2,5-Bis(2,2,2-trifluoroethoxy)phenyl-tethered 1,3,4-Oxadiazoles Derivatives: Synthesis, In Silico Studies, and Biological Assessment as Potential Candidates for Anti-Cancer and Anti-Diabetic Agent
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Pharmacology
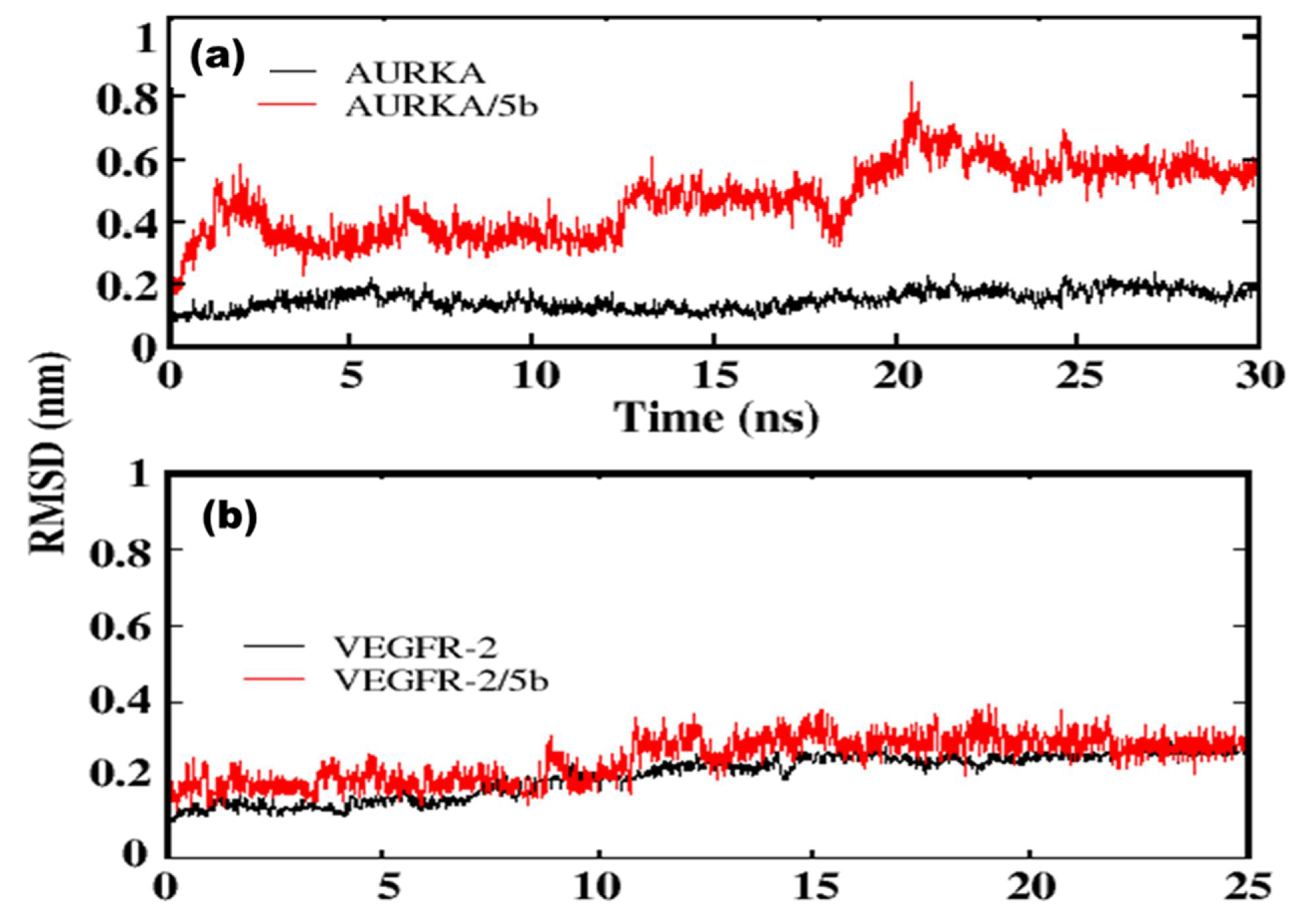
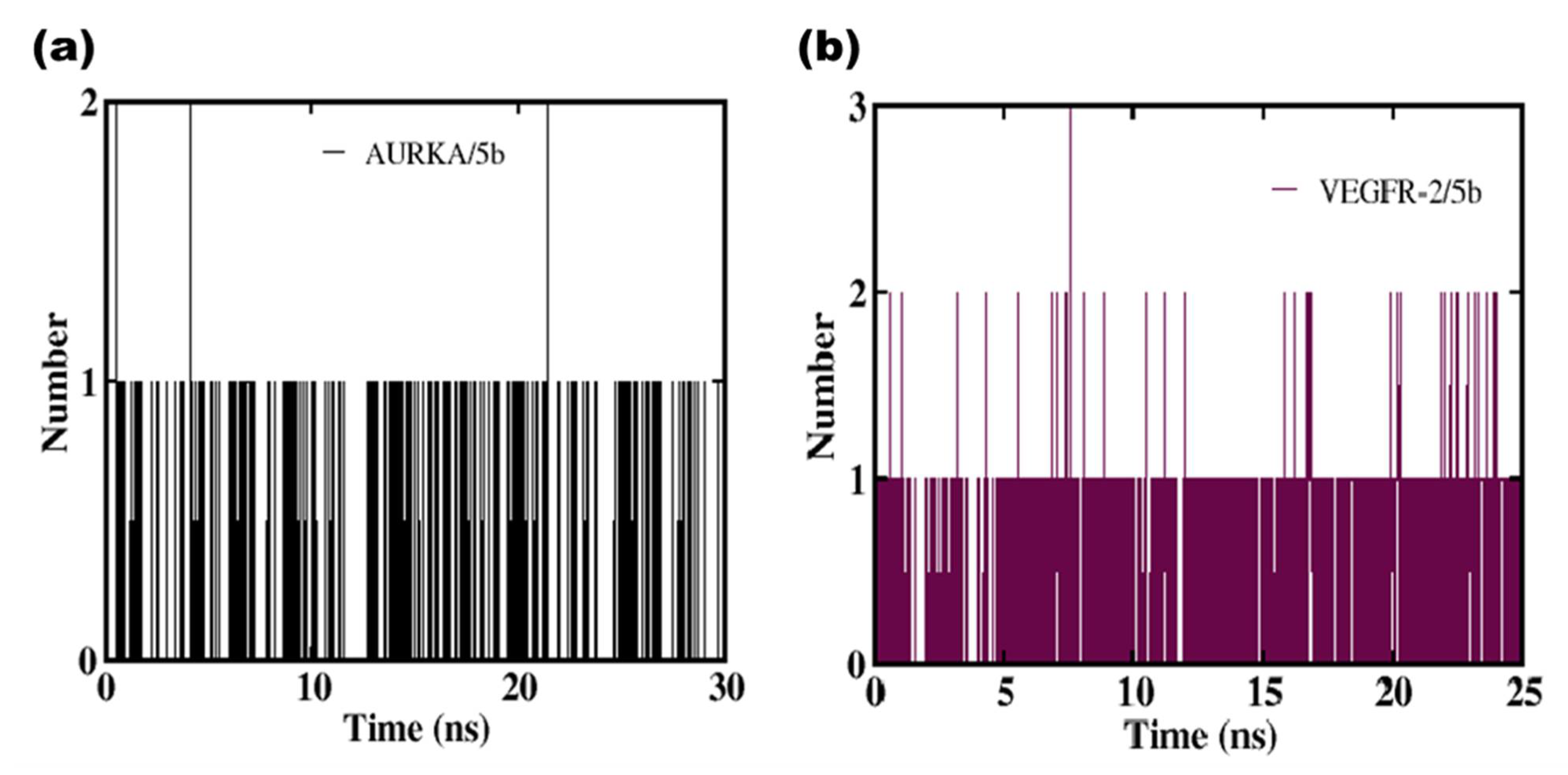
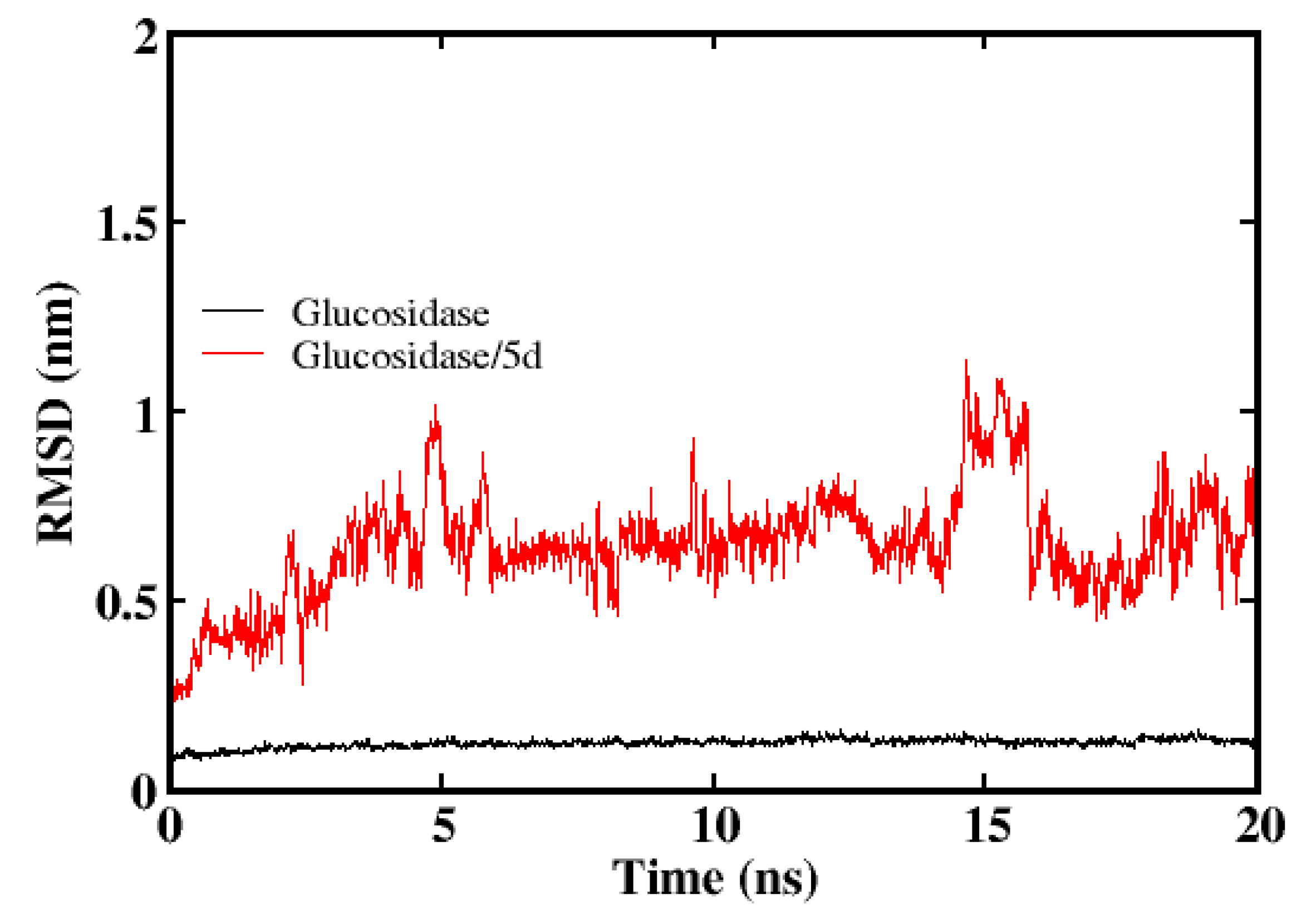
2.2.1. Molecular Docking Studies
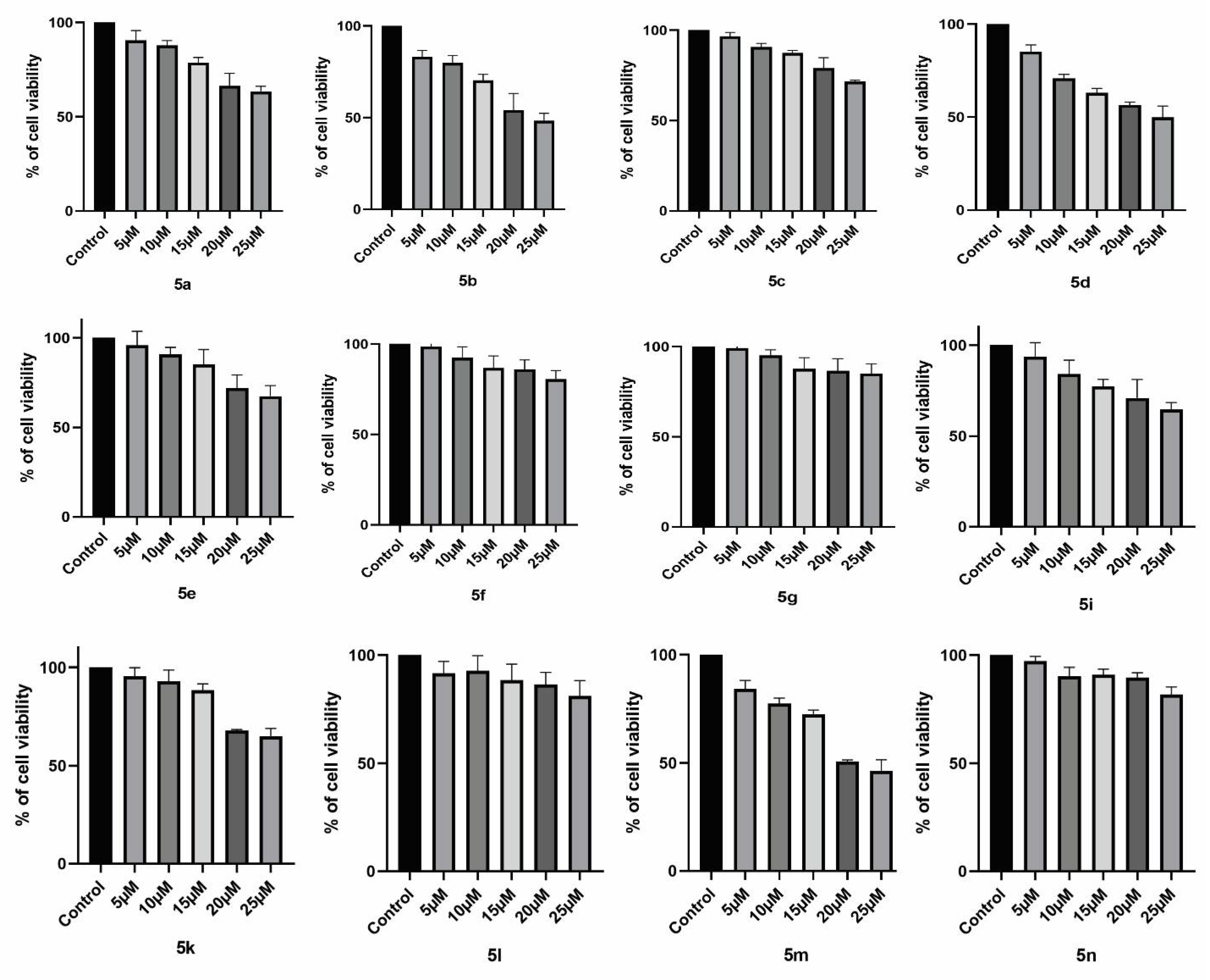
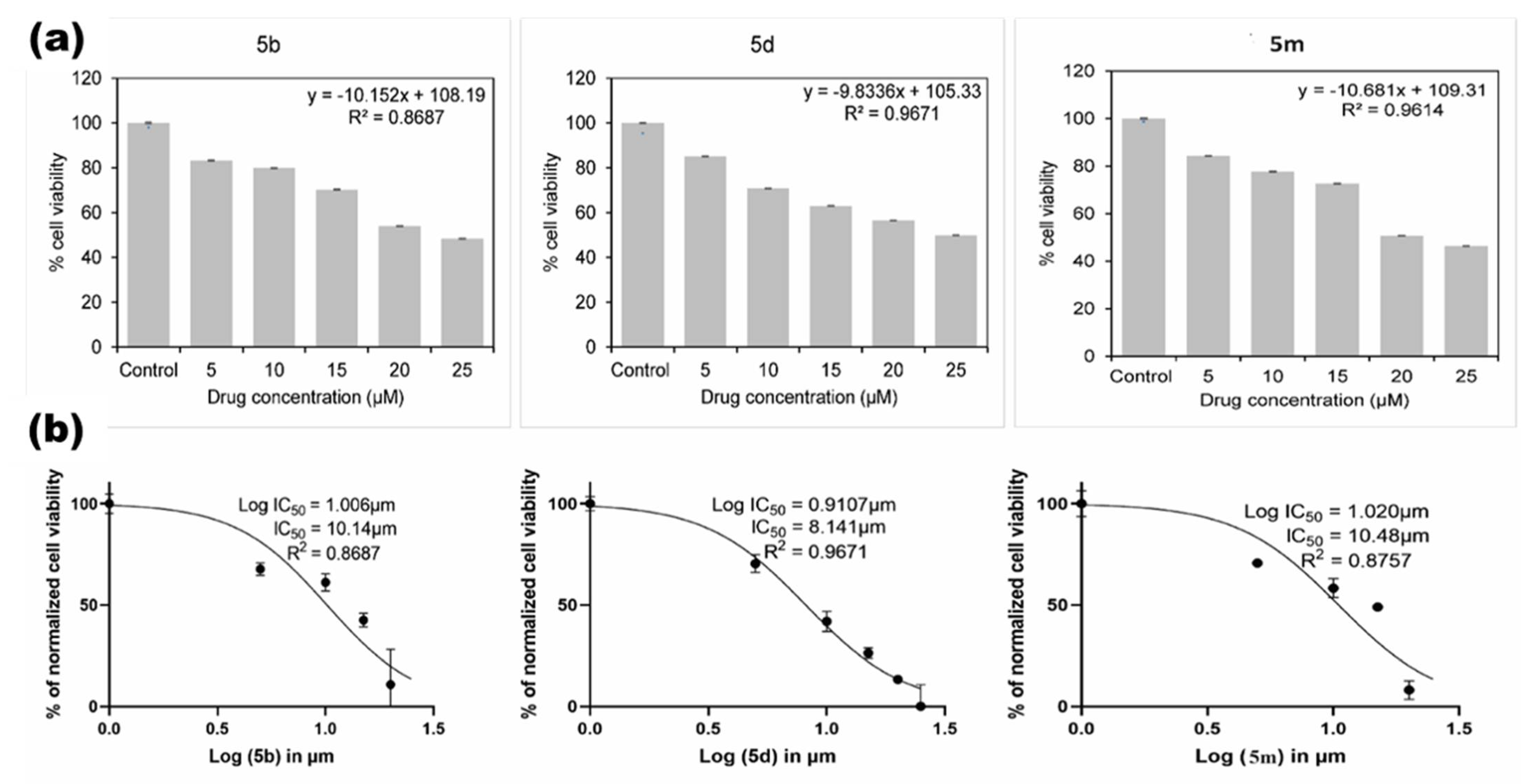
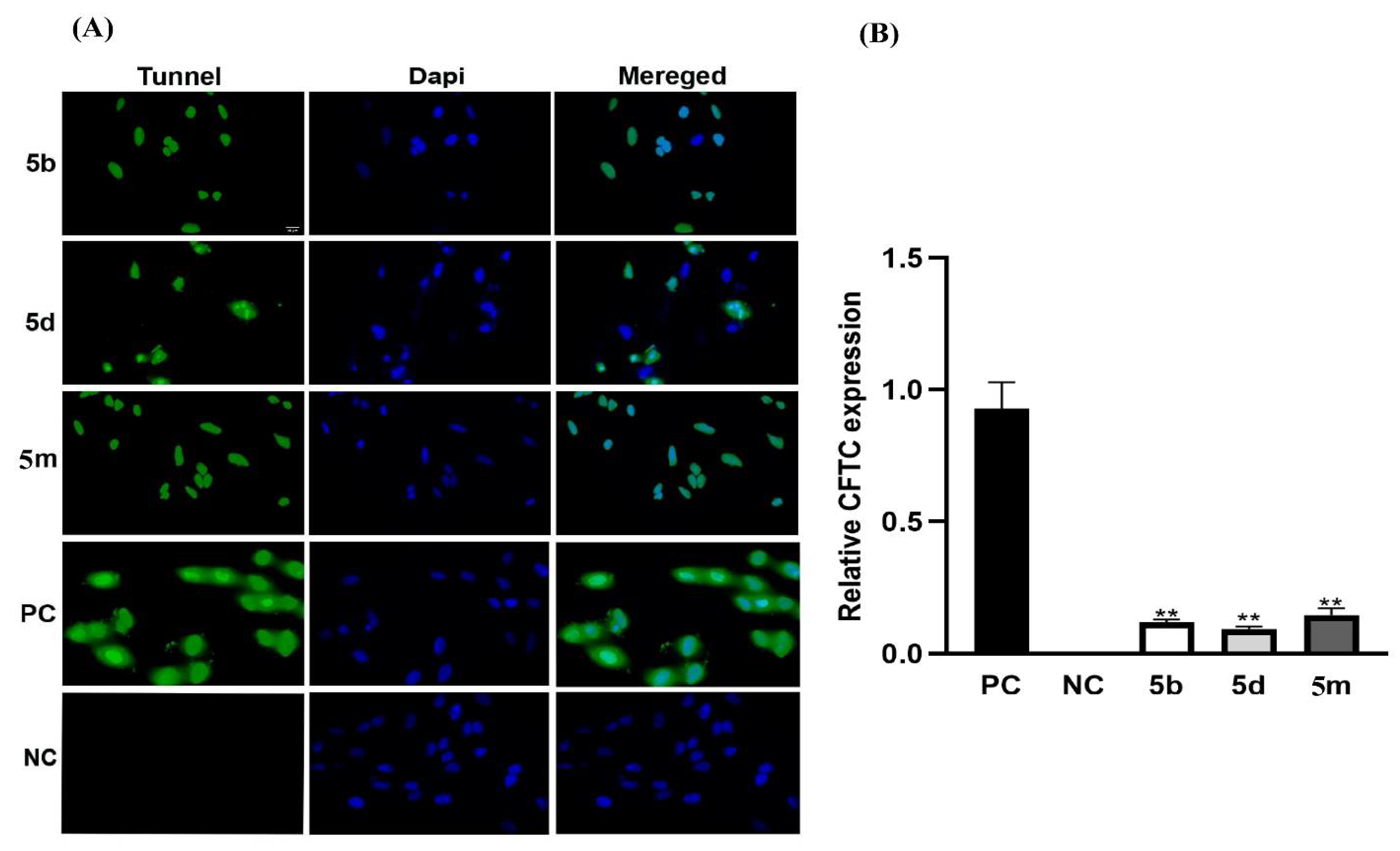
2.2.2. Cytotoxic Studies
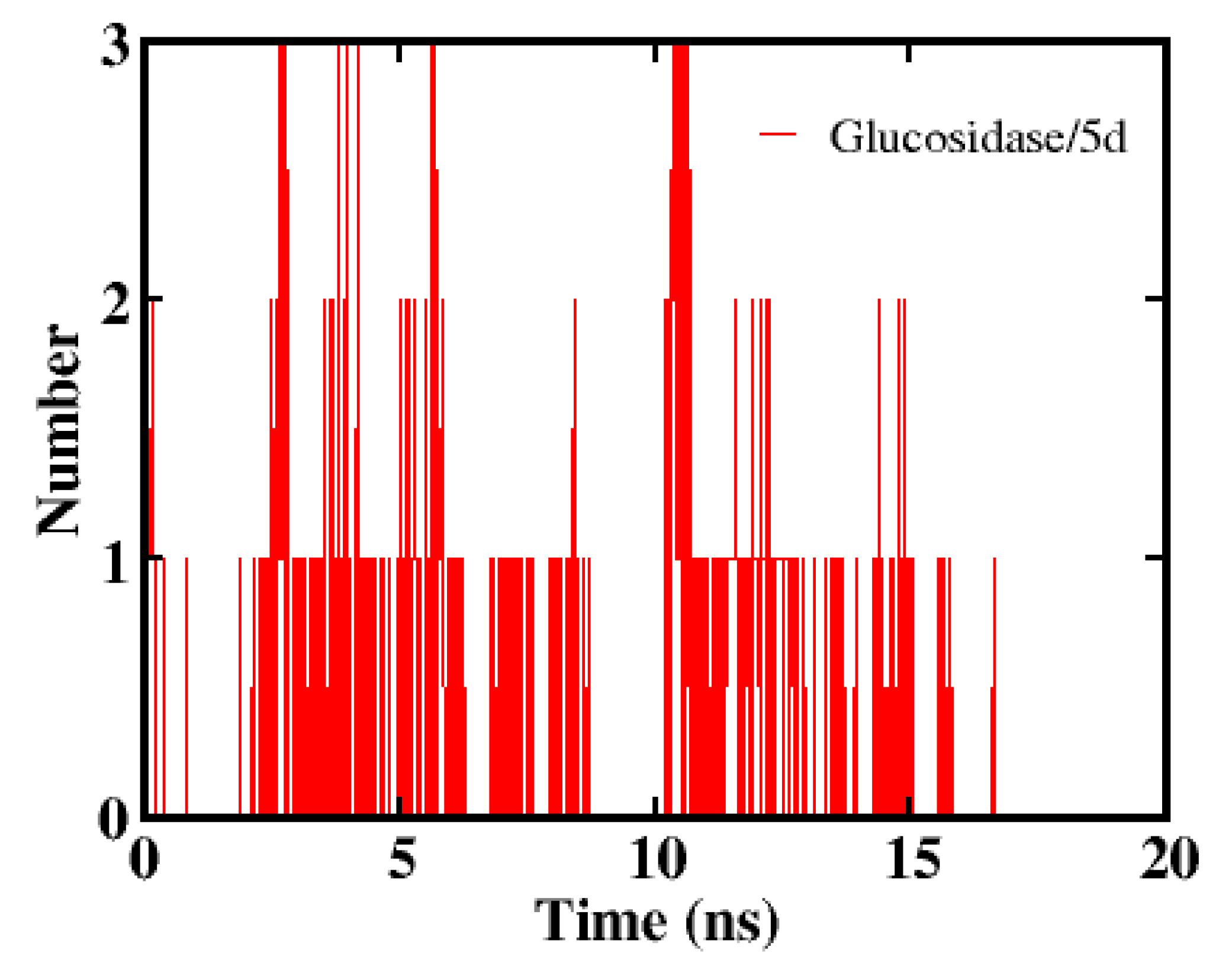
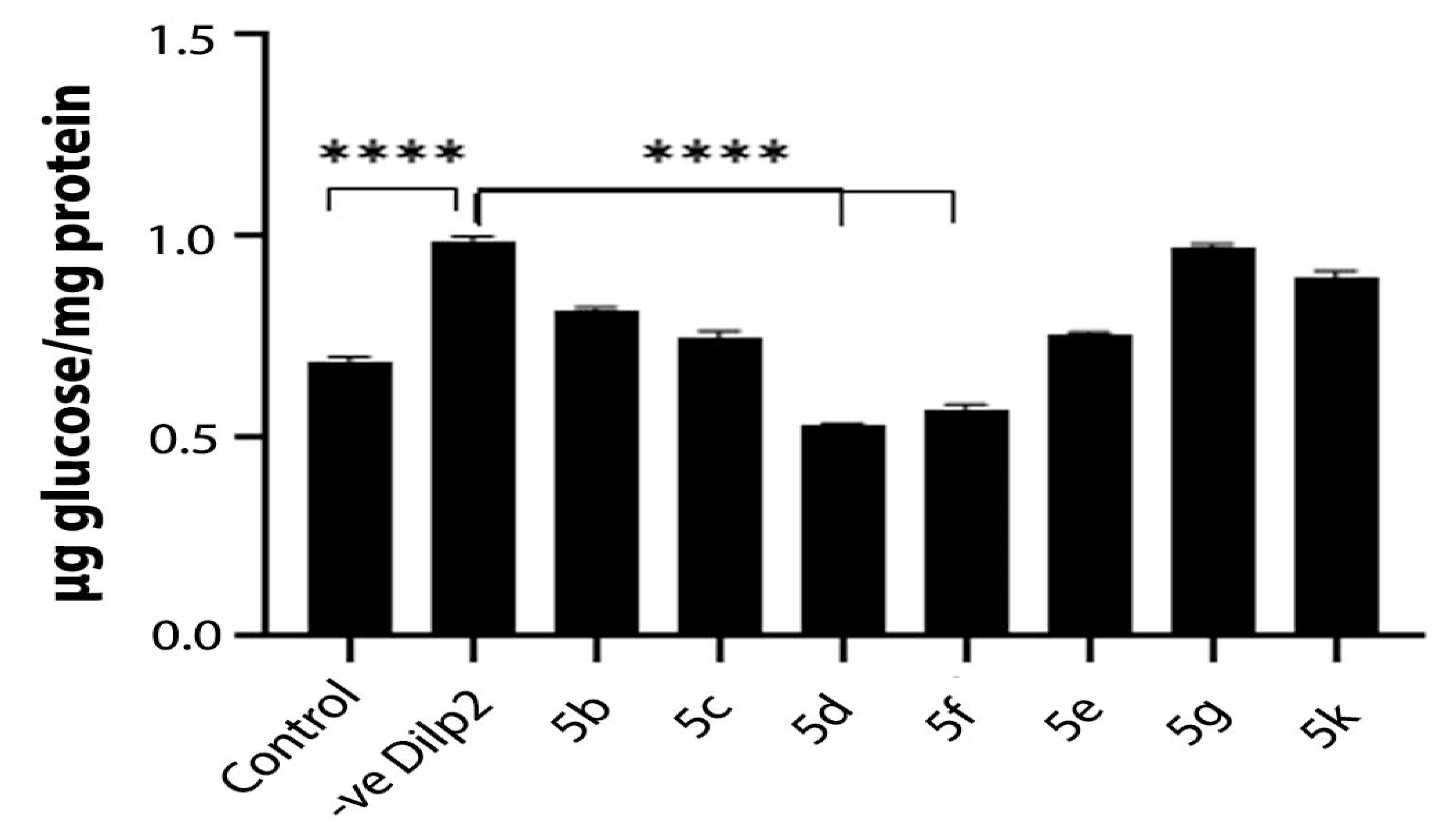
2.2.3. In Vivo Drosophila Study
3. Conclusions
4. Materials and Methods
4.1. Chemistry
4.2. Synthesis of 2,5-Bis(2,2,2-trifluoroethoxy)benzohydrazide (3)
4.3. Synthesis of Hydrazones (4a–n)
4.4. Procedure for the Synthesis of 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy)phenyl]-1,3,4-oxadiazol-3(2H/CH3)-yl}ethanone (5a–h) and (5i–n)
4.4.1. 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy)phenyl]-2-(2,6-dichlorophenyl)-1,3,4-oxadiazol-3(2H)-yl}ethanone (5a)
4.4.2. 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy)phenyl]-2-(6-methoxynapthalyl)-1,3,4-oxadiazol-3(2H)-yl}ethanone (5b)
4.4.3. 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy) phenyl]-2-(biphenyl)-1,3,4-oxadiazol-3(2H)-yl}ethanone (5c)
4.4.4. 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy)phenyl]-2-(4-chlorophenyl)-1,3,4-oxadiazol-3(2H)-yl}ethanone (5d)
4.4.5. 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy)phenyl]-2-(4-furfuryl)-1,3,4-oxadiazol-3(2H)-yl}ethenone (5e)
4.4.6. 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy)phenyl]-2-(2-chlorophenyl)-1,3,4-oxadiazol-3(2H)-yl}ethenone (5f)
4.4.7. 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy)phenyl]-2-(3-flouro-4-methoxyphenyl)-1,3,4-oxadiazol-3(2-methyl)-yl}ethanone (5i)
4.4.8. 1-{5-[2,5-. bis(2,2,2-trifluoroethoxy)phenyl]-2-(4-hydroxy phenyl)-1,3,4-oxadiazol-3(2-methyl)-yl}ethanone (5j)
4.4.9. 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy)phenyl]-2-(2,4-dichlorophenyl)-1,3,4-oxadiazol-3(2-methyl)-yl}ethenone (5k)
4.4.10. 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy)phenyl]-2-(4-florophenyl)-1,3,4-oxadiazol-3(2-methyl)-yl}ethenone (5l)
4.4.11. 1-{5-[2,5-Bis(2,2,2-trifluoroethoxy)phenyl]-2-(2,6-dichlorophenyl)-1,3,4-oxadiazol-3(2-methyl)-yl}ethenone (5m)
4.4.12. 1-{5-[2,5-bis(2,2,2-trifluoroethoxy)phenyl]-2-(4-chlorophenyl)-1,3,4-oxadiazol-3(2-methyl)-yl}ethenone (5n)
4.5. Anticancer Studies
4.5.1. Cell Culture
4.5.2. Cytotoxic Assay
4.5.3. Determination of IC50 Concentration for the Newly Synthesized Compounds
4.5.4. Colony Formation Assay
4.5.5. Tunnel Assay
4.5.6. Statistical Analysis
4.6. Drosophila Stocks and Experimental Design
4.7. In Silico Studies
4.7.1. Docking
4.7.2. Molecular Dynamics Simulation Protocol
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kharb, R.; Sharma, P.C.; Yar, M.S. Pharmacological significance of triazole scaffold. J. Enzym. Inhib. Med. Chem. 2011, 26, 1–21. [Google Scholar] [CrossRef]
- Isloor, A.M.; Kalluraya, B.; Pai, K.S. Synthesis, characterization and biological activities of some new benzo [b] thiophene derivatives. Eur. J. Med. Chem. 2010, 45, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Vijesh, A.; Isloor, A.M.; Shetty, P.; Sundershan, S.; Fun, H.K. New pyrazole derivatives containing 1,2,4-triazoles and benzoxazoles as potent antimicrobial and analgesic agents. Eur. J. Med. Chem. 2013, 62, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Vijesh, A.; Isloor, A.M.; Prabhu, V.; Ahmad, S.; Malladi, S. Synthesis, characterization and anti-microbial studies of some novel 2,4-disubstituted thiazoles. Eur. J. Med. Chem. 2010, 45, 5460–5464. [Google Scholar] [CrossRef] [PubMed]
- Al-Wahaibi, L.H.; Mohamed, A.A.B.; Tawfik, S.S.; Hassan, H.M.; El-Emam, A.A. 1,3,4-Oxadiazole N-Mannich Bases: Synthesis, Antimicrobial, and Anti-Proliferative Activities. Molecules 2021, 26, 2110. [Google Scholar] [CrossRef]
- Peregrym, K.; Szczukowski, Ł.; Wiatrak, B.; Potyrak, K.; Czyżnikowska, Ż.; Świątek, P. In Vitro and In Silico Evaluation of New 1,3,4-Oxadiazole Derivatives of Pyrrolo[3,4-d]pyridazinone as Promising Cyclooxygenase Inhibitors. Int. J. Mol. Sci. 2021, 22, 9130. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.; Rahman, M.U.; Siddiqui, A.A.; Ali, A.; Shaharyar, M. Design and synthesis of benzimidazole analogs endowed with oxadiazole as selective COX-2 inhibitor. Arch. Pharm. 2014, 347, 923–935. [Google Scholar] [CrossRef]
- Isloor, A.M.; Kalluraya, B.; Shetty, P. Regioselective reaction: Synthesis, characterization and pharmacological studies of some new Mannich bases derived from 1,2,4-triazoles. Eur. J. Med. Chem. 2009, 44, 3784–3787. [Google Scholar] [CrossRef]
- Glomb, T.; Wiatrak, B.; Gębczak, K.; Gębarowski, T.; Bodetko, D.; Czyżnikowska, Ż.; Świątek, P. New 1,3,4-Oxadiazole Derivatives of Pyridothiazine-1,1-Dioxide with Anti-Inflammatory Activity. Int. J. Mol. Sci. 2020, 21, 9122. [Google Scholar] [CrossRef]
- Sever, B.; Altıntop, M.D.; Özdemir, A.; Akalın Çiftçi, G.; Ellakwa, D.E.; Tateishi, H.; Radwan, M.O.; Ibrahim, M.A.A.; Otsuka, M.; Fujita, M.; et al. In Vitro and In Silico Evaluation of Anticancer Activity of New Indole-Based 1,3,4-Oxadiazoles as EGFR and COX-2 Inhibitors. Molecules 2020, 25, 5190. [Google Scholar] [CrossRef]
- Verma, G.; Khan, M.F.; Akhtar, W.; Alam, M.M.; Akhter, M.; Shaquiquzzaman, M. A review exploring therapeutic worth of 1,3,4-oxadiazole tailored compounds. Mini Rev. Med. Chem. 2019, 19, 477–509. [Google Scholar] [CrossRef] [PubMed]
- Bharwal, A.; Gupta, M. 1,3,4 Oxadiazole: An emerging scaffold to target different growth factors and kinases. Chem. Biol. Lett. 2020, 7, 225–235. [Google Scholar]
- Kumar, D.; Sundaree, S.; Johnson, E.O.; Shah, K. An efficient synthesis and biological study of novel indolyl-1,3,4-oxadiazoles as potent anticancer agents. Bioorg. Med. Chem. Lett. 2009, 19, 4492–4494. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.; Husain, A.; Mishra, R. Synthesis of benzimidazoles bearing oxadiazole nucleus as anticancer agents. Eur. J. Med. Chem. 2012, 54, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Macaev, F.; Rusu, G.; Pogrebnoi, S.; Gudima, A.; Stingaci, E.; Vlad, L.; Shvets, N.; Kandemirli, F.; Dimoglo, A.; Reynolds, R. Synthesis of novel 5-aryl-2-thio-1,3,4-oxadiazoles and the study of their structure–anti-mycobacterial activities. Bioorg. Med. Chem. 2005, 13, 4842–4850. [Google Scholar] [CrossRef]
- Taha, M.; Ismail, N.H.; Imran, S.; Rokei, M.Q.B.; Saad, S.M.; Khan, K.M. Synthesis of new oxadiazole derivatives as α-glucosidase inhibitors. Bioorg. Med. Chem. 2015, 23, 4155–4162. [Google Scholar] [CrossRef]
- Taha, M.; Ismail, N.H.; Imran, S.; Wadood, A.; Rahim, F.; Saad, S.M.; Khan, K.M.; Nasir, A. Synthesis, molecular docking and α-glucosidase inhibition of 5-aryl-2-(6′-nitrobenzofuran-2′-yl)-1,3,4-oxadiazoles. Bioorg. Chem. 2016, 66, 117–123. [Google Scholar] [CrossRef]
- Gani, R.S.; Kudva, A.K.; Timanagouda, K.; Mujawar, S.B.H.; Joshi, S.D.; Raghu, S.V. Synthesis of novel 5-(2,5-bis (2,2,2-trifluoroethoxy) phenyl)-1,3,4-oxadiazole-2-thiol derivatives as potential glucosidase inhibitors. Bioorg. Chem. 2021, 114, 105046. [Google Scholar] [CrossRef]
- Malladi, S.; Isloor, A.M.; Peethambar, S.; Fun, H.K. Synthesis and biological evaluation of newer analogues of 2,5-disubstituted 1,3,4-oxadiazole containing pyrazole moiety as antimicrobial agents. Arab. J. Chem. 2014, 7, 1185–1191. [Google Scholar] [CrossRef]
- Sangshetti, J.N.; Chabukswar, A.R.; Shinde, D.B. Microwave assisted one pot synthesis of some novel 2,5-disubstituted 1,3,4-oxadiazoles as antifungal agents. Bioorg. Med. Chem. Lett. 2011, 21, 444–448. [Google Scholar] [CrossRef]
- Chandrakantha, B.; Shetty, P.; Nambiyar, V.; Isloor, N.; Isloor, A.M. Synthesis, characterization and biological activity of some new 1,3,4-oxadiazole bearing 2-flouro-4-methoxy phenyl moiety. Eur. J. Med. Chem. 2010, 45, 1206–1210. [Google Scholar] [CrossRef] [PubMed]
- Zarghi, A.; Tabatabai, S.A.; Faizi, M.; Ahadian, A.; Navabi, P.; Zanganeh, V.; Shafiee, A. Synthesis and anticonvulsant activity of new 2-substituted-5-(2-benzyloxyphenyl)-1,3,4-oxadiazoles. Bioorg. Med. Chem. Lett. 2005, 15, 1863–1865. [Google Scholar] [CrossRef] [PubMed]
- Koksal, M.; Ozkan-Dagliyan, I.; Ozyazici, T.; Kadioglu, B.; Sipahi, H.; Bozkurt, A.; Bilge, S.S. Some Novel Mannich Bases of 5-(3,4-Dichlorophenyl)-1,3,4-oxadiazole-2(3H)-one and Their Anti-Inflammatory Activity. Arch. Pharm. 2017, 350, 1700153. [Google Scholar] [CrossRef]
- Narayana, B.; Vijaya Raj, K.K.; Ashalatha, B.V.; Kumari, N.S. Synthesis of Some New 2-(6-Methoxy-2-Naphthyl)-5-Aryl-1,3,4-Oxadiazoles as Possible Non-steroidal Anti-inflammatory and Analgesic Agents. Arch. Pharm. Int. J. Pharm. Med. Chem. 2005, 338, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Mohan, T.P.; Vishalakshi, B.; Bhat, K.S.; Rao, K.S.; Kendappa, G.N. Synthesis and insecticidal activity of some 1,3,4-oxadiazole derivatives containing phenoxyfluorophenyl group. Indian J. Chem. Sect. B Org. Med. Chem. 2004, 43, 1798–1801. [Google Scholar] [CrossRef]
- Garudachari, B.; Isloor, A.M.; Satyanarayana, M.N.; Fun, H.-K.; Hegde, G. Click chemistry approach: Regioselective one-pot synthesis of some new 8-trifluoromethylquinoline based 1,2,3-triazoles as potent antimicrobial agents. Eur. J. Med. Chem. 2014, 74, 324–332. [Google Scholar] [CrossRef]
- Preshlock, S.; Tredwell, M.; Gouverneur, V. 18F-Labeling of arenes and heteroarenes for applications in positron emission tomography. Chem. Rev. 2016, 116, 719–766. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.A.K.; Hoy, V.J.; O’Hagan, D.; Smith, G.T. How good is fluorine as a hydrogen bond acceptor? Tetrahedron 1996, 52, 12613–12622. [Google Scholar] [CrossRef]
- Sunil, D.; Isloor, A.M.; Shetty, P.; Satyamoorthy, K.; Prasad, A.B. 6-[3-(4-Fluorophenyl)-1H-pyrazol-4-yl]-3-[(2-naphthyloxy) methyl][1,2,4] triazolo [3,4-b][1,3,4] thiadiazole as a potent antioxidant and an anticancer agent induces growth inhibition followed by apoptosis in HepG2 cells. Arab. J. Chem. 2010, 3, 211–217. [Google Scholar] [CrossRef]
- Puthiyapurayil, P.; Poojary, B.; Chikkanna, C.; Buridipad, S.K. Design, synthesis and biological evaluation of a novel series of 1,3,4-oxadiazole bearing N-methyl-4-(trifluoromethyl)phenyl pyrazole moiety as cytotoxic agents. Eur. J. Med. Chem. 2012, 53, 203–210. [Google Scholar] [CrossRef]
- Alifieris, C.; Trafalis, D.T. Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol. Ther. 2015, 152, 63–82. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E. Glioblastoma: Overview of disease and treatment. Clin. J. Oncol. Nurs. 2016, 20, S2. [Google Scholar] [CrossRef] [PubMed]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee, S.U. Glioblastoma multiforme: A review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 3. [Google Scholar]
- Rock, K.; McArdle, O.; Forde, P.; Dunne, M.; Fitzpatrick, D.; O’Neill, B.; Faul, C. A clinical review of treatment outcomes in glioblastoma multiforme—The validation in a non-trial population of the results of a randomised Phase III clinical trial: Has a more radical approach improved survival? Br. J. Radiol. 2012, 85, e729–e733. [Google Scholar] [CrossRef]
- Chang, K.-F.; Huang, X.-F.; Chang, J.T.; Huang, Y.-C.; Weng, J.-C.; Tsai, N.-M. Cedrol suppresses glioblastoma progression by triggering DNA damage and blocking nuclear translocation of the androgen receptor. Cancer Lett. 2020, 495, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.-K.; Shen, D.; Chen, Y.-S.; Yang, Q.-Y.; Guo, C.-C.; Feng, B.-H.; Chen, Z.-P. Enhanced MGMT expression contributes to temozolomide resistance in glioma stem-like cells. Chin. J. Cancer 2014, 33, 115. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; De Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Oram, R.A.; Jones, A.G.; Besser, R.E.; Knight, B.A.; Shields, B.M.; Brown, R.J.; Hattersley, A.T.; McDonald, T.J. The majority of patients with long-duration type 1 diabetes are insulin microsecretors and have functioning beta cells. Diabetologia 2014, 57, 187–191. [Google Scholar] [CrossRef]
- Naim, M.J.; Alam, M.J.; Nawaz, F.; Naidu, V.G.M.; Aaghaz, S.; Sahu, M.; Siddiqui, N.; Alam, O. Synthesis, molecular docking and anti-diabetic evaluation of 2,4-thiazolidinedione based amide derivatives. Bioorg. Chem. 2017, 73, 24–36. [Google Scholar] [CrossRef]
- Dowarah, J.; Singh, V.P. Anti-diabetic drugs recent approaches and advancements. Bioorg. Med. Chem. 2020, 28, 115263. [Google Scholar] [CrossRef]
- Avula, S.K.; Khan, A.; Rehman, N.U.; Anwar, M.U.; Al-Abri, Z.; Wadood, A.; Riaz, M.; Csuk, R.; Al-Harrasi, A. Synthesis of 1H-1,2,3-triazole derivatives as new α-glucosidase inhibitors and their molecular docking studies. Bioorg. Chem. 2018, 81, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Kenchappa, R.; Bodke, Y.D.; Chandrashekar, A.; Sindhe, M.A.; Peethambar, S. Synthesis of coumarin derivatives containing pyrazole and indenone rings as potent antioxidant and antihyperglycemic agents. Arab. J. Chem. 2017, 10, S3895–S3906. [Google Scholar] [CrossRef]
- Balan, K.; Ratha, P.; Prakash, G.; Viswanathamurthi, P.; Adisakwattana, S.; Palvannan, T. Evaluation of invitro α-amylase and α-glucosidase inhibitory potential of N2O2 schiff base Zn complex. Arab. J. Chem. 2017, 10, 732–738. [Google Scholar] [CrossRef]
- Raghu, S.V.; Mohammad, F.; Chua, J.Y.; Lam, J.S.W.; Loberas, M.; Sahani, S.; Barros, C.S.; Claridge-Chang, A. A zinc-finger fusion protein refines Gal4-defined neural circuits. Mol. Brain 2018, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Raghu, S.V.; Patil, R. GAL4-UAS system for genetic labeling and visualization of specific regions of brain. In Experiments with Drosophila for Biology Courses; Indian Academy of Sciences: Bengaluru, India, 2021; pp. 233–237. [Google Scholar]
- Brylinski, M. Aromatic interactions at the ligand-protein interface: Implications for the development of docking scoring functions. Chem. Biol. Drug Des. 2018, 91, 380–390. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Cryst. 2002, 40, 82–92. [Google Scholar]
- Husain, A.; Rashid, M.; Mishra, R.; Parveen, S.; Shin, D.-S.; Kumar, D. Benzimidazole bearing oxadiazole and triazolo-thiadiazoles nucleus: Design and synthesis as anticancer agents. Bioorg. Med. Chem. Lett. 2012, 22, 5438–5444. [Google Scholar] [CrossRef]
- Du, Q.-R.; Li, D.-D.; Pi, Y.-Z.; Li, J.-R.; Sun, J.; Fang, F.; Zhong, W.-Q.; Gong, H.-B.; Zhu, H.-L. Novel 1,3,4-oxadiazole thioether derivatives targeting thymidylate synthase as dual anticancer/antimicrobial agents. Bioorg. Med. Chem. 2013, 21, 2286–2297. [Google Scholar] [CrossRef]
- Zhang, S.; Luo, Y.; He, L.-Q.; Liu, Z.-J.; Jiang, A.-Q.; Yang, Y.-H.; Zhu, H.-L. Synthesis, biological evaluation, and molecular docking studies of novel 1, 3, 4-oxadiazole derivatives possessing benzotriazole moiety as FAK inhibitors with anticancer activity. Bioorg. Med. Chem. 2013, 21, 3723–3729. [Google Scholar] [CrossRef]
- Kazmi, M.; Zaib, S.; Ibrar, A.; Amjad, S.T.; Shafique, Z.; Mehsud, S.; Saeed, A.; Iqbal, J.; Khan, I. A new entry into the portfolio of α-glucosidase inhibitors as potent therapeutics for type 2 diabetes: Design, bioevaluation and one-pot multi-component synthesis of diamine-bridged coumarinyl oxadiazole conjugates. Bioorg. Chem. 2018, 77, 190–202. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, H.Y.; Jin, C.H. Mechanistic investigation of anthocyanidin derivatives as α-glucosidase inhibitors. Bioorg. Chem. 2019, 87, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Naveena, C.; Boja, P.; Manjunath, K.; Arulmoli, T.; Shalini, S. Synthesis, characterization and antimicrobial activity of some 2,5-disubstituted-3-acetyl-[1,3,4]-oxadiazoles carrying 2-(aryloxymethyl) phenyl moiety. Der. Pharma Chem. 2011, 3, 247–257. [Google Scholar]
- Shyma, P.; Kalluraya, B.; Peethambar, S.; Telkar, S.; Arulmoli, T. Synthesis, characterization and molecular docking studies of some new 1,3,4-oxadiazolines bearing 6-methylpyridine moiety for antimicrobial property. Eur. J. Med. Chem. 2013, 68, 394–404. [Google Scholar] [CrossRef]
- Buranaamnuay, K. The MTT assay application to measure the viability of spermatozoa: A variety of the assay protocols. Open Vet J. 2021, 11, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Nga, N.T.H.; Ngoc, T.T.B.; Trinh, N.T.M.; Thuoc, T.L.; Thao, D.T.P. Optimization and application of MTT assay in determining density of suspension cells. Anal. Biochem. 2020, 610, 113937. [Google Scholar] [CrossRef]
- Brix, N.; Samaga, D.; Hennel, R.; Gehr, K.; Zitzelsberger, H.; Lauber, K. The clonogenic assay: Robustness of plating efficiency-based analysis is strongly compromised by cellular cooperation. Radiat. Oncol. 2020, 15, 248. [Google Scholar] [CrossRef]
- Georgantzoglou, A.; Merchant, M.J.; Jeynes, J.C.; Mayhead, N.; Punia, N.; Butler, R.E.; Jena, R. Applications of High-Throughput Clonogenic Survival Assays in High-LET Particle Microbeams. Front. Oncol. 2015, 5, 305. [Google Scholar] [CrossRef]
- Ogihara, N.; Haley-Vicente, D. Protein target discovery and characterization-DS modeling and discovery studio streamline target discovery. Genet. Eng. News 2002, 22, 77. [Google Scholar]
- Trott, O.; Olson, A. Vina AutoDock: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Russo, T.V.; Martin, R.L.; Hay, P.J. Density functional calculations on first-row transition metals. J. Chem. Phys. 1994, 101, 7729–7737. [Google Scholar] [CrossRef]
- Lee, B.S. Causal relations among stock returns, interest rates, real activity, and inflation. J. Financ. 1992, 47, 1591–1603. [Google Scholar] [CrossRef]
- Frisch, M.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G. 09, Revision D. 01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Venugopal, P.P.; Chakraborty, D. Theoretical insights into molecular mechanism and energy criteria of PARP-2 enzyme inhibition by benzimidazole analogues. Proteins Struct. Funct. Bioinform. 2021, 89, 988–1004. [Google Scholar] [CrossRef] [PubMed]
- Das, B.K.; Chakraborty, D. Deciphering the competitive inhibition of dihydropteroate synthase by 8 marcaptoguanine analogs: Enhanced potency in phenylsulfonyl fragments. J. Biomol. Struct. Dyn. 2021, 1–20. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]

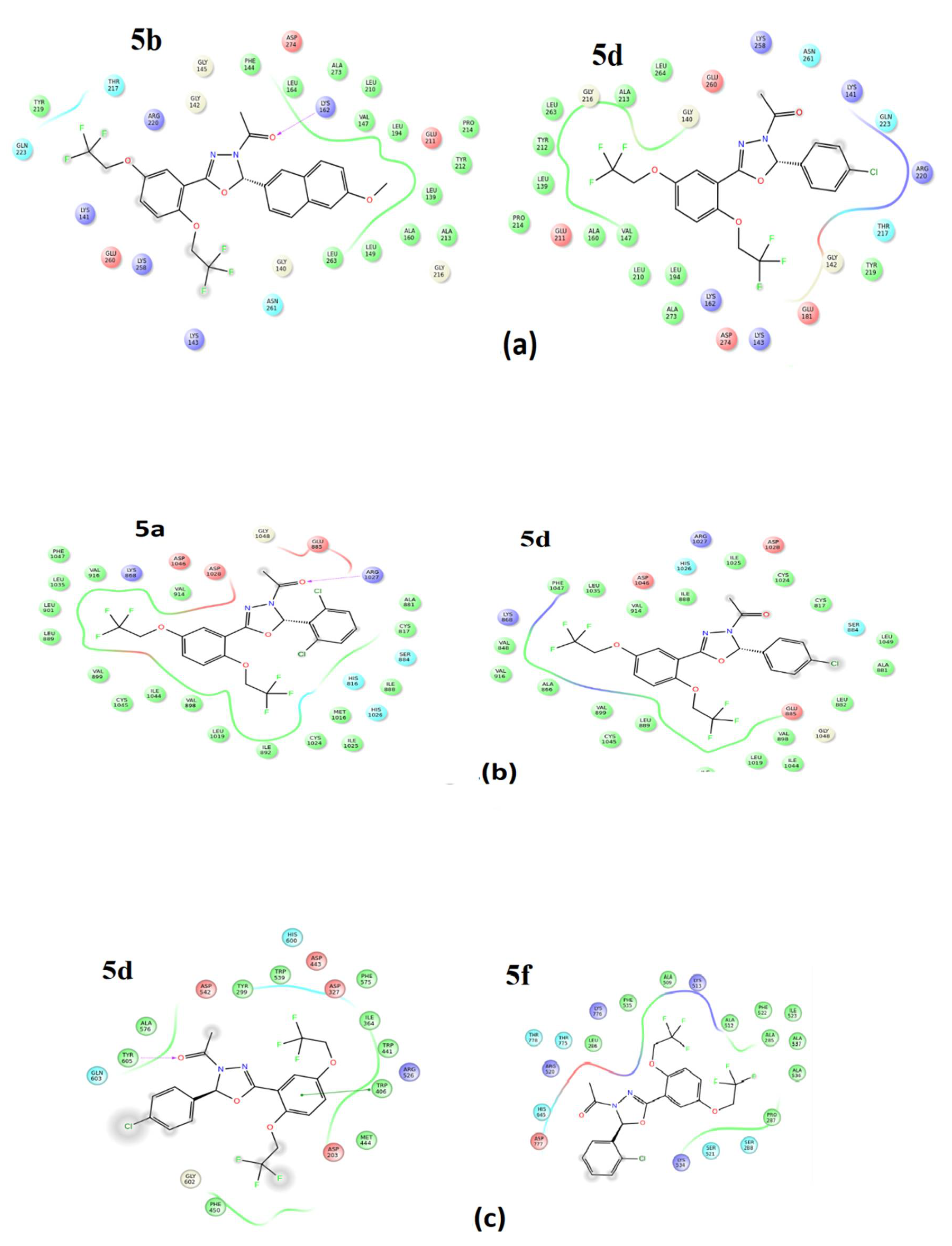

| Compound No | Ar | R |
|---|---|---|
| 5a | 2,6-Dichlorophenyl | H |
| 5b | 6-methoxy naphthyl | H |
| 5c | biphenyl | H |
| 5d | 4-chloro phenyl | H |
| 5e | furfuryl | H |
| 5f | 2-chlorophenyl | H |
| 5g | 3,4-dihydroxy phenyl | H |
| 5h | 3,4-dimethyl phenyl | H |
| 5i | 3-fluro-4-methoxy phenyl | CH3 |
| 5j | 4-hydroxy phenyl | CH3 |
| 5k | 2,4-dichloro phenyl | CH3 |
| 5l | 4-fluoro phenyl | CH3 |
| 5m | 2,6-dichloro phenyl | CH3 |
| 5n | 4-chloro phenyl | CH3 |
| Compound | Binding Affinity (kcal/mol) with | ||
|---|---|---|---|
| AURKA | VEGFR-2 | α-Glucosidase | |
| Temozolomide | −5.7 | −5.9 | --- |
| Acarbose | --- | --- | −7.4 |
| 1,3,4-oxadiazole derivatives | |||
| 5a | −8.6 | −8.8 | −7.9 |
| 5b | −10.0 | −9.9 | −8.8 |
| 5c | −10.1 | −9.0 | −9.2 |
| 5d | −9.4 | −9.2 | −8.2 |
| 5e | −8.8 | −9.0 | −8.8 |
| 5f | −8.7 | −9.3 | −8.5 |
| 5g | −9.2 | −9.2 | −7.9 |
| 5i | −8.8 | −8.9 | −8.3 |
| 5j | −8.6 | −8.8 | −8.4 |
| 5k | −8.8 | −9.1 | −8.8 |
| 5l | −8.5 | −9.1 | −8.5 |
| 5m | −8.5 | −9.5 | −7.8 |
| Compounds | Residues | Distance | Angle | Type |
|---|---|---|---|---|
| 5b | Tyr 212 Tyr 219 | 8.27 6.34 | 14.61 85.36 | face-to-face edge-to-face |
| 5c | Tyr 212 Tyr 219 | 6.14 6.37 | 17.99 82.35 | face-to-face edge-to-face |
| 5d | Tyr 212 Tyr 219 | 9.89 5.60 | 11.61 74.08 | face-to-face edge-to-face |
| 5e | Tyr 212 Tyr 219 | 7.46 6.49 | 22.38 27.86 | face-to-face face-to-face |
| 5i | Tyr 212 Tyr 219 | 8.75 6.36 | 16.72 82.63 | face-to-face edge-to-face |
| 5k | Tyr 219 | 6.51 | 75.07 | edge-to-face |
| 5f | Tyr 219 | 5.23 | 50.33 | edge-to-face |
| 5g | Tyr 212 Tyr 219 | 7.27 6.14 | 12.82 89.12 | face-to-face edge-to-face |
| Compounds | Residues | Distance | Angle | Type |
|---|---|---|---|---|
| 5b | His 816 | 10.89 | 37.06 | face-to-face |
| His 1026 | 9.59, 6.08 | 86.68, 65.42 | edge-to-face | |
| 5c | His 1026 | 6.86, 8.29 | 48.33, 24.93 | face-to-face |
| 6.49 | 66.93 | edge-to-face | ||
| Tyr 1059 | 7.36 | 22.49 | face-to-face | |
| Tyr 1082 | 7.39 | 82.50 | edge-to-face | |
| 5d | His 1026 | 7.76 | 45.03 | face-to-face |
| Phe 1047 | 8.69 | 50.66 | face-to-face | |
| 5e | His 891 | 9.29 | 75.45 | edge-to-face |
| His 1026 | 7.67 | 42.66 | face-to-face | |
| 5.89 | 80.79 | edge-to-face | ||
| Phe 1047 | 8.69 | 52.60 | face-to-face | |
| 5i | His 1026 | 6.57 | 74.77 | edge-to-face |
| 7.10 | 51.63 | face-to-face | ||
| 5k | His 1026 | 7.07 | 39.57 | face-to-face |
| 6.55 | 71.69 | edge-to-face | ||
| 5f | His 1026 | 7.62 | 47.75 | face-to-face |
| Phe 1047 | 8.2 | 48.47 | face-to-face | |
| 5g | His 1026 | 7.75 | 44.92 | face-to-face |
| Phe 1047 | 8.65 | 50.92 | face-to-face |
| Ligand | Hydrogen Bond Occupancy (%) | |
|---|---|---|
| AURKA/5b | 213Ala (H) — 5b (O) | 46.0 |
| 212Tyr (H) — 5b (O) | 2.2 | |
| 137Arg (H) — 5b (O) | 3.9 | |
| VEGFR-2/5b | 1046Asp (H) — 5b (O) | 35.0 |
| 1025Ile (H) — 5b (O) | 3.7 | |
| 816His (H) — 5b (O) | 3.1 | |
| Compounds | MW | DM | Rotor | H Bond Acceptor | H Bond Donor | Molar Refractivity | PSA | Log Po/w | BBB Permeant | Log Kp | Rule of 5 | Bioavailability SCORE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1,3,4-oxadiazole derivatives | ||||||||||||
| 5a | 531.23 | 3.55 | 9 | 11 | 0 | 116.31 | 60.36 | 5.81 | no | −5.02 | No; 2 violations | 0.17 |
| 5b | 542.43 | 5.65 | 10 | 12 | 0 | 130.29 | 69.59 | 5.6 | no | −5.11 | Yes; 1 violation | 0.55 |
| 5c | 538.44 | 4.90 | 10 | 11 | 0 | 131.72 | 60.36 | 6.09 | no | −4.8 | No; 2 violations | 0.17 |
| 5d | 496.79 | 5.55 | 9 | 11 | 0 | 111.3 | 60.36 | 5.31 | no | −5.26 | Yes; 1 violation | 0.55 |
| 5e | 452.3 | 4.65 | 9 | 12 | 0 | 98.56 | 73.50 | 4.17 | no | −6.07 | Yes; 0 violation | 0.55 |
| 5i | 524.39 | 4.97 | 10 | 13 | 0 | 117.43 | 69.59 | 5.3 | no | −5.69 | Yes; 1 violation | 0.55 |
| 5j | 492.37 | 3.42 | 9 | 12 | 1 | 113 | 80.59 | 4.67 | no | −5.80 | Yes; 0 violation | 0.55 |
| 5k | 545.26 | 4.65 | 9 | 11 | 0 | 121 | 60.36 | 6.1 | no | −4.97 | No; 2 violations | 0.17 |
| 5l | 494.36 | 5.99 | 9 | 12 | 0 | 110.94 | 60.36 | 5.38 | no | −5.48 | Yes; 1 violation | 0.55 |
| 5m | 545.26 | 3.13 | 9 | 11 | 0 | 121 | 60.36 | 6.11 | no | −4.97 | No; 2 violations | 0.17 |
| 5f | 496.79 | 2.95 | 9 | 11 | 0 | 111.3 | 60.36 | 5.33 | no | −5.26 | Yes; 1 violation | 0.55 |
| 5g | 494.34 | 2.69 | 9 | 13 | 2 | 110.34 | 100.82 | 4.01 | no | −6.19 | Yes; 0 violation | 0.55 |
| Oxadiazole Compounds | IC50 Concentration in µM | Percentage of Cell Death at 25 µM |
|---|---|---|
| 5a | 11.59 | 36.52 |
| 5b | 10.14 | 51.58 |
| 5c | 14.32 | 28.04 |
| 5d | 8.141 | 50.12 |
| 5e | 13.24 | 32.57 |
| 5f | 12.02 | 19.53 |
| 5g | 11.74 | 15.12 |
| 5i | 10.52 | 35.41 |
| 5k | 15.98 | 35.05 |
| 5l | 12.62 | 16.48 |
| 5m | 10.48 | 53.65 |
| 5n | 11.91 | 17.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shankara, S.D.; Isloor, A.M.; Kudva, A.K.; Raghu, S.V.; Jayaswamy, P.K.; Venugopal, P.P.; Shetty, P.; Chakraborty, D. 2,5-Bis(2,2,2-trifluoroethoxy)phenyl-tethered 1,3,4-Oxadiazoles Derivatives: Synthesis, In Silico Studies, and Biological Assessment as Potential Candidates for Anti-Cancer and Anti-Diabetic Agent. Molecules 2022, 27, 8694. https://doi.org/10.3390/molecules27248694
Shankara SD, Isloor AM, Kudva AK, Raghu SV, Jayaswamy PK, Venugopal PP, Shetty P, Chakraborty D. 2,5-Bis(2,2,2-trifluoroethoxy)phenyl-tethered 1,3,4-Oxadiazoles Derivatives: Synthesis, In Silico Studies, and Biological Assessment as Potential Candidates for Anti-Cancer and Anti-Diabetic Agent. Molecules. 2022; 27(24):8694. https://doi.org/10.3390/molecules27248694
Chicago/Turabian StyleShankara, Sathyanarayana D., Arun M. Isloor, Avinash K. Kudva, Shamprasad Varija Raghu, Pavan K. Jayaswamy, Pushyaraga P. Venugopal, Praveenkumar Shetty, and Debashree Chakraborty. 2022. "2,5-Bis(2,2,2-trifluoroethoxy)phenyl-tethered 1,3,4-Oxadiazoles Derivatives: Synthesis, In Silico Studies, and Biological Assessment as Potential Candidates for Anti-Cancer and Anti-Diabetic Agent" Molecules 27, no. 24: 8694. https://doi.org/10.3390/molecules27248694
APA StyleShankara, S. D., Isloor, A. M., Kudva, A. K., Raghu, S. V., Jayaswamy, P. K., Venugopal, P. P., Shetty, P., & Chakraborty, D. (2022). 2,5-Bis(2,2,2-trifluoroethoxy)phenyl-tethered 1,3,4-Oxadiazoles Derivatives: Synthesis, In Silico Studies, and Biological Assessment as Potential Candidates for Anti-Cancer and Anti-Diabetic Agent. Molecules, 27(24), 8694. https://doi.org/10.3390/molecules27248694







