Visualizing the Spatial Distribution of Arctium lappa L. Root Components by MALDI-TOF Mass Spectrometry Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and Reagents
2.2. Matrix Selection
2.3. Section Preparation of A. lappa Roots and MALDI-TOF MS Imaging
2.4. Statistical Analysis
3. Results and Discussion
3.1. MALDI-TOF MS Matrix Selection
3.2. Spatial Distributions of Components in A. lappa Roots
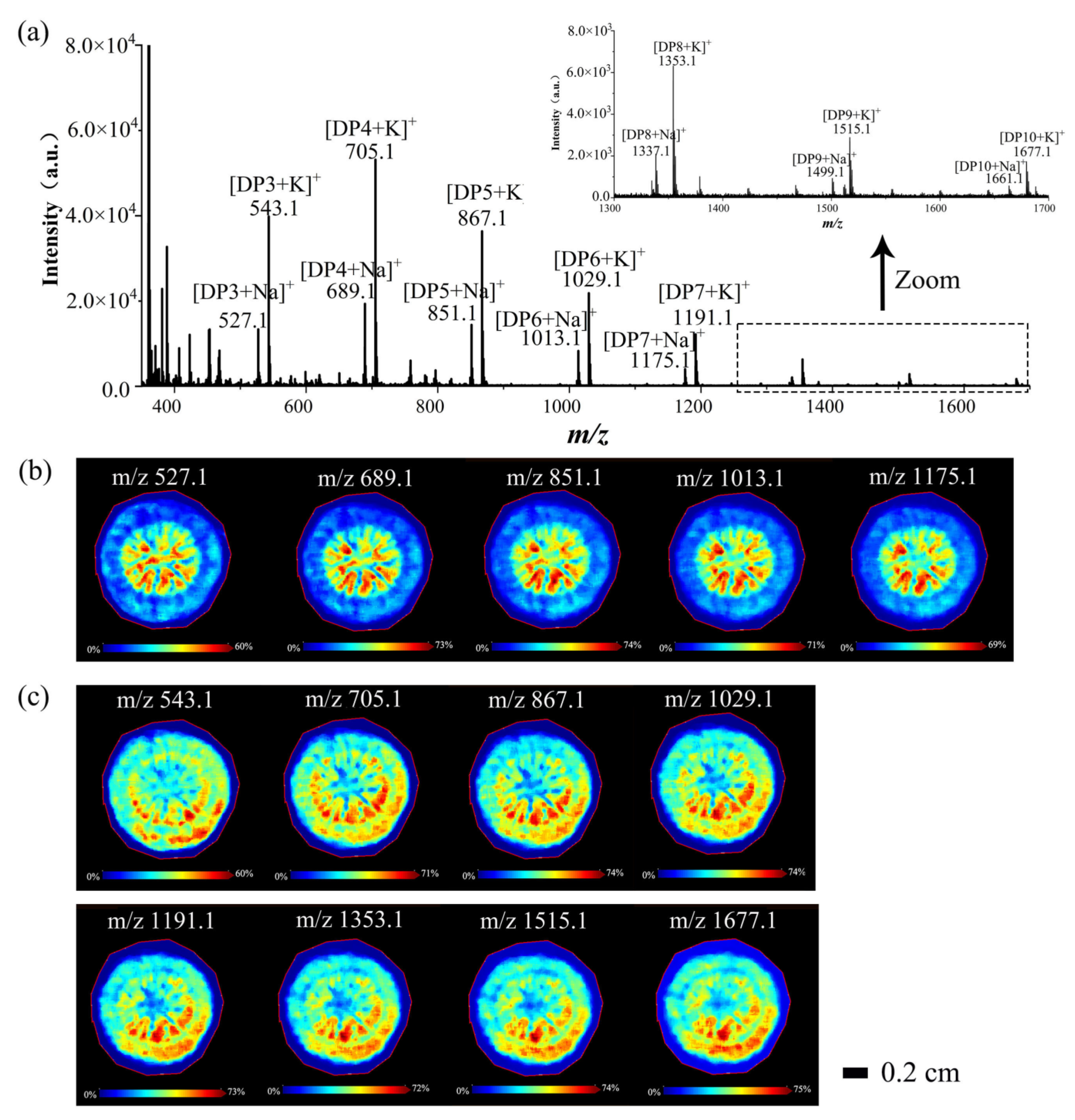
3.2.1. Spatial Distributions of Saccharides in A. lappa Roots
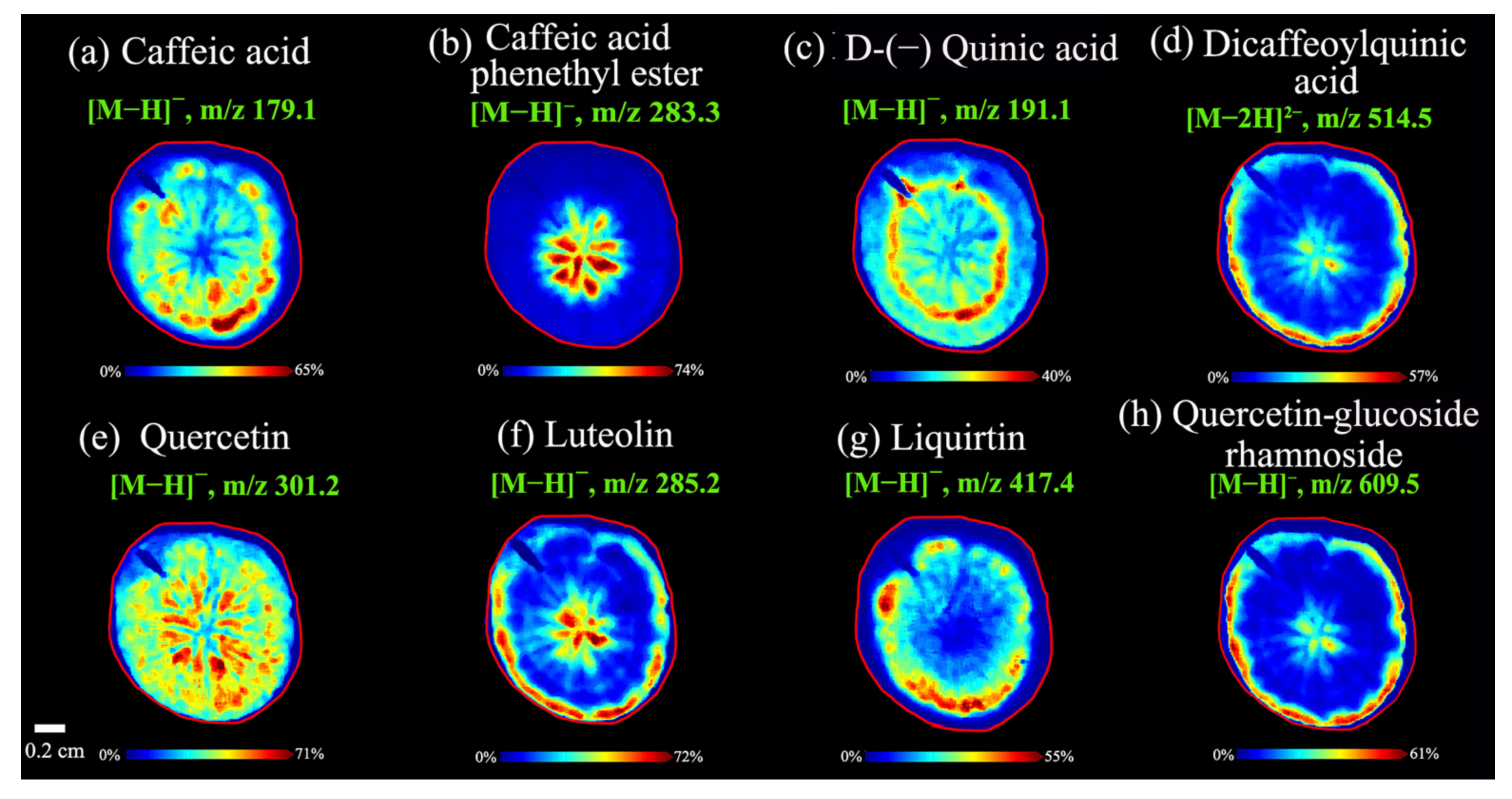
3.2.2. Spatial Distributions of Caffeoylquinic Acids in A. lappa Roots
3.2.3. Spatial Distributions of Flavonoids in A. lappa Roots
3.2.4. Spatial Distributions of Amino Acids in A. lappa Roots
3.2.5. Spatial Distributions of Phospholipids and Choline in A. lappa Roots
3.2.6. Spatial Distributions of Unidentified Compounds in A. lappa Roots
3.3. Validation Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, J.; Cai, Y.Z.; Wong, R.N.; Lee, C.K.; Tang, S.C.; Sze, S.C.; Tong, Y.; Zhang, Y. Comparative analysis of caffeoylquinic acids and lignans in roots and seeds among various burdock (Arctium lappa) genotypes with high antioxidant activity. J. Agric. Food Chem. 2012, 60, 4067–4075. [Google Scholar] [CrossRef] [PubMed]
- Tonea, A.; Badea, M.; Oana, L.; Sava, S.; Vodnar, D. Antibacterial and antifungal activity of endodontic intracanal medications. Clujul Med. 2017, 90, 344–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrera-Balandrano, D.D.; Beta, T.; Chai, Z.; Zhang, X.; Li, Y.; Huang, W. Effect of In Vitro gastro-intestinal digestion on the phenolic composition and antioxidant capacity of burdock roots at different harvest time. Food Chem. 2021, 358, 129897. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, N.; Kan, J.; Sun, R.; Tang, S.; Wang, Z.; Chen, M.; Liu, J.; Jin, C. Anti-inflammatory activity of alkali-soluble polysaccharides from Arctium lappa L. and its effect on gut microbiota of mice with inflammation. Int. J. Biol. Macromol. 2020, 154, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ha, S.J.; Park, J.; Kim, Y.H.; Lee, N.H.; Kim, Y.E.; Hong, Y.S.; Song, K.M. Arctium lappa root extract containing L-arginine prevents TNF-alpha-induced early atherosclerosis in vitro and in vivo. Nutr. Res. 2020, 77, 85–96. [Google Scholar] [CrossRef]
- Li, L.; Qiu, Z.; Dong, H.; Ma, C.; Qiao, Y.; Zheng, Z. Structural characterization and antioxidant activities of one neutral polysaccharide and three acid polysaccharides from the roots of Arctium lappa L.: A comparison. Int. J. Biol. Macromol. 2021, 182, 187–196. [Google Scholar] [CrossRef]
- Tobyn, G.; Denham, A.; Whitelegg, M. Arctium lappa, burdock. In Medical Herbs; Churchill Livingstone: London, UK, 2011; pp. 91–103. [Google Scholar]
- Zheng, Z.; Wang, X.; Liu, P.; Li, M.; Dong, H.; Qiao, X. Semi-preparative separation of 10 caffeoylquinic acid derivatives using high speed counter-current chromatography combined with semi-preparative HPLC from the roots of burdock (Arctium lappa L.). Molecules 2018, 23, 429. [Google Scholar] [CrossRef] [Green Version]
- Lou, Z.; Wang, H.; Zhu, S.; Zhang, M.; Gao, Y.; Ma, C.; Wang, Z. Improved extraction and identification by ultra performance liquid chromatography tandem mass spectrometry of phenolic compounds in burdock leaves. J. Chromatogr. A 2010, 1217, 2441–2446. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhao, X.; Zhu, S.; Dang, J.; Qiao, X.; Qiu, Z.; Tao, Y. Simultaneous determination of oleanolic acid and ursolic acid by in vivo microdialysis via UHPLC-MS/MS using magnetic dispersive solid phase extraction coupling with microwave-assisted derivatization and its application to a pharmacokinetic study of Arctium lappa L. root extract in rats. J. Agric. Food Chem. 2018, 66, 3975–3982. [Google Scholar] [CrossRef]
- Xu, L.; Lu, Y.; Cong, Y.; Zhang, P.; Han, J.; Song, G.; Wang, G.; Chen, K. Polysaccharide produced by Bacillus subtilis using burdock oligofructose as carbon source. Carbohydr. Polym. 2019, 206, 811–819. [Google Scholar] [CrossRef]
- Qin, Z.B.; Ding, L.F.; Wang, X.; Huang, L.J.; Liang, M.J.; Bin, J.; Luo, N.; Deng, L.; Guo, Y.D. Lignans from the seeds of Arctium lappa L. (burdock) and their inhibitory effects on nitric oxide production. Phytochem. Lett. 2019, 34, 43–49. [Google Scholar] [CrossRef]
- Horn, P.J.; Chapman, K.D. Lipidomics in situ: Insights into plant lipid metabolism from high resolution spatial maps of metabolites. Prog. Lipid Res. 2014, 54, 32–52. [Google Scholar] [CrossRef] [PubMed]
- Shiota, M.; Shimomura, Y.; Kotera, M.; Taira, S. Mass spectrometric imaging of localization of fat molecules in water-in-oil emulsions containing semi-solid fat. Food Chem. 2018, 245, 1218–1223. [Google Scholar] [CrossRef]
- Rath, C.M.; Alexandrov, T.; Higginbottom, S.K.; Song, J.; Milla, M.E.; Fischbach, M.A.; Sonnenburg, J.L.; Dorrestein, P.C. Molecular analysis of model gut microbiotas by imaging mass spectrometry and nanodesorption electrospray ionization reveals dietary metabolite transformations. Anal. Chem. 2012, 84, 9259–9267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Fu, X.; Zhou, H.; Rao, W.; Zeng, L.; Yang, Z. Visualized analysis of within-tissue spatial distribution of specialized metabolites in tea (Camellia sinensis) using desorption electrospray ionization imaging mass spectrometry. Food Chem. 2019, 292, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Peukert, M.; Matros, A.; Lattanzio, G.; Kaspar, S.; Abadia, J.; Mock, H.P. Spatially resolved analysis of small molecules by matrix-assisted laser desorption/ionization mass spectrometric imaging (MALDI-MSI). New Phytol. 2012, 193, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, E.; Chaurand, P.; Raghavan, V. Visualizing the distribution of strawberry plant metabolites at different maturity stages by MALDI-TOF imaging mass spectrometry. Food Chem. 2021, 345, 128838. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.; Gopal, J.E.; Muthu, M. A consolidative synopsis of the MALDI-TOF MS accomplishments for the rapid diagnosis of microbial plant disease pathogens. Trends Anal Chem. 2022, 156, 116713. [Google Scholar] [CrossRef]
- Liu, H.; Pan, Y.; Xiong, C.; Han, J.; Wang, X.; Chen, J.; Nie, Z. Matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI MSI) for in situ analysis of endogenous small molecules in biological samples. Trends Anal Chem. 2022, 157, 116809. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, J.; Nie, S.; Xie, M.; Li, S. Rapid profiling strategy for oligosaccharides and polysaccharides by MALDI TOF mass spectrometry. Food Hydrocoll. 2022, 124, 107237. [Google Scholar] [CrossRef]
- Sun, C.; Liu, W.; Ma, S.; Zhang, M.; Geng, Y.; Wang, X. Development of a high-coverage matrix-assisted laser desorption/ionization mass spectrometry imaging method for visualizing the spatial dynamics of functional metabolites in Salvia miltiorrhiza Bge. J. Chromatogr. A 2020, 1614, 460704. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Qiu, Z.; Qi, Y.; Zhao, D.; Ali, I.; Sun, C.; Xu, L.; Zheng, Z.; Ma, C. AuNPs/NiFe-LDHs-assisted laser desorption/ionization mass spectrometry for efficient analysis of metronidazole and its metabolites in water samples. J. Hazard. Mater. 2022, 423, 126893. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y. MALDI matrix research for biopolymers. Mass Spectrom. 2015, 4, A0037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.; Zhang, M.; Dong, H.; Liu, W.; Guo, L.; Wang, X. A spatially-resolved approach to visualize the distribution and biosynthesis of flavones in Scutellaria baicalensis Georgi. J. Pharm. Biomed. Anal. 2020, 179, 113014. [Google Scholar] [CrossRef]
- Moro, T.M.; Clerici, M.T. Burdock (Arctium lappa L) roots as a source of inulin-type fructans and other bioactive compounds: Current knowledge and future perspectives for food and non-food applications. Food Res. Int. 2021, 141, 109889. [Google Scholar] [CrossRef]
- Zhang, B.; Li, M.; Qiao, Y.; Gao, P.; Li, L.; Zheng, Z. Potential use of low-field nuclear magnetic resonance to determine the drying characteristics and quality of Arctium lappa L. in hot-blast air. LWT Food Sci. Technol. 2020, 132, 109829. [Google Scholar] [CrossRef]
- Štikarovská, M.; Chmelík, J. Determination of neutral oligosaccharides in vegetables by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chim. Acta 2004, 520, 47–55. [Google Scholar] [CrossRef]
- Twaij, B.M.; Taha, A.J.; Bhuiyan, F.H.; Hasan, M.N. Effect of saccharides on secondary compounds production from stem derived callus of Datura inoxia. Biotechnol. Rep. 2022, 33, e00701. [Google Scholar] [CrossRef]
- Qiu, Z.; Qiao, Y.; Zhang, B.; Sun-Waterhouse, D.; Zheng, Z. Bioactive polysaccharides and oligosaccharides from garlic (Allium sativum L.): Production, physicochemical and biological properties, and structure–function relationships. Compr. Rev. Food Sci. Food Saf. 2022, 21, 3033–3095. [Google Scholar] [CrossRef]
- Feenstra, A.D.; O’Neill, K.C.; Yagnik, G.B.; Lee, Y.J. Organic-Inorganic binary mixture matrix for comprehensive laser-desorption ionization mass spectrometric analysis and imaging of medium-size molecules including phospholipids, glycerolipids, and oligosaccharides. RSC Adv. 2016, 6, 99260–99268. [Google Scholar] [CrossRef]
- Sarabia, L.D.; Boughton, B.A.; Rupasinghe, T.; van de Meene, A.M.L.; Callahan, D.L.; Hill, C.B.; Roessner, U. High-mass-resolution MALDI mass spectrometry imaging reveals detailed spatial distribution of metabolites and lipids in roots of barley seedlings in response to salinity stress. Metabolomics 2018, 14, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki, M.; Fujii, K.; Kitayama, K. Environmental control of root exudation of low-molecular weight organic acids in tropical rainforests. Ecosystems 2012, 15, 1194–1203. [Google Scholar] [CrossRef]
- Pandino, G.; Bonomo, A.; Scavo, A.; Mauromicale, G.; Lombardo, S. Caffeoylquinic acids and flavones profile in Cynara cardunculus L. seedlings under controlled conditions as affected by light and water-supply treatments. Sci. Hortic. 2022, 302, 111180. [Google Scholar] [CrossRef]
- Chen, L.; Teng, H.; Cao, H. Chlorogenic acid and caffeic acid from Sonchus oleraceus Linn synergistically attenuate insulin resistance and modulate glucose uptake in HepG2 cells. Food Chem. Toxicol. 2019, 127, 182–187. [Google Scholar] [CrossRef]
- Xiang, C.; Liu, M.; Lu, Q.; Fan, C.; Lu, H.; Feng, C.; Yang, X.; Li, H.; Tang, W. Blockade of TLRs-triggered macrophage activation by caffeic acid exerted protective effects on experimental ulcerative colitis. Cell Immunol. 2021, 365, 104364. [Google Scholar] [CrossRef]
- Schnarr, L.; Segatto, M.L.; Olsson, O.; Zuin, V.G.; Kummerer, K. Flavonoids as biopesticides - Systematic assessment of sources, structures, activities and environmental fate. Sci. Total Environ. 2022, 824, 153781. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Yuan, R.; Li, C.; Pan, Y.; Zhang, Z.; Zhu, Y.; Nie, Y. Antibacterial and hypoglycemic activity of flavonoids in etiolated tissues from fresh-cut Chinese water-chestnuts (Eleocharis tuberosa). Sci. Hortic. 2022, 297, 110925. [Google Scholar] [CrossRef]
- Ferracane, R.; Graziani, G.; Gallo, M.; Fogliano, V.; Ritieni, A. Metabolic profile of the bioactive compounds of burdock (Arctium lappa) seeds, roots and leaves. J. Pharm. Biomed. Anal. 2010, 51, 399–404. [Google Scholar] [CrossRef]
- Bozzo, G.G.; Unterlander, N. In through the out door: Biochemical mechanisms affecting flavonoid glycoside catabolism in plants. Plant Sci. 2021, 308, 110904. [Google Scholar] [CrossRef]
- Zeng, T.; Sun, X.; Miao, Y.; Gu, S.; Tian, L.; Zheng, Y.; Jiang, Y.; Zhang, X.; Feng, Z.; Pei, J.; et al. Integrating bioclimatic factors and secondary metabolism to predict the suitable producing area of plants with high specific metabolite content in a real-world environment - a case of Carthamus tinctorius L. Ind. Crop. Prod. 2022, 177, 114545. [Google Scholar] [CrossRef]
- Ren, G.Q.; Luan, X.N.; Dou, D.Q. Analysis of functional components of burdock (Arctium lappa root) standard water extract and its aphrodisiac effect in experimental rats. J. Chem. Soc. Pak. 2020, 42, 94–102. [Google Scholar] [CrossRef]
- Guo, N.; Zhang, S.; Gu, M.; Xu, G. Function, transport, and regulation of amino acids: What is missing in rice? Crop. J. 2021, 9, 530–542. [Google Scholar] [CrossRef]
- Xing, J.; Zhang, L.; Duan, Z.; Lin, J. Coordination of phospholipid-based signaling and membrane trafficking in plant immunity. Trends Plant Sci. 2021, 26, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Vincent, P.; Maneta-Peyret, L.; Sturbois-Balcerzak, B.; Duvert, M.; Cassagne, C.; Moreau, P. One of the origins of plasma membrane phosphatidylserine in plant cells is a local synthesis by a serine exchange activity. FEBS Lett. 1999, 464, 80–84. [Google Scholar] [CrossRef] [Green Version]
- De Rudder, K.E.; Sohlenkamp, C.; Geiger, O. Plant-exuded choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J. Biol. Chem. 1999, 274, 20011–20016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niculescu, M.D. Choline and phosphatidylcholine. In Encyclopedia of Human Nutrition, 3rd ed.; Academic Press: New York, NY, USA, 2013; pp. 346–351. [Google Scholar] [CrossRef]
- Zeisel, S.H.; da Costa, K.A. Choline: An essential nutrient for public health. Nutr. Rev. 2009, 67, 615–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rongsawat, T.; Peltier, J.B.; Boyer, J.C.; Véry, A.A.; Sentenac, H. Looking for root hairs to overcome poor soils. Trends Plant Sci. 2021, 26, 83–94. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Qiu, Z.; Jiang, M.; Zhang, B.; Chen, Q.; Zhang, C.; Zheng, Z.; Qiao, X. Visualizing the Spatial Distribution of Arctium lappa L. Root Components by MALDI-TOF Mass Spectrometry Imaging. Foods 2022, 11, 3957. https://doi.org/10.3390/foods11243957
Li L, Qiu Z, Jiang M, Zhang B, Chen Q, Zhang C, Zheng Z, Qiao X. Visualizing the Spatial Distribution of Arctium lappa L. Root Components by MALDI-TOF Mass Spectrometry Imaging. Foods. 2022; 11(24):3957. https://doi.org/10.3390/foods11243957
Chicago/Turabian StyleLi, Lingyu, Zhichang Qiu, Mingdi Jiang, Bin Zhang, Qiang Chen, Chaojie Zhang, Zhenjia Zheng, and Xuguang Qiao. 2022. "Visualizing the Spatial Distribution of Arctium lappa L. Root Components by MALDI-TOF Mass Spectrometry Imaging" Foods 11, no. 24: 3957. https://doi.org/10.3390/foods11243957
APA StyleLi, L., Qiu, Z., Jiang, M., Zhang, B., Chen, Q., Zhang, C., Zheng, Z., & Qiao, X. (2022). Visualizing the Spatial Distribution of Arctium lappa L. Root Components by MALDI-TOF Mass Spectrometry Imaging. Foods, 11(24), 3957. https://doi.org/10.3390/foods11243957