCT) for advanced non-small cell lung cancer (NSCLC) patients. Lancet 2016, 387, 1415–1426. [Google Scholar] [CrossRef] [PubMed]
- Melosky, B.; Blais, N.; Cheema, P.; Couture, C.; Juergens, R.; Kamel-Reid, S.; Tsao, M.; Wheatley-Price, P.; Xu, Z.; Ionescu, D. Standardizing Biomarker Testing for Canadian Patients with Advanced Lung Cancer. Curr. Oncol. 2018, 25, 73–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, K.M.; Sheffield, B.S.; Yip, S.; Lakzadeh, P.; Qian, C.; Nam, J. Comprehensive Genomic Profiling for Non-Small-Cell Lung Cancer: Health and Budget Impact. Curr. Oncol. 2020, 27, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Kopetz, S.; Shaw, K.R.M.; Lee, J.J.; Zhang, J.; Litzenburger, B.; Holla, V.; Kinyua, W.; Broaddus, E.; Daniels, M.S.; Meric-Bernstam, F.; et al. Use of a Targeted Exome Next-Generation Sequencing Panel Offers Therapeutic Opportunity and Clinical Benefit in a Subset of Patients With Advanced Cancers. JCO Precis. Oncol. 2019, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Dahlman, K.H.; Knol, J.; Gilbert, J.; Puzanov, I.; Means-Powell, J.; Balko, J.M.; Lovly, C.M.; Murphy, B.A.; Goff, L.W.; et al. Enabling a Genetically Informed Approach to Cancer Medicine: A Retrospective Evaluation of the Impact of Comprehensive Tumor Profiling Using a Targeted Next-Generation Sequencing Panel. Oncologist 2014, 19, 616–622. [Google Scholar] [CrossRef] [Green Version]
- Drilon, A.; Wang, L.; Arcila, M.E.; Balasubramanian, S.; Greenbowe, J.R.; Ross, J.S.; Stephens, P.; Lipson, D.; Miller, V.A.; Kris, M.G.; et al. Broad, hybrid capture–based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin. Cancer Res. 2015, 21, 3631–3639. [Google Scholar] [CrossRef] [Green Version]
- Pennell, N.A.; Arcila, M.E.; Gandara, D.R.; West, H. Biomarker Testing for Patients With Advanced Non–Small Cell Lung Cancer: Real-World Issues and Tough Choices. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 531–542. [Google Scholar] [CrossRef]
- Yip, S.; Christofides, A.; Banerji, S.; Downes, M.R.; Izevbaye, I.; Lo, B.; MacMillan, A.; McCuaig, J.; Stockley, T.; Yousef, G.M.; et al. A Canadian Guideline on the Use of Next-Generation Sequencing in Oncology. Curr. Oncol. 2019, 26, 241–254. [Google Scholar] [CrossRef] [Green Version]
- Pennell, N.; Mutebi, A.; Zhou, Z.-Y.; Ricculli, M.L.; Tang, W.; Wang, H.; Guerin, A.; Arnhart, T.; Dalal, A.; Sasane, M.; et al. Economic Impact of Next-Generation Sequencing Versus Single-Gene Testing to Detect Genomic Alterations in Metastatic Non–Small-Cell Lung Cancer Using a Decision Analytic Model. JCO Precis. Oncol. 2019, 3, 1–9. [Google Scholar] [CrossRef]
- Dalal, A.; Guerin, A.; Mutebi, A.; Culver, K. Economic analysis of BRAF gene mutation testing in real world practive using claims data: Costs of single gene versus panel tests in patients with lung cancer. J. Med. Econ. 2018, 21, 649–655. [Google Scholar] [CrossRef]
- Vanderpoel, J.; Stevens, A.L.; Emond, B.; Lafeuille, M.-H.; Hilts, A.; Lefebvre, P.; Morrison, L. Total cost of testing for genomic alterations associated with next-generation sequencing versus polymerase chain reaction testing strategies among patients with metastatic non-small cell lung cancer: Total cost of NGS vs. PCR genomic testing in mNSCLC. J. Med. Econ. 2022, 25, 457–468. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, C.; Pataky, R.; Bremner, K.E.; Rangrej, J.; Chan, K.K.; Cheung, W.Y.; Hoch, J.S.; Peacock, S.; Krahn, M.D. Estimating the cost of cancer care in British Columbia and Ontario: A Canadian inter-provincial comparison. Healthc. Policy 2017, 12, 95. [Google Scholar] [CrossRef] [Green Version]
- Bruno, D.S.; Hess, L.M.; Li, X.; Su, E.W.; Zhu, Y.E.; Patel, M. Racial disparities in biomarker testing and clinical trial enrollment in non-small cell lung cancer (NSCLC). J. Clin. Oncol. 2021, 39, 9005. [Google Scholar] [CrossRef]
- Robert, N.J.; Espirito, J.L.; Chen, L.; Nwokeji, E.; Karhade, M.; Evangelist, M.; Spira, A.; Neubauer, M.; Bullock, S.; Walberg, J.; et al. Biomarker testing and tissue journey among patients with metastatic non-small cell lung cancer receiving first-line therapy in The US Oncology Network. Lung Cancer 2022, 166, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Martos, G.; Pabani, A.; Bebb, D.G.; Gibson, A.W.; Dean, M.L.; Petersen, L. Molecular characteristics of BRAF mutated non-small cell lung cancer and therapeutic outcomes: Multi-institution study. J. Clin. Oncol. 2021, 39, e21029. [Google Scholar] [CrossRef]
- Zer, A.; Ding, K.; Lee, S.M.; Goss, G.D.; Seymour, L.; Ellis, P.M.; Hackshaw, A.; Bradbury, P.A.; Han, L.; O’Callaghan, C.J.; et al. Pooled Analysis of the Prognostic and Predictive Value of KRAS Mutation Status and Mutation Subtype in Patients with Non–Small Cell Lung Cancer Treated with Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors. J. Thorac. Oncol. 2016, 11, 312–323. [Google Scholar] [CrossRef] [Green Version]
- Laskin, J.; Liu, S.; Tolba, K.; Heining, C.; Schlenk, R.; Cheema, P.; Cadranel, J.; Jones, M.; Drilon, A.; Cseh, A. NRG1 fusion-driven tumors: Biology, detection, and the therapeutic role of afatinib and other ErbB-targeting agents. Ann. Oncol. 2020, 31, 1693–1703. [Google Scholar] [CrossRef]
- Ou, S.-H.I.; Sokol, E.; Trabucco, S.; Jin, D.; Frampton, G.; Graziano, S.; Elvin, J.; Vergilio, J.-A.; Killian, J.; Ngo, N.; et al. NTRK1-3 genomic fusions in non-small cell lung cancer (NSCLC) determined by comprehensive genomic profiling. Ann. Oncol. 2019, 30, v638. [Google Scholar] [CrossRef]
- Chu, Q.; Agha, A.; Devost, N.; Walton, R.N.; Ghosh, S.; Ho, C. Biopsy on Progression in Patients with EGFR Mutation–Positive Advanced Non-Small-Cell Lung Cancer—A Canadian Experience. Curr. Oncol. 2020, 27, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Stargardter, M.; McBride, A.; Tosh, J.; Sachdev, R.; Yang, M.; Ambavane, A.; Mittal, M.; Vioix, H.; Liu, F.X. Budget impact of tepotinib in the treatment of adult patients with metastatic non-small cell lung cancer harboring METex14 skipping alterations in the United States. J. Med. Econ. 2021, 24, 816–827. [Google Scholar] [CrossRef]
- Harvey, M.J.; Cunningham, R.; Sawchyn, B.; Montesion, M.; Reddy, P.; McBride, A.; Chawla, A.J. Budget Impact Analysis of Comprehensive Genomic Profiling in Patients With Advanced Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2021, 5, 1611–1624. [Google Scholar] [CrossRef]
- Signorovitch, J.; Zhou, Z.; Ryan, J.; Anhorn, R.; Chawla, A. Budget impact analysis of comprehensive genomic profiling in patients with advanced non-small cell lung cancer. J. Med. Econ. 2018, 22, 140–150. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Hovelson, D.H.; Suga, J.M.; Anderson, D.M.; Koh, H.A.; Dees, E.C.; McNulty, B.; Burkard, M.E.; Guarino, M.; Khatri, J.; et al. Real-World Performance of a Comprehensive Genomic Profiling Test Optimized for Small Tumor Samples. JCO Precis. Oncol. 2021, 5, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- VanderLaan, P.A.; Rangachari, D.; Majid, A.; Parikh, M.S.; Gangadharan, S.P.; Kent, M.S.; McDonald, D.C.; Huberman, M.S.; Kobayashi, S.S.; Costa, D.B. Tumor biomarker testing in non-small-cell lung cancer: A decade of change. Lung Cancer 2018, 116, 90–95. [Google Scholar] [CrossRef]
- Lim, C.; Sekhon, H.; Cutz, J.; Hwang, D.; Kamel-Reid, S.; Carter, R.; Santos, G.D.C.; Waddell, T.; Binnie, M.; Patel, M.; et al. Improving Molecular Testing and Personalized Medicine in Non-Small-Cell Lung Cancer in Ontario. Curr. Oncol. 2017, 24, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Makarem, M.; Ezeife, D.A.; Smith, A.C.; Li, J.J.N.; Law, J.H.; Tsao, M.-S.; Leighl, N.B. Reflex ROS1 IHC Screening with FISH Confirmation for Advanced Non-Small Cell Lung Cancer—A Cost-Efficient Strategy in a Public Healthcare System. Curr. Oncol. 2021, 28, 3268–3279. [Google Scholar] [CrossRef]
- Johnston, K.M.; Sheffield, B.S.; Yip, S.; Lakzadeh, P.; Qian, C.; Nam, J. Costs of in-house genomic profiling and implications for economic evaluation: A case example of non-small cell lung cancer (NSCLC). J. Med. Econ. 2020, 23, 1123–1129. [Google Scholar] [CrossRef]
- Ontario Ministry of Health. Ontario Health Insurance Plan Schedule of Benefits and Fees for Physician Services; Ontario Ministry of Health: Toronto, ON, Canada, 2021.
- Canadian Institute for Health Information. Patient Cost Estimates by Jurisdiction, Case Mix Group and Age Group, 2014–2015 to 2018–2019; Canadian Institute for Health Information: Ottawa, ON, Canada, 2021. [Google Scholar]
- Statistics Canada. Consumer Price Index, Monthly, Not Seasonally Adjusted; Statistics Canada: Ottawa, ON, Canada, 2021.
- Forsythe, M.L.; Alwithenani, A.; Bethune, D.; Castonguay, M.; Drucker, A.; Flowerdew, G.; French, D.; Fris, J.; Greer, W.; Henteleff, H.; et al. Molecular profiling of non-small cell lung cancer. PLoS ONE 2020, 15, e0236580. [Google Scholar] [CrossRef] [PubMed]
- Shiau, C.J.; Babwah, J.P.; da Cunha Santos, G.; Sykes, J.R.; Boerner, S.L.; Geddie, W.R.; Leighl, N.B.; Wei, C.; Kamel-Reid, S.; Hwang, D.M. Sample features associated with success rates in population-based EGFR mutation testing. J. Thora. Oncol. 2014, 9, 947–956. [Google Scholar] [CrossRef] [Green Version]
- Alwithenani, A.; Bethune, D.; Castonguay, M.; Drucker, A.; Flowerdew, G.; Forsythe, M.; French, D.; Fris, J.; Greer, W.; Henteleff, H.; et al. Profiling non-small cell lung cancer reveals that PD-L1 is associated with wild type EGFR and vascular invasion, and immunohistochemistry quantification of PD-L1 correlates weakly with RT-qPCR. PLoS ONE 2021, 16, e0251080. [Google Scholar] [CrossRef] [PubMed]
- Statistics Canada. Population Estimates on July 1st, by Age and Sex; Statistics Canada: Ottawa, ON, Canada, 2022. [Google Scholar]
- Zheng, Y.; Vioix, H.; Liu, F.X.; Singh, B.; Sharma, S.; Sharda, D. Diagnostic and economic value of biomarker testing for targetable mutations in non-small-cell lung cancer: A literature review. Futur. Oncol. 2022, 18, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Perdrizet, K.; Stockley, T.; Law, J.H.; Shabir, M.; Zhang, T.; Le, L.W.; Lau, A.; Tsao, M.S.; Pal, P.; Cabanero, M.; et al. Non-small cell lung cancer (NSCLC) next generation sequencing (NGS) using the Oncomine Comprehensive Assay (OCA) v3: Integrating expanded genomic sequencing into the Canadian publicly funded health care model. J. Clin. Oncol. 2019, 37, 2620. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Z.; Zhan, J.; Zhao, X.; Chen, X.; Xiao, L.; Wu, K.; Ma, Y.; Li, M.; Yang, Y.; et al. Utility of comprehensive genomic profiling in directing treatment and improving patient outcomes in advanced non-small cell lung cancer. BMC Med. 2021, 19, 223. [Google Scholar] [CrossRef] [PubMed]
- Loong, H.H.; Wong, C.K.; Chan, C.P.; Chang, A.; Zhou, Z.-Y.; Tang, W.; Gibbs, M. Clinical and Economic Impact of Upfront Next-Generation Sequencing for Metastatic NSCLC in East Asia. JTO Clin. Res. Rep. 2022, 3, 100290. [Google Scholar] [CrossRef]
- Cheung, W.Y.; Butler, J.R.; Kliewer, E.V.; Demers, A.A.; Musto, G.; Welch, S.; Sivananthan, G.; Navaratnam, S. Analysis of wait times and costs during the peri-diagnostic period for non-small cell lung cancer. Lung Cancer 2011, 72, 125–131. [Google Scholar] [CrossRef] [PubMed]
- David, E.A.; Daly, M.E.; Li, C.-S.; Chiu, C.-L.; Cooke, D.T.; Brown, L.M.; Melnikow, J.; Kelly, K.; Canter, R.J. Increasing Rates of No Treatment in Advanced-Stage Non–Small Cell Lung Cancer Patients: A Propensity-Matched Analysis. J. Thorac. Oncol. 2017, 12, 437–445. [Google Scholar] [CrossRef] [Green Version]
- Khozin, S.; Blumenthal, G.M.; Jiang, X.; He, K.; Boyd, K.; Murgo, A.; Justice, R.; Keegan, P.; Pazdur, R.U.S. Food and Drug Administration Approval Summary: Erlotinib for the First-Line Treatment of Metastatic Non-Small Cell Lung Cancer with Epidermal Growth Factor Receptor Exon 19 Deletions or Exon 21 (L858R) Substitution Mutations. Oncologist 2014, 19, 774–779. [Google Scholar] [CrossRef] [Green Version]
- Ramalingam, S.S.; Vansteenkiste, J.; Planchard, D.; Cho, B.C.; Gray, J.E.; Ohe, Y.; Zhou, C.; Reungwetwattana, T.; Cheng, Y.; Chewaskulyong, B.; et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 2020, 382, 41–50. [Google Scholar] [CrossRef]
- Mok, T.; Camidge, D.; Gadgeel, S.; Rosell, R.; Dziadziuszko, R.; Kim, D.-W.; Pérol, M.; Ou, S.-H.; Ahn, J.; Shaw, A.; et al. Updated overall survival and final progression-free survival data for patients with treatment-naive advanced ALK-positive non-small-cell lung cancer in the ALEX study. Ann. Oncol. 2020, 31, 1056–1064. [Google Scholar] [CrossRef]
- Stewart, D.J.; Maziak, D.E.; Moore, S.M.; Brule, S.Y.; Gomes, M.; Sekhon, H.; Dennie, C.; Lo, B.; Fung-Kee-Fung, M.; Bradford, J.; et al. The need for speed in advanced non-small cell lung cancer: A population kinetics assessment. Cancer Med. 2021, 10, 9040–9046. [Google Scholar] [CrossRef]
- Bartlett, J.; Amemiya, Y.; Arts, H.; Bayani, J.; Eng, B.; Grafodatskaya, D.; Reid, S.K.; Lariviere, M.; Lo, B.; McClure, R.; et al. Multisite verification of the accuracy of a multi-gene next generation sequencing panel for detection of mutations and copy number alterations in solid tumours. PLoS ONE 2021, 16, e0258188. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.M.; Hensing, T.; Schrock, A.B.; Allen, J.; Sanford, E.; Gowen, K.; Kulkarni, A.; He, J.; Suh, J.H.; Lipson, D.; et al. Comprehensive Genomic Profiling Identifies a Subset of Crizotinib-Responsive ALK-Rearranged Non-Small Cell Lung Cancer Not Detected by Fluorescence In Situ Hybridization. Oncologist 2016, 21, 762–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrock, A.B.; Frampton, G.M.; Herndon, D.; Greenbowe, J.R.; Wang, K.; Lipson, D.; Yelensky, R.; Chalmers, Z.R.; Chmielecki, J.; Elvin, J.A. Comprehensive genomic profiling identifies frequent drug-sensitive EGFR exon 19 deletions in NSCLC not identified by prior molecular testing. Clin. Cancer Res. 2016, 22, 3281–3285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauml, J.; Viteri, S.; Minchom, A.; Bazhenova, L.; Ou, S.; Schaffer, M.; Le Croy, N.; Riley, R.; Mahadevia, P.; Girard, N. FP07.12 Underdiagnosis of EGFR Exon 20 Insertion Mutation Variants: Estimates from NGS-based Real-World Datasets. J. Thorac. Oncol. 2021, 16, S208–S209. [Google Scholar] [CrossRef]
- Yuan, M.; Huang, L.-L.; Chen, J.-H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef] [Green Version]
- Ai, X.; Guo, X.; Wang, J.; Stancu, A.L.; Joslin, P.M.; Zhang, D.; Zhu, S. Targeted therapies for advanced non-small cell lung cancer. Oncotarget 2018, 9, 37589–37607. [Google Scholar] [CrossRef] [Green Version]
- Weymann, D.; Dragojlovic, N.; Pollard, S.; Regier, D.A. Allocating healthcare resources to genomic testing in Canada: Latest evidence and current challenges. J. Community Genet. 2019, 13, 467–476. [Google Scholar] [CrossRef]
- Hirsh, V.; Melosky, B.; Goss, G.; Morris, D.; Morzycki, W. A Personalized Approach to Treatment: Use of egfr Tyrosine Kinase Inhibitors for the Treatment of Non-Small-Cell Lung Cancer in Canada. Curr. Oncol. 2012, 19, 78–90. [Google Scholar] [CrossRef] [Green Version]
- Orlov-Slavu, M.C.; Popa, A.M.; Tulin, A.; Stoian, A.P.; Poiana, C.; Paleru, C.; Calu, V.; Nitipir, C. The Utility of Next-Generation Sequencing in the Treatment Decision-Making for Metastatic Non-Small-Cell Lung Cancer. Cureus 2021, 13, e16919. [Google Scholar] [CrossRef]


| Population and Clinical Inputs | Value | Source | |
|---|---|---|---|
| Population | Number of covered lives | 1,000,000 | Assumption |
| % of adults with LC | 0.066% | Canadian Cancer Statistics, 2020 [2] | |
| % with NSCLC among LC population | 88.0% | Canadian Cancer Statistics, 2020 [2] | |
| % diagnosed metastatic, Stage III and Stage IV, among NSCLC population | 65.7% | Canadian Cancer Statistics, 2020 [2] | |
| Testing strategy distribution | % undergoing testing with tissue NGS | 50.0% | [6,16,17] |
| % undergoing sequential testing | 5.0% | Expert Opinion | |
| % undergoing exclusionary testing | 10.0% | Expert Opinion | |
| % undergoing non-comprehensive testing | 25.0% | Expert Opinion | |
| % undergoing rapid panel testing | 10.0% | Expert Opinion | |
| Frequency of genomic alterations | % testing positive for EGFR mutation a | 17.0% | Johnston et al., 2020 [6] |
| % testing positive for ALK mutation a | 3.0% | Johnston et al., 2020 [6] | |
| % testing positive for ROS1 mutation a | 1.0% | Johnston et al., 2020 [6] | |
| % testing positive for BRAF mutation | 2.0% | Johnston et al., 2020 [6] | |
| % testing positive for V600E mutation among those with BRAF mutation a | 64.5% | Martos et al., 2021 [18] | |
| % testing positive for KRAS mutation | 25.0% | Johnston et al., 2020 [6] | |
| % testing positive for G12C mutation among those with KRAS mutation a | 41.0% | Zer et al., 2015 [19] | |
| % testing positive for MET mutation a | 1.0% | Johnston et al., 2020 [6] | |
| % testing positive for HER2/ERBB2 mutation a | 3.0% | Johnston et al., 2020 [6] | |
| % testing positive for RET mutation a | 1.0% | Johnston et al., 2020 [6] | |
| % testing positive for NRG1 mutation a | 0.3% | Laskin et al., 2020 [20] | |
| % testing positive for NTRK1 mutation a | 0.1% | Ou et al., 2019 [21] | |
| % testing positive for NTRK2 mutation a | 0.01% | Ou et al., 2019 [21] | |
| % testing positive for NTRK3 mutation a | 0.01% | Ou et al., 2019 [21] | |
| Rebiopsies | % needing rebiopsy after each test (NGS) | 12.1% | Tomlins et al., 2021 [26] |
| % needing rebiopsy after each test (single-gene testing strategies) | 5.5% | VanderLaan et al., 2018 [27] | |
| % receiving rebiopsy after each test (NGS and single-gene testing strategies) | 30.0% | Chu et al., 2020 [22] | |
| % failed rebiopsy (of each rebiopsy attempted) | 12.0% | Lim et al., 2017 [28] | |
| % with rebiopsy complications | 9.0% | Chu et al., 2020 [22] | |
| Time-to-test and clinical turnaround time (weeks) | Time-to-test result, NGS | 3.0 | Makarem et al., 2021 [29] |
| Time-to-test result, EGFR, ALK, ROS1 mutations, per mutation | 1.0 | Makarem et al., 2021 [29] | |
| Time-to-test result, KRAS | 3.0 | Makarem et al., 2021 [29] | |
| Time-to-test result, all other mutations, per mutation | 1.5 | Pennell et al., 2019 [12] | |
| Time-to-oncologist visit | 2.0 | Makarem et al., 2021 [29] | |
| Time-to-rebiopsy | 2.0 | Makarem et al., 2021 [29] | |
| Cost inputs | (2021 CAD) b | Source | |
| Testing | NGS (tissue) | CAD 1000 | Makarem et al., 2021 [29] |
| EGFR c | CAD 240 | Makarem et al., 2021 [29] | |
| ALK, NRTK1/2/3, per mutation d | CAD 100 | Makarem et al., 2021 [29] | |
| ROS1, MET, RET, NRG1, per mutation e | CAD 400 | Makarem et al., 2021 [29] | |
| BRAF, KRAS, HER2/ERBB2, per mutation c | CAD 200 | Makarem et al., 2021 [29] | |
| PD-L1 d | CAD 138 | Johnston et al., 2020 [30] | |
| Medical | Rebiopsy f | CAD 368 | Ontario Schedule of Benefits, 2021 [31] |
| Rebiopsy complication | CAD 5411 | Canadian Institute for Health Information, 2019 [32] | |
| Interventional radiology visit | CAD 73 | Ontario Schedule of Benefits, 2021 [31] | |
| Oncologist visit | CAD 157 | Ontario Schedule of Benefits, 2021 [31] | |
| Oncologist follow-up visit | CAD 105 | Ontario Schedule of Benefits, 2021 [31] | |
| Estimated cost associated with delaying care (per week) | CAD 406 | De Oliveira et al., 2017 [15] | |
| NGS | Sequential | Exclusionary | Non-Comprehensive | Rapid Panel | All Four Single-Gene Strategies Combined a | |
|---|---|---|---|---|---|---|
| Clinical outputs | ||||||
| Proportion of patients tested (%) | 50.0 | 5.0 | 10.0 | 25.0 | 10.0 | 50.0 |
| Total patients tested (n) | 190.8 | 19.1 | 38.2 | 95.4 | 38.2 | 190.9 |
| Proportion of patients tested positive for a mutation with an approved targeted therapy b (%) | 38.0 | 31.7 | 29.3 | 20.3 | 34.4 | 26.1 |
| Patients tested positive for a mutation with an approved targeted therapy b (n) | 72.4 | 6.1 | 11.2 | 19.4 | 13.1 | 49.7 |
| Number of visits c (n) | 384.8 | 55.9 | 103.7 | 271.1 | 107.4 | 538.2 |
| Number of visits per patient c (n) | 2.0 | 2.9 | 2.7 | 2.8 | 2.8 | 2.8 |
| Number of rebiopsies (n) | 3.2 | 2.4 | 4.4 | 3.8 | 0.8 | 11.5 |
| Number of rebiopsies per patient (n) | 0.02 | 0.13 | 0.12 | 0.04 | 0.02 | 0.06 |
| Time to initiation of appropriate targeted therapy, per patient d (weeks) | 5.1 | 16.1 | 15.3 | 6.4 | 7.0 | 9.2 |
| Cost outputs | ||||||
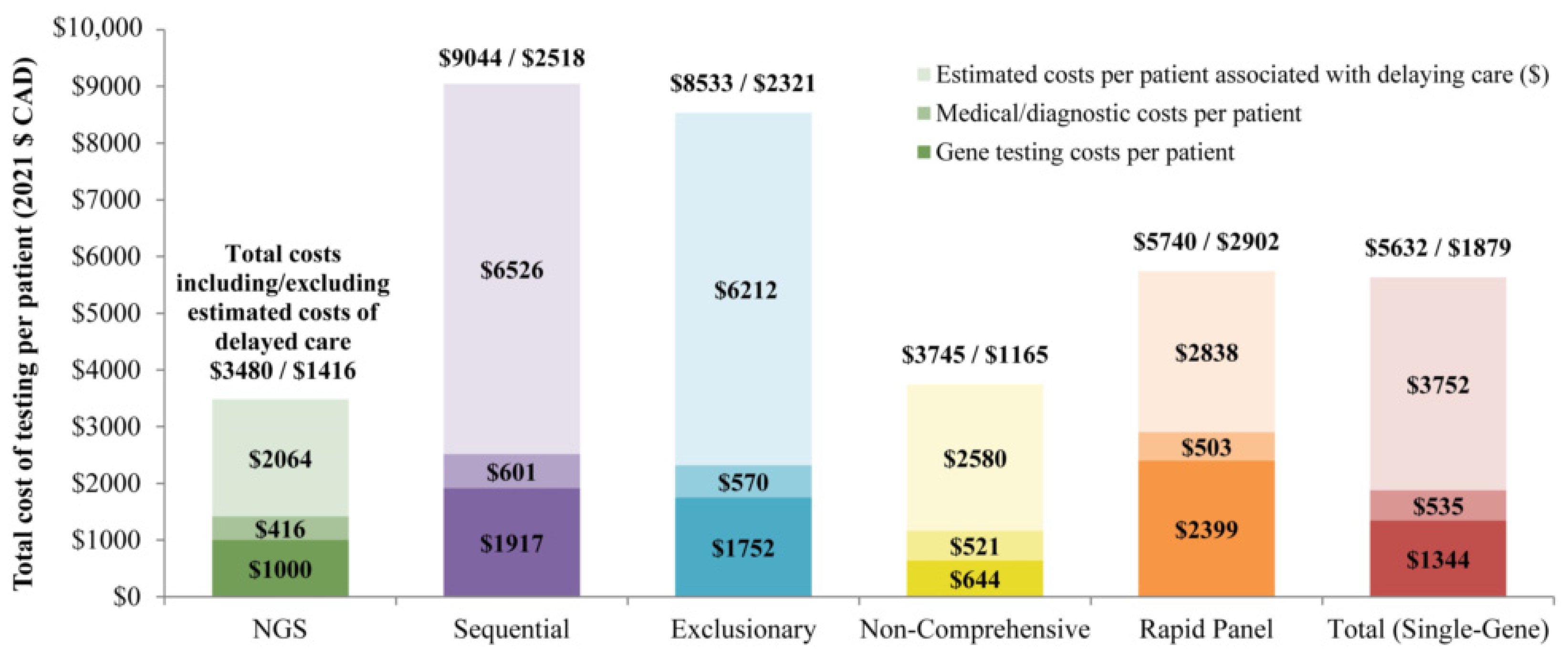
| Total costs e (CAD) | 270,117 | 48,088 | 88,680 | 111,135 | 110,869 | 358,773 |
| Total costs per patient (CAD) | 1416 | 2518 | 2321 | 1165 | 2902 | 1879 |
| Total costs including estimated costs associated with delaying care (CAD) | 663,998 | 172,740 | 325,963 | 357,245 | 219,275 | 1,075,222 |
| Estimated costs per patient associated with delaying care f (CAD) | 2064 | 6526 | 6212 | 2580 | 2838 | 3752 |
| Total costs per patient including estimated costs associated with delaying care (CAD) | 3480 | 9044 | 8533 | 3745 | 5740 | 5632 |
| Input | Low Case | High Case | Sources | |
|---|---|---|---|---|
| Clinical Event Rates | Proportion of patients needing a rebiopsy | 9.7% | 14.5% | Assumption b |
| Proportion of patients with rebiopsy complications | 7.2% | 10.8% | Assumption c | |
| Rate of successful identification of EGFR mutation | 7.0% | 20.6% | Forsyth et al., 2020 [34]; Shiau et al., 2014 [35] | |
| Rate of successful identification of KRAS mutation | 20.0% | 36.1% | Assumption c; Alwithenani et al., 2021 [36] | |
| Testing Costs | NGS (tissue) | CAD 750 | CAD1250 | Assumption d |
| EGFR | CAD 200 | CAD288 | Makarem et al., 2021 [29]; Assumption e | |
| KRAS | CAD 160 | CAD 240 | Assumption b | |
| Diagnostic Testing/Medical Costs | PD-L1 | CAD 104 | CAD 166 | Makarem et al., 2021 [29]; Assumption f |
| Rebiopsy | CAD 294 | CAD 442 | Assumption b | |
| Rebiopsy complication | CAD 4329 | CAD 6493 | Assumption b | |
| Interventional radiology visit | CAD 58 | CAD 88 | Assumption b | |
| Oncologist visit for consultation | CAD 126 | CAD 188 | Assumption b | |
| Oncologist follow-up visit | CAD 84 | CAD 126 | Assumption b | |
| Cost of delaying treatment (per week) | CAD 325 | CAD 487 | Assumption b |
| NGS | Sequential | Exclusionary | Non-Comprehensive Sequential | Rapid Panel | Total (All Strategies) a | |
|---|---|---|---|---|---|---|
| Base case with 50% of patients tested by NGS | ||||||
| Proportion of patients tested (%) | 50.0 | 5.0 | 10.0 | 25.0 | 10.0 | 100.0 |
| Number of patients tested by strategy (n) | 190.8 | 19.1 | 38.2 | 95.4 | 38.2 | 381.7 |
| Proportion of patients tested positive for a mutation with an approved targeted therapy2 (%) | 38.0 | 32.1 | 29.3 | 20.3 | 34.4 | 32.0 |
| Patients tested positive for a mutation with an approved targeted therapy b (n) | 72.4 | 6.1 | 11.2 | 19.4 | 13.1 | 122.3 |
| Time to initiation of appropriate targeted therapy, per patient c (weeks) | 5.1 | 16.1 | 15.3 | 6.4 | 7.0 | 7.2 |
| Total annual cost at the plan level d (CAD) | 270,117 | 48,088 | 88,680 | 111,135 | 110,869 | 628,890 |
| Total annual cost at the plan level including estimated costs associated with delaying care e (CAD) | 663,998 | 172,740 | 325,963 | 357,245 | 219,275 | 1,739,220 |
| Increased proportion with 70% of patients tested by NGS | ||||||
| Increased proportion of patients tested by NGS (%) | 70.0 | 5.0 | 5.0 | 15.0 | 5.0 | 100.0 |
| Number of patients tested by strategy (n) | 267.1 | 19.1 | 19.1 | 57.2 | 19.1 | 381.7 |
| Proportion of patients tested positive for a mutation with an approved targeted therapy b (%) | 38.0 | 31.7 | 29.3 | 20.3 | 34.4 | 34.4 |
| Patients tested positive for a mutation with an approved targeted therapy b (n) | 101.4 | 6.1 | 5.6 | 11.6 | 6.6 | 131.3 |
| Time to initiation of appropriate targeted therapy, per patient c (weeks) | 5.1 | 16.1 | 15.3 | 6.4 | 7.0 | 6.5 |
| Total annual cost at the plan level d (CAD) | 378,135 | 48,240 | 44,498 | 66,641 | 55,435 | 592,796 |
| Total annual cost at the plan level, including estimated costs associated with delaying care e (CAD) | 929,528 | 172,740 | 162,981 | 214,197 | 109,637 | 1,589,084 |
| Incremental impact | ||||||
| Incremental patients tested positive for a mutation with an approved targeted therapy b (n) | 29.0 | 0.0 | −5.6 | −7.8 | −6.5 | 9.1 |
| Incremental annual budget impact d, total (CAD) | 108,017 | 0 | −44,183 | −44,494 | −55,435 | −36,094 |
| Incremental budget impact, PMPM (CAD) | 0.009 | 0.000 | −0.004 | −0.004 | −0.005 | −0.003 |
| Incremental annual budget impact, including estimated costs associated with delaying care e, total (CAD) | 265,530 | 0 | −162,981 | −143,048 | −109,637 | −150,137 |
| Incremental budget impact, including estimated costs associated with delaying care, PMPM (CAD) | 0.022 | 0.000 | −0.014 | −0.012 | −0.009 | −0.013 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheffield, B.S.; Eaton, K.; Emond, B.; Lafeuille, M.-H.; Hilts, A.; Lefebvre, P.; Morrison, L.; Stevens, A.L.; Ewara, E.M.; Cheema, P. Cost Savings of Expedited Care with Upfront Next-Generation Sequencing Testing versus Single-Gene Testing among Patients with Metastatic Non-Small Cell Lung Cancer Based on Current Canadian Practices. Curr. Oncol. 2023, 30, 2348-2365. https://doi.org/10.3390/curroncol30020180
Sheffield BS, Eaton K, Emond B, Lafeuille M-H, Hilts A, Lefebvre P, Morrison L, Stevens AL, Ewara EM, Cheema P. Cost Savings of Expedited Care with Upfront Next-Generation Sequencing Testing versus Single-Gene Testing among Patients with Metastatic Non-Small Cell Lung Cancer Based on Current Canadian Practices. Current Oncology. 2023; 30(2):2348-2365. https://doi.org/10.3390/curroncol30020180
Chicago/Turabian StyleSheffield, Brandon S., Kiefer Eaton, Bruno Emond, Marie-Hélène Lafeuille, Annalise Hilts, Patrick Lefebvre, Laura Morrison, Andrea L. Stevens, Emmanuel M. Ewara, and Parneet Cheema. 2023. "Cost Savings of Expedited Care with Upfront Next-Generation Sequencing Testing versus Single-Gene Testing among Patients with Metastatic Non-Small Cell Lung Cancer Based on Current Canadian Practices" Current Oncology 30, no. 2: 2348-2365. https://doi.org/10.3390/curroncol30020180
APA StyleSheffield, B. S., Eaton, K., Emond, B., Lafeuille, M.-H., Hilts, A., Lefebvre, P., Morrison, L., Stevens, A. L., Ewara, E. M., & Cheema, P. (2023). Cost Savings of Expedited Care with Upfront Next-Generation Sequencing Testing versus Single-Gene Testing among Patients with Metastatic Non-Small Cell Lung Cancer Based on Current Canadian Practices. Current Oncology, 30(2), 2348-2365. https://doi.org/10.3390/curroncol30020180





