Potential of New Bacterial Strains for a Multiproduct Bioprocess Application: A Case Study Using Isolates of Lactic Acid Bacteria from Pineapple Silage of Costa Rican Agro-Industrial Residues
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.1.1. Lactic Acid Bacteria
2.1.2. LAB Growth Curves
2.2. Production of Metabolites
2.3. Bioreactor Fermentation
2.4. Chemical Analytical Methods
2.4.1. HPLC Analysis
2.4.2. LTA Analysis
2.4.3. Polyacrylamide Gel Electrophoresis (PAGE) Analysis of LTA
2.5. Microbiological Analysis
2.5.1. Surrogate Bacteria and Growth Conditions
2.5.2. Antimicrobial Activity Determination
2.6. Data Analysis and Statistical Design
2.6.1. Bacterial Growth
2.6.2. Antimicrobial Activity
3. Results and Discussion
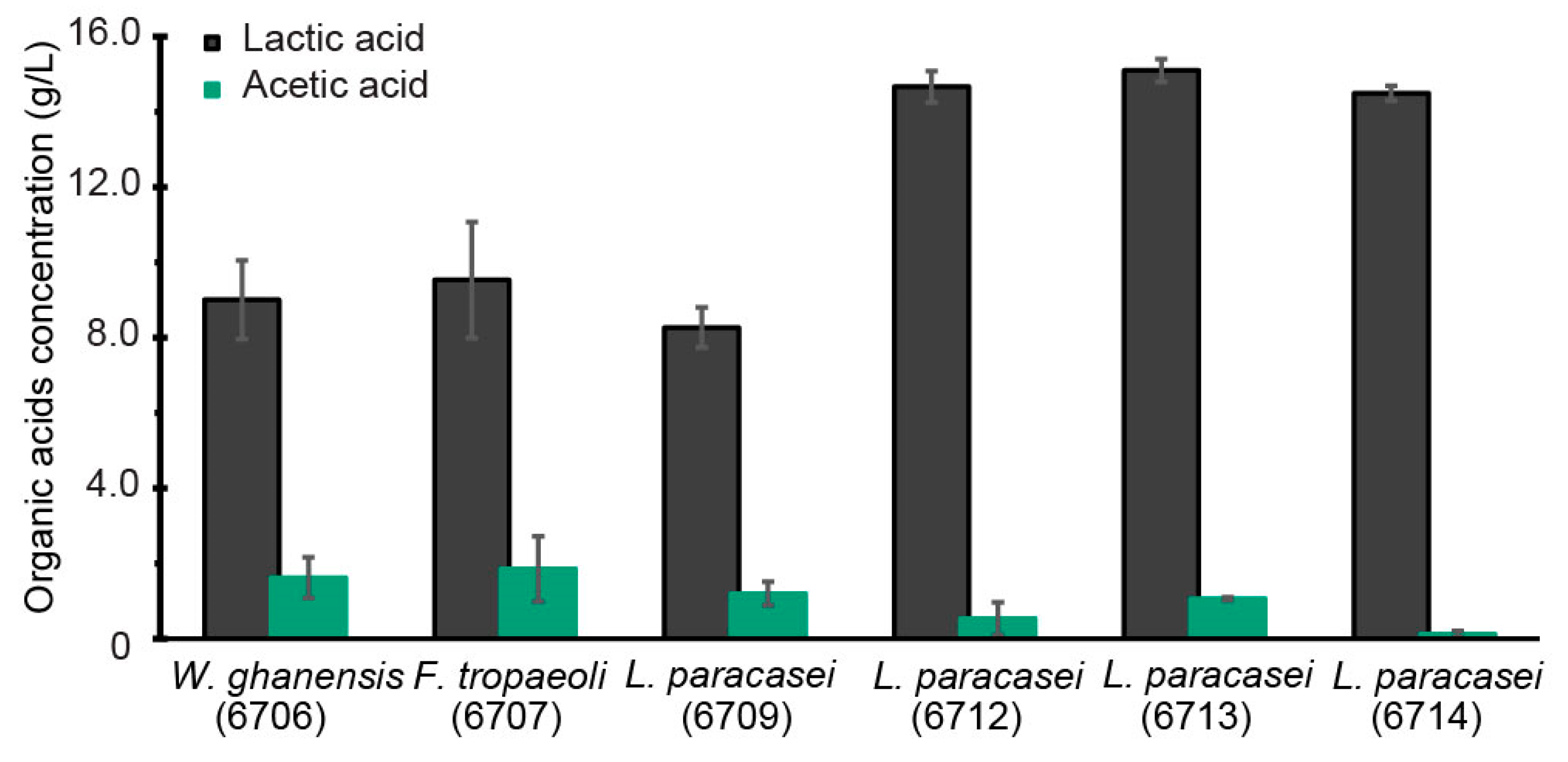
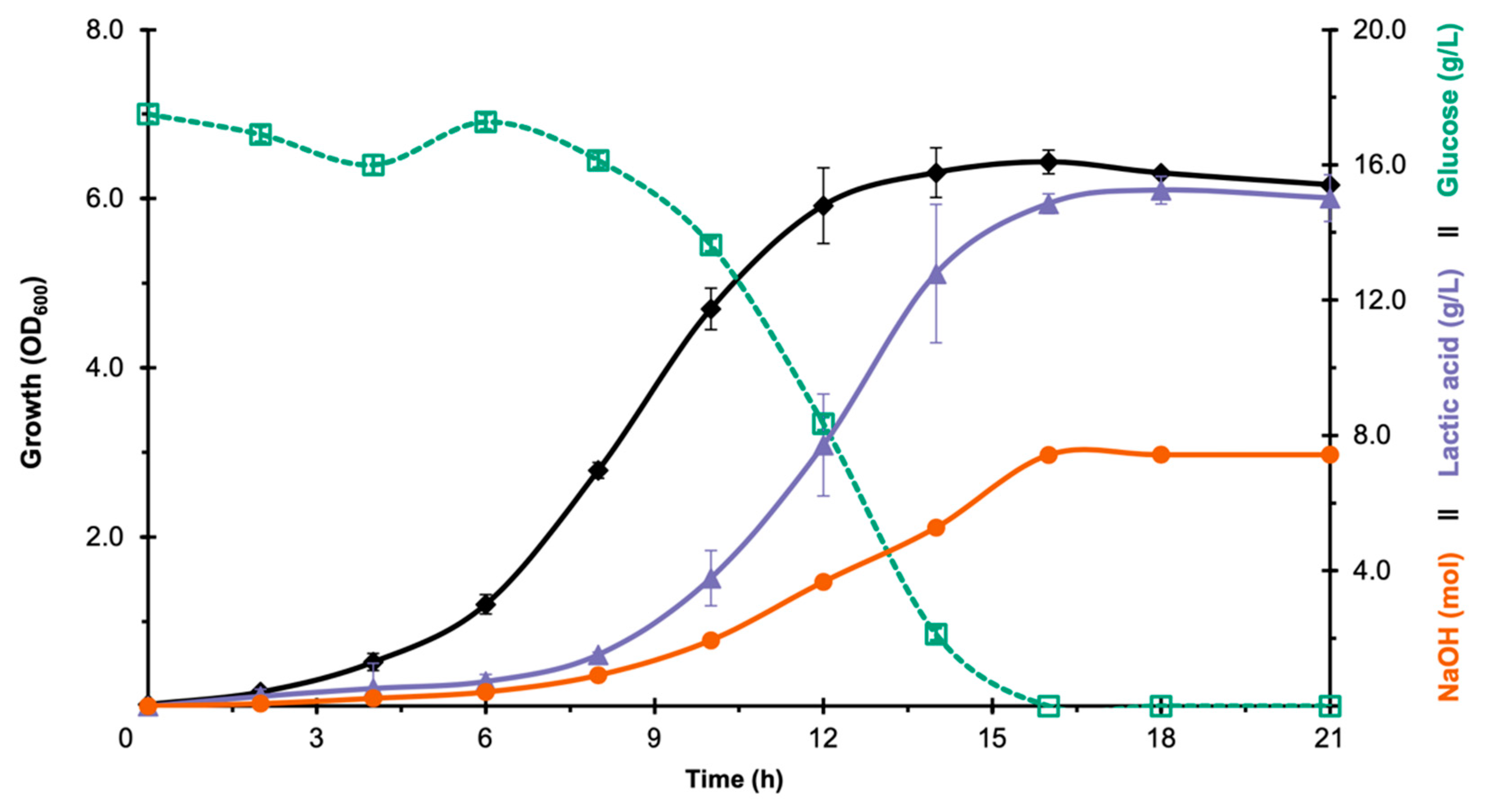
3.1. Selection of LAB Strain for Bioreactor Fermentation
3.2. Assessment of Potential for LTA Synthesis
3.3. Assessment of Antagonistic Activity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salvetti, E.; Harris, H.M.B.; Felis, G.E.; O’Toole, P.W. Comparative genomics of the genus lactobacillus reveals robust phylogroups that provide the basis for reclassification. Appl. Environ. Microbiol. 2018, 84, e00993-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, A.; Tanizawa, Y.; Arita, M. Isolation and identification of lactic acid bacteria from environmental samples. In Lactic Acid Bacteria; Kanauchi, M., Ed.; Springer: New York, NY, USA, 2019; pp. 3–13. [Google Scholar]
- Vinderola, G. (Ed.) Lactic Acid Bacteria: Microbiological and Functional Aspects, 5th ed.; Taylor & Francis: Boca Raton, FL, USA, 2019. [Google Scholar]
- Eş, I.; Mousavi Khaneghah, A.; Barba, F.J.; Saraiva, J.A.; Sant’Ana, A.S.; Hashemi, S.M.B. Recent advancements in lactic acid production—A review. Food Res. Int. 2018, 107, 763–770. [Google Scholar] [CrossRef]
- Manandhar, A.; Shah, A. Techno-Economic analysis of bio-based lactic acid production utilizing corn grain as feedstock. Processes 2020, 8, 199. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Yadav, N.; Nain, L.; Khare, S.K. A simple downstream processing protocol for the recovery of lactic acid from the fermentation broth. Bioresour. Technol. 2020, 318, 124260. [Google Scholar] [CrossRef] [PubMed]
- Mora-Villalobos, A.; Montero-Zamora, J.; Barboza, N.; Rojas-Garbanzo, C.; Usaga, J.; Redondo-Solano, M.; Schroedter, L.; Olszewska-Widdrat, A.; López-Gómez, J.P. Multi-product lactic acid bacteria fermentations: A review. Fermentation 2020, 6, 23. [Google Scholar] [CrossRef] [Green Version]
- van Pijkeren, J.P.; Barrangou, R. Genome editing of food-grade lactobacilli to develop therapeutic probiotics. Microbiol. Spectr. 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Gómez, J.P.; Unger, P.; Schneider, R.; Venus, J. From upstream to purification: Production of lactic acid from the organic fraction of municipal solid waste. Waste Biomass Valorization 2020, 11, 5247–5254. [Google Scholar] [CrossRef] [Green Version]
- Kusumoto, S.; Fukase, K.; Fujimoto, Y. Synthesis of lipopolysaccharide, peptidoglycan, and lipoteichoic acid fragments. In Comprehensive Glycoscience; Elsevier: Amsterdam, The Netherlands, 2007; pp. 685–711. [Google Scholar]
- Malanovic, N.; Lohner, K. Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. Biochim. Biophys. Acta BBA–Biomembr. 2016, 1858, 936–946. [Google Scholar] [CrossRef] [Green Version]
- Reichmann, N.T.; Gründling, A. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes: Synthesis and function of glycolipid and PGP-LTA. FEMS Microbiol. Lett. 2011, 319, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, S.; Santa Maria, J.P.; Walker, S. Wall Teichoic Acids of Gram-Positive Bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef] [Green Version]
- Villéger, R.; Saad, N.; Grenier, K.; Falourd, X.; Foucat, L.; Urdaci, M.C.; Bressollier, P.; Ouk, T.S. Characterization of lipoteichoic acid structures from three probiotic Bacillus strains: Involvement of d-alanine in their biological activity. Antonie Van Leeuwenhoek 2014, 106, 693–706. [Google Scholar] [CrossRef] [Green Version]
- Weill, F.S.; Cela, E.M.; Paz, M.L.; Ferrari, A.; Leoni, J.; Maglio, D.H.G. Lipoteichoic acid from Lactobacillus rhamnosus GG as an oral photoprotective agent against UV-induced carcinogenesis. Br. J. Nutr. 2013, 109, 457–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzoli, R.; Bosco, F.; Mizrahi, I.; Bayer, E.A.; Pessione, E. Towards lactic acid bacteria-based biorefineries. Biotechnol. Adv. 2014, 32, 1216–1236. [Google Scholar] [CrossRef] [PubMed]
- Zondervan, E.; Nawaz, M.; de Haan, A.B.; Woodley, J.M.; Gani, R. Optimal design of a multi-product biorefinery system. Comput. Chem. Eng. 2011, 35, 1752–1766. [Google Scholar] [CrossRef]
- Kamm, B.; Kamm, M. Biorefineries–multi product processes. In White Biotechnology; Ulber, R., Sell, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 175–204. [Google Scholar]
- Champagne, C.P. The lactic acid bacteria. Int. Dairy J. 1994, 4, 665–666. [Google Scholar] [CrossRef]
- Börner, R.A.; Kandasamy, V.; Axelsen, A.M.; Nielsen, A.T.; Bosma, E.F. Genome editing of lactic acid bacteria: Opportunities for food, feed, pharma and biotech. FEMS Microbiol. Lett. 2019, 366, fny291. [Google Scholar] [CrossRef] [PubMed]
- WingChing Jones, R.; Redondo Solano, M.; Usaga, J.; Uribe, L.; Barboza, N. Tipificación con secuencias multilocus en Lactobacillus casei procedentes de ensilados de cáscara de piña. Agron. Mesoam. 2021, 32, 508–522. [Google Scholar] [CrossRef]
- Montero-Zamora, J.; Cortés-Muñoz, M.; Esquivel, P.; Mora-Villalobos, J.A.; Velázquez, C. Growth conditions and survival kinetics during storage of Lactobacillus rhamnosus GG for the design of a sustainable probiotic whey-based beverage containing Costa Rican guava fruit pulp. J. Food Sci. 2020, 85, 3478–3486. [Google Scholar] [CrossRef] [PubMed]
- Chan-Blanco, Y.; Bonilla-Leiva, A.R.; Velázquez, A.C. Using banana to generate lactic acid through batch process fermentation. Appl. Microbiol. Biotechnol. 2003, 63, 147–152. [Google Scholar] [CrossRef]
- Toldrá, F.; Marshall, R.J. Food and nutritional analysis—dairy products. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Gambelli, L. Milk and its sugar-lactose: A picture of evaluation methodologies. Beverages 2017, 3, 35. [Google Scholar] [CrossRef]
- Morath, S.; Geyer, A.; Hartung, T. Structure–function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J. Exp. Med. 2001, 193, 393–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cela, E.M.; Weill, F.S.; Pazm, M.L.; Leoni, J.; Gonzalez Maglio, D.H. Lipoteichoic acid challenge induces higher inflammatory responses than lipopolysaccharide in UV-irradiated keratinocytes: Letter to the Editor. Photodermatol. Photoimmunol. Photomed. 2015, 31, 111–114. [Google Scholar] [CrossRef]
- Meredith, T.C.; Swoboda, J.G.; Walker, S. Late-stage polyribitol phosphate wall teichoic acid biosynthesis in Staphylococcus aureus. J. Bacteriol. 2008, 190, 3046–3056. [Google Scholar] [CrossRef] [Green Version]
- Xia, G.; Maier, L.; Sanchez-Carballo, P.; Otto, M.; Holst, O.; Peschel, A. Glycosylation of wall teichoic acid in Staphylococcus aureus by tarm. J. Biol. Chem. 2010, 285, 13405–13415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lourenço, F.; Pinto, T.D.J.A. Antibiotic microbial assay using kinetic-reading microplate system. Braz. J. Pharm. Sci. 2011, 47, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Tarrah, A.; Pakroo, S.; Corich, V.; Giacomini, A. Whole-genome sequence and comparative genome analysis of Lactobacillus paracasei DTA93, a promising probiotic lactic acid bacterium. Arch. Microbiol. 2020, 202, 1997–2003. [Google Scholar] [CrossRef] [PubMed]
- Simsek, D.; Kiymaci, M.E.; Tok, K.C.; Gumustas, M.; Altanlar, N. Investigation of the probiotic and metabolic potential of Fructobacillus tropaeola and Apilactobacillus kunkeei from apiaries. Arch. Microbiol. 2022, 204, 432. [Google Scholar] [CrossRef] [PubMed]
- Vinay-Lara, E.; Hamilton, J.J.; Stahl, B.; Broadbent, J.R.; Reed, J.L.; Steele, J.L. Genome –Scale Reconstruction of Metabolic Networks of Lactobacillus casei ATCC 334 and 12A. PLoS ONE 2014, 9, e110785. [Google Scholar] [CrossRef]
- Ha, G.; Kim, J.; Im, S.; Shin, S.-J.; Yang, H.-J.; Jeong, D.-Y. Application of response surface methodology in medium optimization to improve lactic acid production by Lactobacillus paracasei SRCM201474. J. Life Sci. 2020, 30, 522–531. [Google Scholar] [CrossRef]
- Mladenović, D.; Pejin, J.; Kocić-Tanackov, S.; Djukić-Vuković, A.; Mojović, L. Enhanced lactic acid production by adaptive evolution of Lactobacillus paracasei on agro-industrial substrate. Appl. Biochem. Biotechnol. 2019, 187, 753–769. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.-K.; Wee, Y.-J.; Choi, G.-W. A novel lactic acid bacterium for the production of high purity l-lactic acid, Lactobacillus paracasei subsp. paracasei CHB2121. J. Biosci. Bioeng. 2012, 114, 155–159. [Google Scholar] [CrossRef]
- Vieco-Saiz, N.; Belguesmia, Y.; Raspoet, R.; Auclair, E.; Gancel, F.; Kempf, I.; Drider, D. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 2019, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Pengxin, Y.; Jiawen, W.; Manman, L.; Ping, L.; Qing, G. Purification and characterization of a novel bacteriocin from Lactobacillus paracasei ZFM54. LWT 2021, 143, 111125. [Google Scholar] [CrossRef]
- Belguesmia, Y.; Bendjeddou, K.; Kempf, I.; Boukherroub, R.; Drider, D. Heterologous Biosynthesis of five new class II bacteriocins from Lactobacillus paracasei CNCM I-5369 with antagonistic activity against pathogenic Escherichia coli strains. Front. Microbiol. 2020, 11, 1189. [Google Scholar] [CrossRef]


| Strain | Maximal OD600 * | Growth Rate (h−1) * |
|---|---|---|
| F. tropaeoli 6705 | 0.54 7 ± 0.149 a | 0.129 ± 0.049 a |
| F. tropaeoli 6707 | 1.081 ± 0.239 b | 0.250 ± 0.100 b,c,d,e |
| L. paracasei 6709 | 1.099 ± 0.224 b | 0.250 ± 0.104 c,d,e |
| L. paracasei 6710 | 1.600 ± 0.215 c | 0.366 ± 0.046 e |
| L. paracasei 6711 | 0.555 ± 0.052 a | 0.145 ± 0.043 a,b |
| L. paracasei 6712 | 1.015 ± 0.165 b | 0.191 ± 0.029 a,b,c,d |
| L. paracasei 6713 | 0.703 ± 0.152 a | 0.242 ± 0.051 a,b,c,d,e |
| L. paracasei 6714 | 1.602 ± 0.248 c | 0.266 ± 0.115 d,e |
| L. paracasei 6715 | 0.504 ± 0.047 a | 0.141 ± 0.040 a |
| L. fermentum 6702 | 0.484 ± 0.066 a | 0.137 ± 0.052 a |
| L. fermentum 6704 | 1.196 ± 0.226 b | 0.306 ± 0.060 d,e |
| L. parafarraginis 6717 | 0.470 ± 0.043 a | 0.147 ± 0.044 a,b,c |
| L. parafarraginis 6719 | 0.501 ± 0.034 a | 0.147 ± 0.045 a,b,c |
| W. ghanensis 6706 | 1.107 ± 0.164 b | 0.283 ± 0.009 d,e |
| Strain | Supernatant Ratio | Absorbance at 600 nm | |||
|---|---|---|---|---|---|
| E. coli 25922 | P. fluorescens 6.2 | L. innocua 4.1 | L. innocua 5.1 | ||
| No Supernatant | 1.214 ± 0.077 a | 0.609 ± 0.026 a | 0.467 ± 0.040 a | 0.469 ± 0.028 a | |
| L. paracasei (6712) | 1:1 | 0.453 ± 0.023 b | 0.359 ± 0.048 c | 0.136 ± 0.003 b | 0.237 ± 0.011 b |
| 1:2 | 0.465 ± 0.035 b | 0.431 ± 0.119 b,c | 0.182 ± 0.007 c | 0.244 ± 0.010 b | |
| 1:4 | 0.486 ± 0.013 b | 0.438 ± 0.049 b,c | 0.196 ± 0.002 d | 0.257 ± 0.012 b | |
| 1:8 | 0.694 ± 0.028 b | 0.569 ± 0.055 a,b | 0.210±0.001 e | 0.262 ± 0.004 b | |
| L. paracasei (6713) | 1:1 | 0.426 ± 0.052 b | 0.378 ± 0.021 a | 0.183 ± 0.025 b | 0.207 ± 0.025 b |
| 1:2 | 0.429 ± 0.009 b | 0.424 ± 0.091 a | 0.203 ± 0.006 b,c | 0.241 ± 0.024 b,c | |
| 1:4 | 0.469 ± 0.017 b | 0.537 ± 0.187 a | 0.216 ± 0.007 b,c | 0.239 ± 0.013 b,c | |
| 1:8 | 0.504 ± 0.017 b | 0.562 ± 0.176 a | 0.227 ± 0.012 c | 0.270 ± 0.006 c | |
| L. paracasei (6714) | 1:1 | 0.407 ± 0.045 b | 0.318 ± 0.037 b | 0.085 ± 0.004 b | 0.207 ± 0.013 b |
| 1:2 | 0.443 ± 0.055 b | 0.340 ± 0.046 b | 0.188 ± 0.004 c | 0.227 ± 0.010 b,c | |
| 1:4 | 0.487 ± 0.036 b | 0.363 ± 0.028 b | 0.190 ± 0.008 c | 0.239 ± 0.003 c | |
| 1:8 | 0.694 ± 0.028 b | 0.394 ± 0.068 b | 0.226 ± 0.008 d | 0.358 ± 0.010 b | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montero-Zamora, J.; Rojas-Vargas, M.D.; Barboza, N.; López-Gómez, J.P.; Mora-Villalobos, J.A.; Redondo-Solano, M. Potential of New Bacterial Strains for a Multiproduct Bioprocess Application: A Case Study Using Isolates of Lactic Acid Bacteria from Pineapple Silage of Costa Rican Agro-Industrial Residues. Fermentation 2022, 8, 361. https://doi.org/10.3390/fermentation8080361
Montero-Zamora J, Rojas-Vargas MD, Barboza N, López-Gómez JP, Mora-Villalobos JA, Redondo-Solano M. Potential of New Bacterial Strains for a Multiproduct Bioprocess Application: A Case Study Using Isolates of Lactic Acid Bacteria from Pineapple Silage of Costa Rican Agro-Industrial Residues. Fermentation. 2022; 8(8):361. https://doi.org/10.3390/fermentation8080361
Chicago/Turabian StyleMontero-Zamora, Jéssica, María Daniela Rojas-Vargas, Natalia Barboza, José Pablo López-Gómez, José Aníbal Mora-Villalobos, and Mauricio Redondo-Solano. 2022. "Potential of New Bacterial Strains for a Multiproduct Bioprocess Application: A Case Study Using Isolates of Lactic Acid Bacteria from Pineapple Silage of Costa Rican Agro-Industrial Residues" Fermentation 8, no. 8: 361. https://doi.org/10.3390/fermentation8080361
APA StyleMontero-Zamora, J., Rojas-Vargas, M. D., Barboza, N., López-Gómez, J. P., Mora-Villalobos, J. A., & Redondo-Solano, M. (2022). Potential of New Bacterial Strains for a Multiproduct Bioprocess Application: A Case Study Using Isolates of Lactic Acid Bacteria from Pineapple Silage of Costa Rican Agro-Industrial Residues. Fermentation, 8(8), 361. https://doi.org/10.3390/fermentation8080361







