Evaluation of Feasibility of Using the Bacteriophage T4 Lysozyme to Improve the Hydrolysis and Biochemical Methane Potential of Secondary Sludge
Abstract
:1. Introduction
2. Materials and Methods
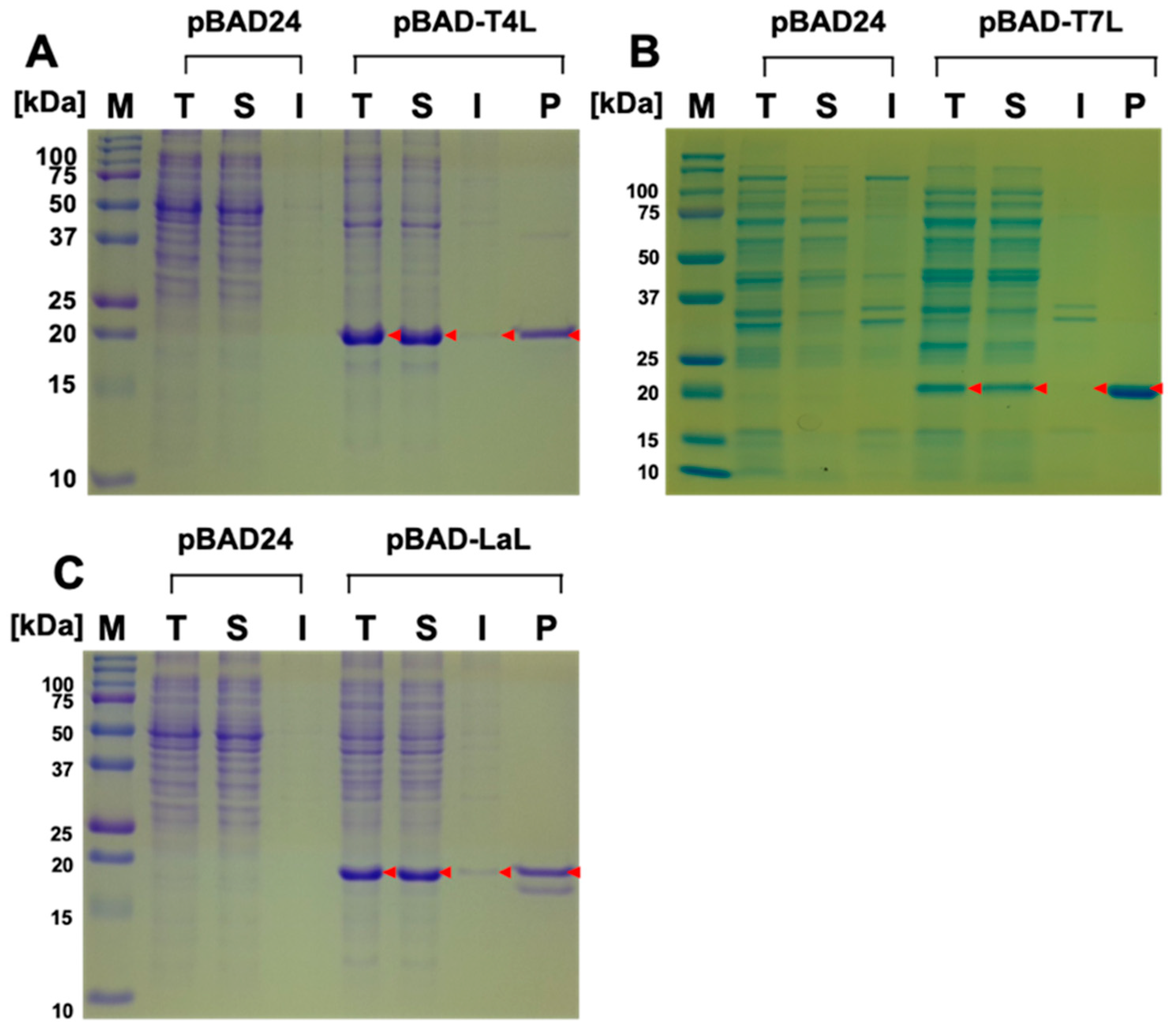
2.1. Expression and Purification of Bacteriophage Lysozymes
2.2. Secondary Sludge as Substrates
2.3. Investigation of the Hydrolysis Efficiency and Biochemical Methane Potential (BMP)
2.4. Analytical Protocols
3. Results and Discussion
3.1. Characteristics of the Sludge and Bacteriophage T4 Lysozyme Treatment
3.2. Effect of Bacteriophage T4 Lysozymes on Biogas Production
3.3. Differences in Bacterial Communities
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TS | Total solid |
| VS | Volatile solid |
| TSS | Total suspended solid |
| VSS | Volatile suspended solid |
| COD | Chemical oxygen demand |
| T4L | Bacteriophage T4 lysozyme |
| T7L | Bacteriophage T7 lysozyme |
| LaL | Bacteriophage lambda lysozyme |
| AD | Anaerobic digestion |
| OLR | Organic loading rate |
| BMP | Biochemical methane potential |
| WWTPs | Wastewater treatment plants |
| GlcNAc | N-acetyl-D-glucosamine |
| MurNAc | N-acetylmuramic acid |
| CSTR | Completely stirred tank reactor |
| SBR | Sequencing batch reactor |
| MLE | Modified Ludzack-Ettinger |
| MBR | Membrane bioreactor |
| A2O | Anaerobic-anoxic/oxic |
References
- Pilli, S.; More, T.T.; Yan, S.; Tyagi, R.D.; Surampalli, R.Y. Fenton pre-treatment of secondary sludge to enhance anaerobic digestion: Energy balance and greenhouse gas emissions. Chem. Eng. J. 2016, 283, 285–292. [Google Scholar] [CrossRef]
- Shin, J.; Cho, S.-K.; Lee, J.; Hwang, K.; Chung, J.W.; Jang, H.-N.; Shin, S.G. Performance and Microbial Community Dynamics in Anaerobic Digestion of Waste Activated Sludge: Impact of Immigration. Energies 2019, 12, 573. [Google Scholar] [CrossRef]
- Hagos, K.; Zong, J.; Li, D.; Liu, C.; Lu, X. Anaerobic co-digestion process for biogas production: Progress, challenges and perspectives. Renew. Sust. Energ. Rev. 2017, 76, 1485–1496. [Google Scholar] [CrossRef]
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of anaerobic digestion process: A review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef] [PubMed]
- Ganesh Saratale, R.; Kumar, G.; Banu, R.; Xia, A.; Periyasamy, S.; Dattatraya Saratale, G. A critical review on anaerobic digestion of microalgae and macroalgae and co-digestion of biomass for enhanced methane generation. Bioresour. Technol. 2018, 262, 319–332. [Google Scholar] [CrossRef]
- Lee, J.; Han, G.; Shin, S.G.; Koo, T.; Cho, K.; Kim, W.; Hwang, S. Seasonal monitoring of bacteria and archaea in a full-scale thermophilic anaerobic digester treating food waste-recycling wastewater: Correlations between microbial community characteristics and process variables. Chem. Eng. J. 2016, 300, 291–299. [Google Scholar] [CrossRef]
- Rabii, A.; Aldin, S.; Dahman, Y.; Elbeshbishy, E. A review on anaerobic co-digestion with a focus on the microbial populations and the effect of multi-stage digester configuration. Energies 2019, 12, 1106. [Google Scholar] [CrossRef]
- Mahdy, A.; Mendez, L.; Ballesteros, M.; González-Fernández, C. Algaculture integration in conventional wastewater treatment plants: Anaerobic digestion comparison of primary and secondary sludge with microalgae biomass. Bioresour. Technol. 2015, 184, 236–244. [Google Scholar] [CrossRef]
- Carlsson, M.; Lagerkvist, A.; Morgan-Sagastume, F. The effects of substrate pre-treatment on anaerobic digestion systems: A review. Waste Manag. 2012, 32, 1634–1650. [Google Scholar] [CrossRef]
- Cerrillo, M.; Viñas, M.; Bonmatí, A. Removal of volatile fatty acids and ammonia recovery from unstable anaerobic digesters with a microbial electrolysis cell. Bioresour. Technol. 2016, 219, 348–356. [Google Scholar] [CrossRef]
- Mutungwazi, A.; Mukumba, P.; Makaka, G. Biogas digester types installed in South Africa: A review. Renew. Sust. Energ. Rev. 2018, 81, 172–180. [Google Scholar] [CrossRef]
- Liu, G.; Wang, K.; Li, X.; Ma, L.; Ma, X.; Chen, H. Enhancement of excess sludge hydrolysis and decomposition with different lysozyme dosage. J. Hazard. Mater. 2019, 366, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Hendriks, A.T.W.M.; van Lier, J.B.; de Kreuk, M. Pre-treatments to enhance the biodegradability of waste activated sludge: Elucidating the rate limiting step. Biotechnol. Adv. 2018, 36, 1434–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Qiao, W.; Wang, X.; Takayanagi, K.; Shofie, M.; Li, Y.-Y. Kinetic characterization of thermophilic and mesophilic anaerobic digestion for coffee grounds and waste activated sludge. Waste Manag. 2015, 36, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Lee, J.; Loh, K.-C.; Dai, Y.; Tong, Y.W. Enhancement of biogas production in anaerobic co-digestion of food waste and waste activated sludge by biological co-pretreatment. Energy 2017, 137, 479–486. [Google Scholar] [CrossRef]
- Pilli, S.; Yan, S.; Tyagi, R.D.; Surampalli, R.Y. Thermal pretreatment of sewage sludge to enhance anaerobic digestion: A review. Crit. Rev. Environ. Sci. Technol. 2015, 45, 669–702. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, Y.-J.; Meng, S.-J.; Kiran, E.U.; Liu, Y. Enzymatic pretreatment of activated sludge, food waste and their mixture for enhanced bioenergy recovery and waste volume reduction via anaerobic digestion. Appl. Energy 2016, 179, 1131–1137. [Google Scholar] [CrossRef]
- Ennouri, H.; Miladi, B.; Diaz, S.Z.; Güelfo, L.A.F.; Solera, R.; Hamdi, M.; Bouallagui, H. Effect of thermal pretreatment on the biogas production and microbial communities balance during anaerobic digestion of urban and industrial waste activated sludge. Bioresour. Technol. 2016, 214, 184–191. [Google Scholar] [CrossRef]
- Bougrier, C.; Delgenès, J.P.; Carrère, H. Combination of Thermal Treatments and Anaerobic Digestion to Reduce Sewage Sludge Quantity and Improve Biogas Yield. Process Saf. Environ. Prot. 2006, 84, 280–284. [Google Scholar] [CrossRef]
- Fang, S.; Gu, W.; Chen, L.; Yu, Z.; Dai, M.; Lin, Y.; Liao, Y.; Ma, X. Ultrasonic pretreatment effects on the co-pyrolysis of municipal solid waste and paper sludge through orthogonal test. Bioresour. Technol. 2018, 258, 5–11. [Google Scholar] [CrossRef]
- Carrère, H.; Dumas, C.; Battimelli, A.; Batstone, D.J.; Delgenès, J.P.; Steyer, J.P.; Ferrer, I. Pretreatment methods to improve sludge anaerobic degradability: A review. J. Hazard. Mater. 2010, 183, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; de Vries, H.; Rouau, X. Dry fractionation process as an important step in current and future lignocellulose biorefineries: A review. Bioresour. Technol. 2013, 134, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Liu, C.; Liu, J.; Guo, X. Evaluation of the microbial cell structure damages in alkaline pretreatment of waste activated sludge. Bioresour. Technol. 2015, 196, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; Chuetor, S.; Monlau, F.; Solhy, A.; Rouau, X. Eco-friendly dry chemo-mechanical pretreatments of lignocellulosic biomass: Impact on energy and yield of the enzymatic hydrolysis. Appl. Energy 2014, 113, 97–105. [Google Scholar] [CrossRef]
- Yu, S.; Zhang, G.; Li, J.; Zhao, Z.; Kang, X. Effect of endogenous hydrolytic enzymes pretreatment on the anaerobic digestion of sludge. Bioresour. Technol. 2013, 146, 758–761. [Google Scholar] [CrossRef] [PubMed]
- Hosseini Koupaie, E.; Dahadha, S.; Bazyar Lakeh, A.A.; Azizi, A.; Elbeshbishy, E. Enzymatic pretreatment of lignocellulosic biomass for enhanced biomethane production-A review. J. Environ. Manag. 2019, 233, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Porfírio, S.; Carlson, R.W.; Azadi, P. Elucidating Peptidoglycan Structure: An Analytical Toolset. Trends. Microbiol. 2019, 27, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef]
- Bilej, M. Mucosal Immunity in Invertebrates. In Mucosal Immunology, 4th ed.; Mestecky, J., Strober, W., Russell, M.W., Kelsall, B.L., Cheroutre, H., Lambrecht, B.N., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 135–144. [Google Scholar]
- Wu, T.; Jiang, Q.; Wu, D.; Hu, Y.; Chen, S.; Ding, T.; Ye, X.; Liu, D.; Chen, J. What is new in lysozyme research and its application in food industry? A review. Food Chem. 2019, 274, 698–709. [Google Scholar] [CrossRef]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell Wall Hydrolases in Bacteria: Insight on the Diversity of Cell Wall Amidases, Glycosidases and Peptidases toward Peptidoglycan. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Zaloba, P.; Bailey-Elkin, B.A.; Derksen, M.; Mark, B.L. Structural and Biochemical Insights into the Peptidoglycan Hydrolase Domain of FlgJ from Salmonella typhimurium. PLoS ONE 2016, 11, e0149204. [Google Scholar] [CrossRef] [PubMed]
- Rivera, I.; Molina, R.; Lee, M.; Mobashery, S.; Hermoso, J.A. Orthologous and Paralogous AmpD Peptidoglycan Amidases from Gram-Negative Bacteria. Microb. Drug Resist. 2016, 22, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Lamppa, J.W.; Tanyos, S.A.; Griswold, K.E. Engineering Escherichia coli for soluble expression and single step purification of active human lysozyme. J. Biotechnol. 2013, 164, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Angelidaki, I.; Alves, M.; Bolzonella, D.; Borzacconi, L.; Campos, J.; Guwy, A.; Kalyuzhnyi, S.; Jenicek, P.; Van Lier, J. Defining the biomethane potential (BMP) of solid organic wastes and energy crops: A proposed protocol for batch assays. Water Sci. Technol. 2009, 59, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi-Nik, M.; Heidari, A.; Ramezani Azghandi, S.; Asadi Mohammadi, F.; Younesi, H. Drinking water treatment sludge as an effective additive for biogas production from food waste; kinetic evaluation and biomethane potential test. Bioresour. Technol. 2018, 260, 421–426. [Google Scholar] [CrossRef] [PubMed]
- APHA-AWWA-WEF. Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association: Washington, DC, USA, 2005. [Google Scholar]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Zhang, J.; Lv, C.; Tong, J.; Liu, J.; Liu, J.; Yu, D.; Wang, Y.; Chen, M.; Wei, Y. Optimization and microbial community analysis of anaerobic co-digestion of food waste and sewage sludge based on microwave pretreatment. Bioresour. Technol. 2016, 200, 253–261. [Google Scholar] [CrossRef]
- Grosser, A. Determination of methane potential of mixtures composed of sewage sludge, organic fraction of municipal waste and grease trap sludge using biochemical methane potential assays. A comparison of BMP tests and semi-continuous trial results. Energy 2018, 143, 488–499. [Google Scholar] [CrossRef]
- Bayr, S.; Kaparaju, P.; Rintala, J. Screening pretreatment methods to enhance thermophilic anaerobic digestion of pulp and paper mill wastewater treatment secondary sludge. Chem. Eng. J. 2013, 223, 479–486. [Google Scholar] [CrossRef] [Green Version]
- Ju, F.; Zhang, T. Bacterial assembly and temporal dynamics in activated sludge of a full-scale municipal wastewater treatment plant. ISME J. 2014, 9, 683. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Luo, K.; Li, X.-M.; Wang, D.-B.; Zheng, W.; Zeng, G.-M.; Liu, J.-J. Enhanced efficiency of biological excess sludge hydrolysis under anaerobic digestion by additional enzymes. Bioresour. Technol. 2010, 101, 2924–2930. [Google Scholar] [CrossRef] [PubMed]
- Marzorati, M.; Wittebolle, L.; Boon, N.; Daffonchio, D.; Verstraete, W. How to get more out of molecular fingerprints: Practical tools for microbial ecology. Environ. Microbiol. 2008, 10, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.-D.; He, J.-G.; Qiu, W.; Tang, J.; Liu, T.-T. Microbial community related to lysozyme digestion process for boosting waste activated sludge biodegradability. Bioresour. Technol. 2015, 175, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Eddy, M. Wastewater Engineering: Treatment and Resource Recovery, 5th ed.; McGraw-Hill Education: New York, NY, USA, 2013. [Google Scholar]
- Liu, G.; Li, X.; Ma, L.; Ma, X.; Chen, H. Enhancement of excess sludge hydrolysis and decomposition by combined lysozyme and rhamnolipid pretreatment. Bioresour. Technol. 2019, 289, 121703. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yu, Z.; Zhao, J.; Zhang, H. Do microbial communities in an anaerobic bioreactor change with continuous feeding sludge into a full-scale anaerobic digestion system? Bioresour. Technol. 2018, 249, 89–98. [Google Scholar] [CrossRef] [PubMed]
- He, Z.-W.; Liu, W.-Z.; Gao, Q.; Tang, C.-C.; Wang, L.; Guo, Z.-C.; Zhou, A.-J.; Wang, A.-J. Potassium ferrate addition as an alternative pre-treatment to enhance short-chain fatty acids production from waste activated sludge. Bioresour. Technol. 2018, 247, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, N.; Tewari, R.; Hoondal, G.S. Biotechnological aspects of chitinolytic enzymes: A review. Appl. Microbiol. Biotechnol. 2006, 71, 773–782. [Google Scholar] [CrossRef] [PubMed]



| Plasmids | Description | Sources |
|---|---|---|
| pBAD/Myc-His/lacZ | Expression vector, PBAD:lacZ, pBR322 ori, AmpR | Invitrogen |
| pBAD24 | Expression vector, PBAD, pBR322 ori, AmpR | Invitrogen |
| pBAD-T7L | Plasmid expressing T7 lysozyme with C-terminal 6xHis tag, AmpR | This study |
| pBAD-T4L, LaL | Plasmid expressing T4, λ lysozyme with N-terminal 6xHis tag, AmpR | This study |
| pLysS | Plasmid containing T7 lysozyme gene | Novagen |
| pSyn-T4L, LaL | Plasmid containing synthesized T4, λ lysozyme gene | This study |
| Oligonucleotides | 5′ to 3′ | Reference |
| T4L-F | gctaacaggaggaattaacc | This study |
| T4L-R | tgagtttttgttcgggccca | This study |
| LaL-F | gctaacaggaggaattaacc | This study |
| LaL-R | tgagtttttgttcgggccca | This study |
| T7L-F | ggctaacaggaggaattaaccatggctcgtgtacagtttaa | This study |
| T7L-R | tgagtttttgttcgggcccaagctttccacggtcagaagtgacca | This study |
| Parameters | Busan | Pohang | Ulsan |
|---|---|---|---|
| Daily flow rates (m3/day) | 450,000 | 232,000 | 250,000 |
| Treatment process | MLE | MBR | A2O |
| Operational modes | CSTR | SBR | SBR |
| Aerobic reactor size (m3) | 2821 | 3221 | 3593 |
| pH in aerobic reactor | 7.3 | 6.8 | 6.9 |
| Alkalinity (mg CaCO3 eq./L) in aerobic reactor | 257 | 288 | 430 |
| Hydrolysis experiment | |
|---|---|
| Parameters | Description |
| Substrate (S) | Secondary sludges from Busan, Pohang, Ulsan WWTPs |
| Enzyme (E) | Bacteriophage T4 lysozyme (T4L) |
| pH | 7.0 |
| Temperature (°C) | 45.0 |
| S-E ratio | 1 g sludge VSS/g VS of T4L |
| Working Volume (mL) | 50 |
| BMP test | |
| Parameters | Description |
| pH | 7.5 |
| Temperature (°C) | 37°C |
| F/M ratio | 1 g VS/g VSS |
| Working Volume (mL) | 70 |
| Medium A (g/L) | NH4Cl, 100; NaCl, 10; MgCl2 6H2O, 10; CaCl2 2H2O, 5 |
| Medium B (g/L) | K2HPO4 3H2O, 200 |
| Medium C (g/L) | Resazurin, 0.5 |
| Medium D (g/L) | FeCl2 4H2O, 2; H3BO3, 0.05; ZnCl2, 0.05; CuCl2 2H2O, 0.038; MnCl2 4H2O, 0.05; (NH4)6Mo7O24 4H2O, 0.05; AlCl3, 0.05; CoCl2 6H2O, 0.05; NiCl2 6H2O, 0.092; ethylenediaminetetraacetate, 0.5; concentrated HCl, 1 mL; Na2SeO3 5H2O, 0.1 |
| Medium E (g/L) | Biotin, 2; folic acid, 2; pyridoxine acid, 10; ridoflavin, 5; thiamine hydrochloride, 5; cyanocobalamine, 0.1; nicotinic acid, 5; P-aminobenzoic acid, 5; lipoic acid, 5; DL-pantothenic acid |
| Analytical Items (g/L) | Raw Secondary Sludges | |||||
|---|---|---|---|---|---|---|
| Busan | Pohang | Ulsan | ||||
| Average | Stdev. * | Average | Stdev. | Average | Stdev. | |
| pH | 7.3 | 6.8 | 6.9 | |||
| TS | 12.3 | 0.1 | 6.1 | 0.0 | 4.3 | 0.2 |
| VS | 10.3 | 0.0 | 5.4 | 0.1 | 3.4 | 0.1 |
| TSS | 9.6 | 0.9 | 5.7 | 0.1 | 3.6 | 0.0 |
| VSS | 8.2 | 0.7 | 4.3 | 0.2 | 3.0 | 0.1 |
| COD | 14.7 | 1.0 | 7.3 | 0.4 | 5.5 | 0.0 |
| sCOD | 0.1 | 0.0 | 0.0 | 0.0 | 0.4 | 0.0 |
| Total carbohydrate | 1.5 | 0.2 | 0.8 | 0.0 | 0.5 | 0.0 |
| Protein | 7.7 | 0.3 | 3.5 | 0.2 | 2.1 | 0.0 |
| Lipid | 1.0 | 0.3 | 0.9 | 0.0 | 0.6 | 0.2 |
| VS/TS (%) | 83.5 | 89.1 | 78.5 | |||
| VSS/TS (%) | 66.6 | 70.9 | 68.9 | |||
| VSS/VS (%) | 79.7 | 79.6 | 87.8 | |||
| Busan | Pohang | Ulsan | ||||
|---|---|---|---|---|---|---|
| Raw | T4L | Raw | T4L | Raw | T4L | |
| Biogas production (NmL/g VSin) | 58.2 | 111.0 | 106.2 | 130.3 | 94.8 | 106.2 |
| Methane content (%) | 64.7 | 65.4 | 60.6 | 66.1 | 58.9 | 64.1 |
| t75(d) * | 12.7. | 6.8 | 6.8 | 5.5 | 4.7 | 4.7 |
| k (d) | 0.37 | 0.54 | 0.50 | 0.57 | 0.47 | 0.58 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Woo, S.-G.; Lee, J.; Lee, D.-H.; Hwang, S. Evaluation of Feasibility of Using the Bacteriophage T4 Lysozyme to Improve the Hydrolysis and Biochemical Methane Potential of Secondary Sludge. Energies 2019, 12, 3644. https://doi.org/10.3390/en12193644
Kim S, Woo S-G, Lee J, Lee D-H, Hwang S. Evaluation of Feasibility of Using the Bacteriophage T4 Lysozyme to Improve the Hydrolysis and Biochemical Methane Potential of Secondary Sludge. Energies. 2019; 12(19):3644. https://doi.org/10.3390/en12193644
Chicago/Turabian StyleKim, Sangmin, Seung-Gyun Woo, Joonyeob Lee, Dae-Hee Lee, and Seokhwan Hwang. 2019. "Evaluation of Feasibility of Using the Bacteriophage T4 Lysozyme to Improve the Hydrolysis and Biochemical Methane Potential of Secondary Sludge" Energies 12, no. 19: 3644. https://doi.org/10.3390/en12193644
APA StyleKim, S., Woo, S.-G., Lee, J., Lee, D.-H., & Hwang, S. (2019). Evaluation of Feasibility of Using the Bacteriophage T4 Lysozyme to Improve the Hydrolysis and Biochemical Methane Potential of Secondary Sludge. Energies, 12(19), 3644. https://doi.org/10.3390/en12193644






