Anaerobic Digestion of Agricultural Waste Using Microbial Inocula: Performance and Characterization of Bacterial Communities Using 16S rRNA Sequencing Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Substrate and Inocula
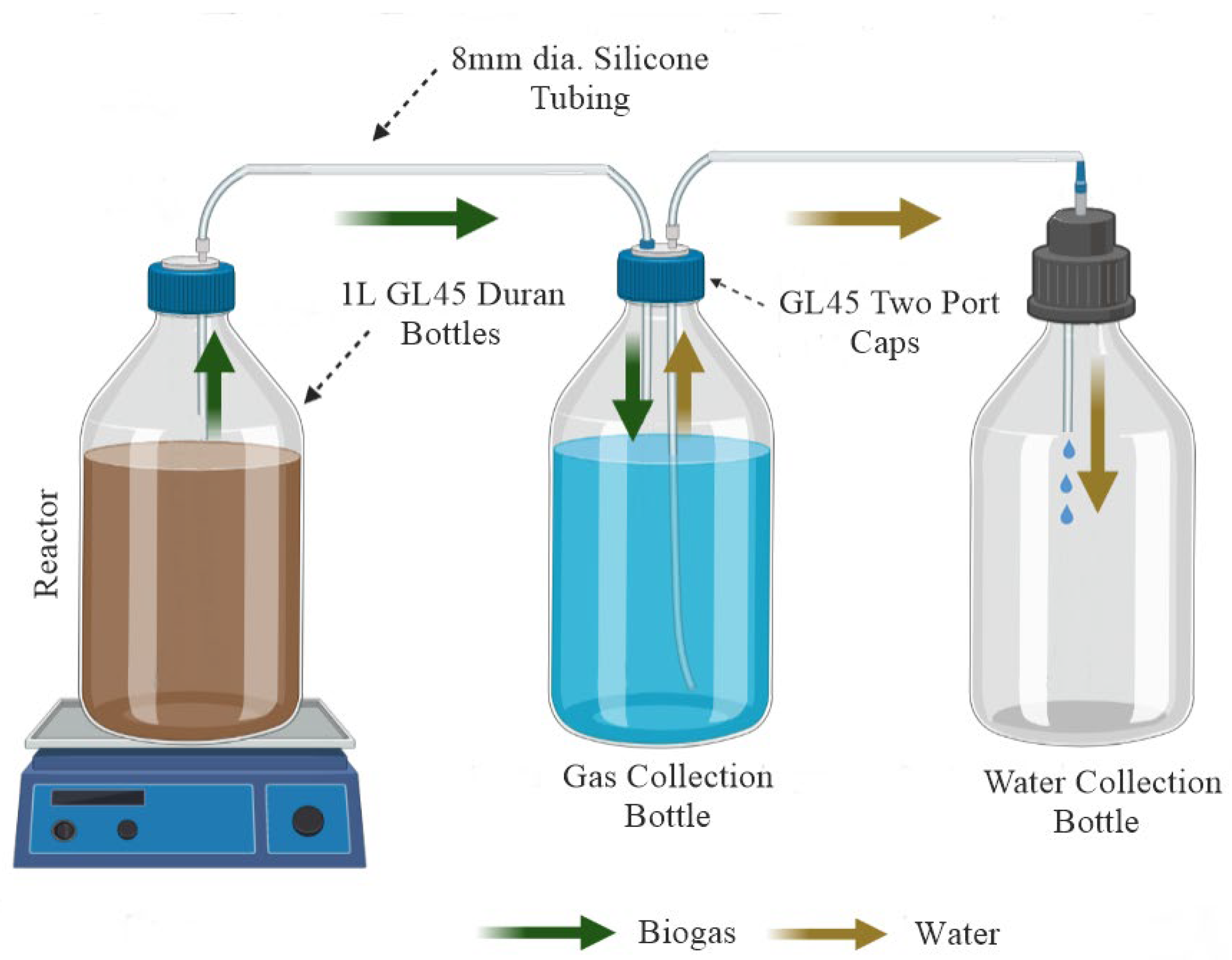
2.2. Anaerobic Digestion Batches Set-Up
2.3. Biochemical Characterization of Substrate and Inocula
2.4. Analytical Methods
2.5. Data Analysis
3. Results
3.1. Characterization of the Raw Substrate and Used Inocula
3.1.1. Total Solids and Volatile Solids
3.1.2. Biochemical Characterization
3.2. Monitoring of the BMP Experiments
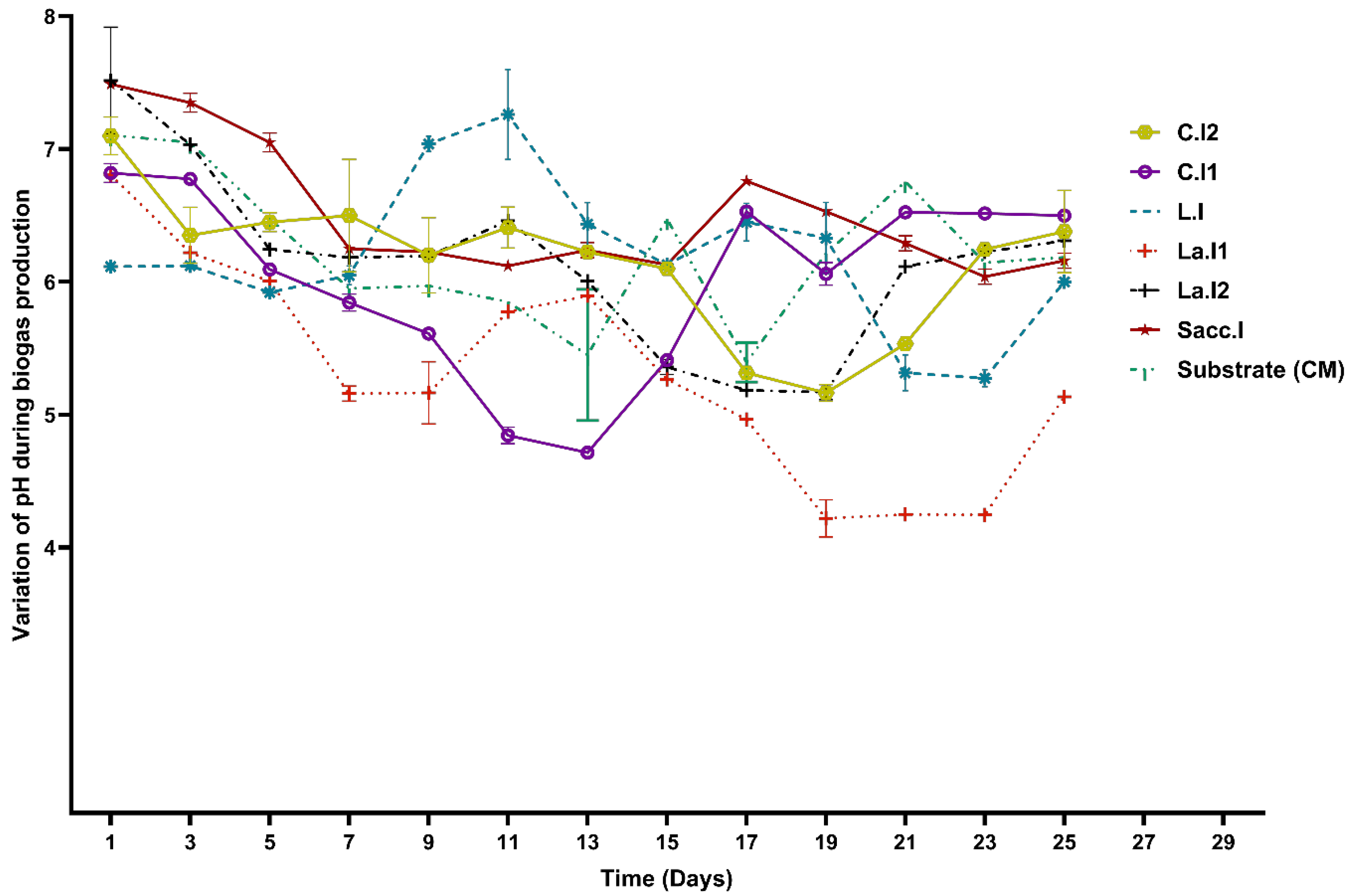
3.3. Daily Monitoring of pH Variation during BMP Experiments
3.4. Biogas Production in Different BMP Experiments
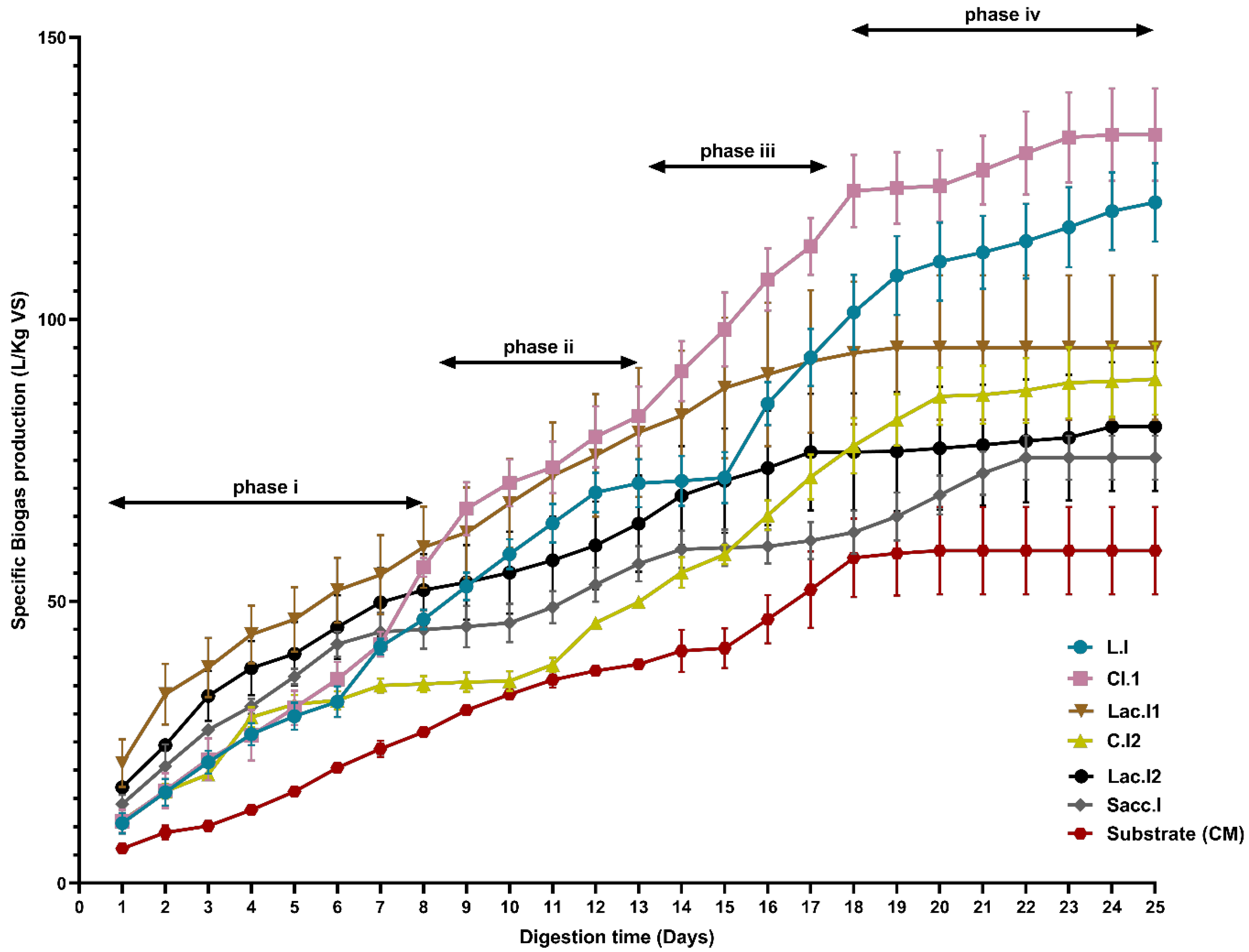
3.4.1. Daily Monitoring of Biogas Production
3.4.2. Cumulative Biogas Yield of Different BMP Experiments
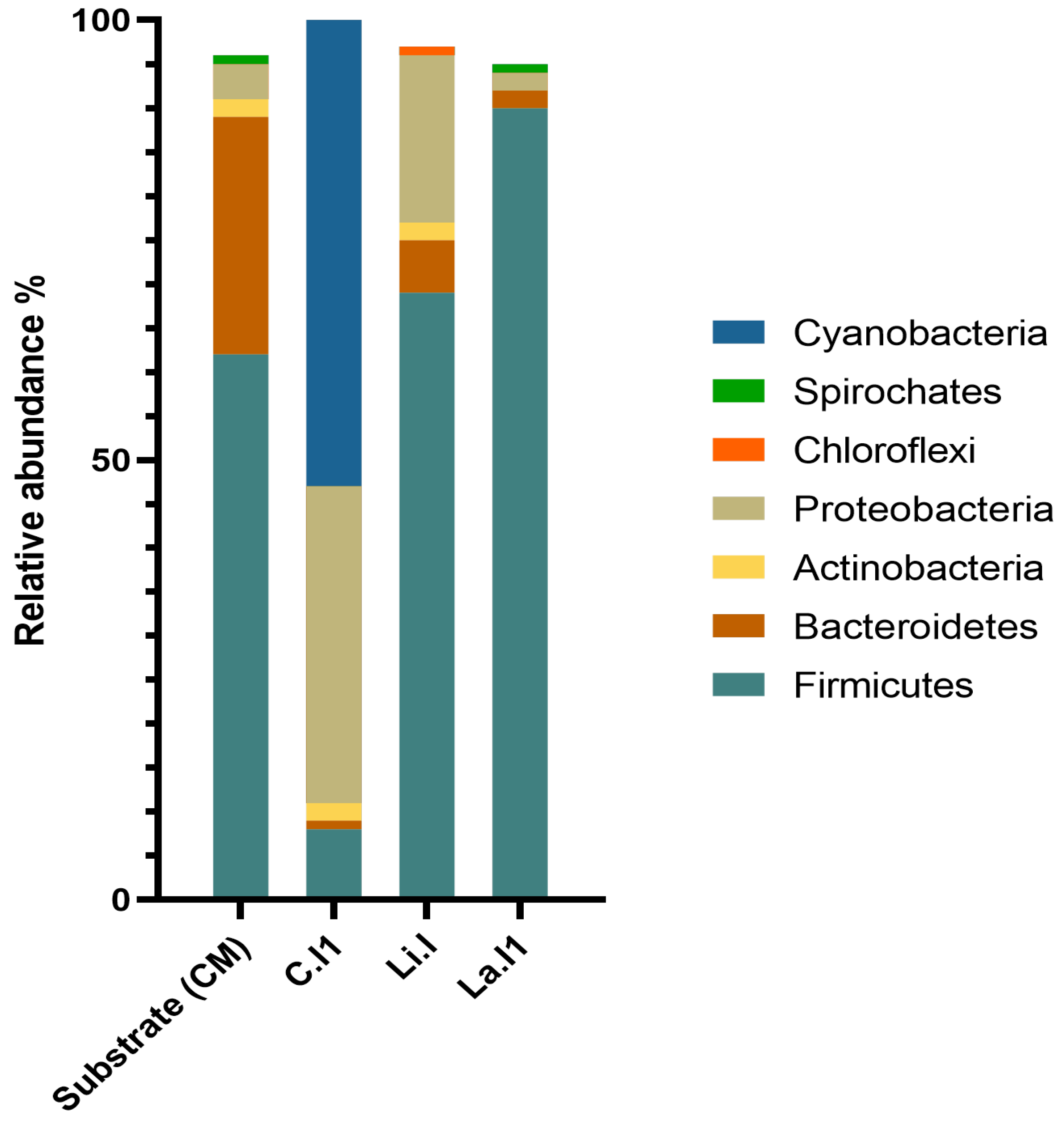
3.5. 16S rRNA Sequencing Analysis of Bacterial Compositions
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, A.; Sah, B.; Singh, A.R.; Deng, Y.; He, X.; Kumar, P.; Bansal, R.C. A Review of Multi Criteria Decision Making (MCDM) towards Sustainable Renewable Energy Development. Renew. Sustain. Energy Rev. 2017, 69, 596–609. [Google Scholar] [CrossRef]
- Bouallagui, H.; Torrijos, M.; Godon, J.J.; Moletta, R.; Ben Cheikh, R.; Touhami, Y.; Delgenes, J.P.; Hamdi, M. Two-Phases Anaerobic Digestion of Fruit and Vegetable Wastes: Bioreactors Performance. Biochem. Eng. J. 2004, 21, 193–197. [Google Scholar] [CrossRef]
- Yao, Y.; Wei, Y.; An, L.; Zhou, J. Effect of Inoculum on Anaerobic Co-Digestion of Vegetable Processing Wastes and Cattle Manure at High Solids Concentration. Waste Biomass Valoriz. 2018, 9, 2091–2098. [Google Scholar] [CrossRef]
- Lahboubi, N.; Kerrou, O.; Karouach, F.; Bakraoui, M.; Schüch, A.; Schmedemann, K.; Stinner, W.; El Bari, H.; Essamri, A. Methane Production from Mesophilic Fed-Batch Anaerobic Digestion of Empty Fruit Bunch of Palm Tree. Biomass Convers. Biorefinery 2020, 12, 3751–3760. [Google Scholar] [CrossRef]
- Abbas, Y.; Yun, S.; Wang, Z.; Zhang, Y.; Zhang, X.; Wang, K. Recent Advances in Bio-Based Carbon Materials for Anaerobic Digestion: A Review. Renew. Sustain. Energy Rev. 2021, 135, 110378. [Google Scholar] [CrossRef]
- Wang, K.; Yun, S.; Xing, T.; Li, B.; Abbas, Y.; Liu, X. Binary and Ternary Trace Elements to Enhance Anaerobic Digestion of Cattle Manure: Focusing on Kinetic Models for Biogas Production and Digestate Utilization. Bioresour. Technol. 2021, 323, 124571. [Google Scholar] [CrossRef]
- Wu, D.; Li, L.; Zhao, X.; Peng, Y.; Yang, P.; Peng, X. Anaerobic Digestion: A Review on Process Monitoring. Renew. Sustain. Energy Rev. 2019, 103, 1–12. [Google Scholar] [CrossRef]
- Kowalczyk-Juśko, A. Agricultural Biogas—Characteristics, Substrates and Its Use. Siln. Spalinowe 2012, 51, 148. [Google Scholar]
- Dahlgren, S.; Dahlgren, S. Biogas-Based Fuels as Renewable Energy in the Transport Sector: An Overview of the Potential of Using CBG, LBG and Other Vehicle Fuels Produced from Biogas from Biogas. Biofuels 2022, 13, 587–599. [Google Scholar] [CrossRef]
- Alatriste-Mondragón, F.; Samar, P.; Cox, H.H.J.; Ahring, B.K.; Iranpour, R. Anaerobic Codigestion of Municipal, Farm, and Industrial Organic Wastes: A Survey of Recent Literature. Water Environ. Res. 2006, 78, 607–636. [Google Scholar] [CrossRef]
- Coarita Fernandez, H.; Teixeira Franco, R.; Bayard, R.; Buffiere, P. Mechanical Pre-Treatments Evaluation of Cattle Manure Before Anaerobic Digestion. Waste Biomass Valoriz. 2020, 11, 5175–5184. [Google Scholar] [CrossRef]
- Romero-Güiza, M.S.; Vila, J.; Mata-Alvarez, J.; Chimenos, J.M.; Astals, S. The Role of Additives on Anaerobic Digestion: A Review. Renew. Sustain. Energy Rev. 2016, 58, 1486–1499. [Google Scholar] [CrossRef]
- Ghaleb, A.A.S.; Kutty, S.R.M.; Ho, Y.C.; Jagaba, A.H.; Noor, A.; Al-Sabaeei, A.M.; Almahbashi, N.M.Y. Response Surface Methodology to Optimize Methane Production from Mesophilic Anaerobic Co-Digestion of Oily-Biological Sludge and Sugarcane Bagasse. Sustainability 2020, 12, 2116. [Google Scholar] [CrossRef] [Green Version]
- Szilágyi, Á.; Bodor, A.; Tolvai, N.; Kovács, K.L.; Bodai, L.; Wirth, R.; Bagi, Z.; Szepesi, Á.; Markó, V.; Kakuk, B.; et al. A Comparative Analysis of Biogas Production from Tomato Bio-Waste in Mesophilic Batch and Continuous Anaerobic Digestion Systems. PLoS ONE 2021, 16, e0248654. [Google Scholar] [CrossRef]
- Rodríguez, E.; García-Encina, P.A.; Stams, A.J.M.; Maphosa, F.; Sousa, D.Z. Meta-Omics Approaches to Understand and Improve Wastewater Treatment Systems. Rev. Environ. Sci. Biotechnol. 2015, 14, 385–406. [Google Scholar] [CrossRef]
- Carabeo-Pérez, A.; Guerra-Rivera, G.; Ramos-Leal, M.; Jiménez-Hernández, J. Metagenomic Approaches: Effective Tools for Monitoring the Structure and Functionality of Microbiomes in Anaerobic Digestion Systems. Appl. Microbiol. Biotechnol. 2019, 103, 9379–9390. [Google Scholar]
- Kim, N.-K.; Lee, S.-H.; Kim, Y.; Park, H.-D. Current Understanding and Perspectives in Anaerobic Digestion Based on Genome-Resolved Metagenomic Approaches. Bioresour. Technol. 2022, 344, 126350. [Google Scholar] [CrossRef] [PubMed]
- Stolze, Y.; Zakrzewski, M.; Maus, I.; Eikmeyer, F.; Jaenicke, S.; Rottmann, N.; Siebner, C.; Pühler, A.; Schlüter, A. Comparative Metagenomics of Biogas-Producing Microbial Communities from Production-Scale Biogas Plants Operating under Wet or Dry Fermentation Conditions. Biotechnol. Biofuels 2015, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Wang, H.; Qiu, Y.; Ren, L.; Jiang, B. Microbial Characteristics in Anaerobic Digestion Process of Food Waste for Methane Production—A Review. Bioresour. Technol. 2018, 248, 29–36. [Google Scholar] [CrossRef]
- Wang, H.; Fotidis, I.A.; Yan, Q.; Angelidaki, I. Feeding Strategies of Continuous Biomethanation Processes during Increasing Organic Loading with Lipids or Glucose for Avoiding Potential Inhibition. Bioresour. Technol. 2021, 327, 124812. [Google Scholar] [CrossRef]
- ISO 11734; Water Quality—Evaluation of the Anaerobic Biodegradability of Organic Compounds in Digested Sludge—Method by Measurement of the Biogas Production, ISO Guideline 11734. European Committee for Standardization: Brussels, Belgium, 1995.
- VDI 4630; Fermentation of Organic Materials—Characterisation of the Substrate, Sampling, Collection of Material Data, Fermentation Tests, VDI Guideline 4630. Verlag des Vereins Deutscher: Düsseldorf, Germany, 2006.
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Dubois, M.; Dang, T.T.; Berrin, J.G.; Haltrich, D.; To, K.A.; Sigoillot, J.C.; Yamabhai, M. Cloning, Expression in Pichia Pastoris, and Characterization of a Thermostable GH5 Mannan Endo-1,4-Beta-Mannosidase from Aspergillus Niger BK01. Microb. Cell Fact. 1956, 8, 59. [Google Scholar] [CrossRef]
- Marion, M.; Bradford, Z.; Siadat, S.A.; Bakhshandeh, A.; Ghasemi Pirbalouti, A.; Hashemi, M. Interactive Effects of Drought Stress and Chitosan Application on Physiological Characteristics and Essential Oil Yield of Thymus Daenensis Celak. Crop J. 1976, 5, 407–415. [Google Scholar] [CrossRef]
- APHA Part 3000 Metals; 3111 Metals by Flame Atomic Absorption Spectrometry. 3111 B Direct Air—Acetylene Flame Method. Standard Methods for the Examination of Water and Wastewater. American Public Health Association: Washington, DC, USA, 1992; pp. 3–104.
- Brandt, C.; Bongcam-Rudloff, E.; Müller, B. Abundance Tracking by Long-Read Nanopore Sequencing of Complex Microbial Communities in Samples from 20 Different Biogas/Wastewater Plants. Appl. Sci. 2020, 10, 7518. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A Tool for Visualizing High-Throughput Microbial Community Data. Gigascience 2013, 2, 2–5. [Google Scholar] [CrossRef] [Green Version]
- Elango, D.; Pulikesi, M.; Baskaralingam, P.; Ramamurthi, V.; Sivanesan, S. Production of Biogas from Municipal Solid Waste with Domestic Sewage. J. Hazard. Mater. 2007, 141, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, M.; Diab, A.; Soliman, M. Methane Production from Anaerobic Co-Digestion of Sludge with Fruit and Vegetable Wastes: Effect of Mixing Ratio and Inoculum Type. Biomass Convers. Biorefinery 2021, 11, 989–998. [Google Scholar] [CrossRef]
- Wang, C.; Hong, F.; Lu, Y.; Li, X.; Liu, H. Improved Biogas Production and Biodegradation of Oilseed Rape Straw by Using Kitchen Waste and Duck Droppings as Co-Substrates in Two-Phase Anaerobic Digestion. PLoS ONE 2017, 12, e0182361. [Google Scholar] [CrossRef] [Green Version]
- Rico, C.; García, H.; Rico, J.L. Physical-Anaerobic-Chemical Process for Treatment of Dairy Cattle Manure. Bioresour. Technol. 2011, 102, 2143–2150. [Google Scholar] [CrossRef]
- Suhartini, S.; Nurika, I.; Paul, R.; Melville, L. Estimation of Biogas Production and the Emission Savings from Anaerobic Digestion of Fruit-Based Agro-Industrial Waste and Agricultural Crops Residues. Bioenergy Res. 2021, 14, 844–859. [Google Scholar] [CrossRef]
- Castrillón, L.; Marañón, E.; Fernández-Nava, Y.; Ormaechea, P.; Quiroga, G. Thermophilic Co-Digestion of Cattle Manure and Food Waste Supplemented with Crude Glycerin in Induced Bed Reactor (IBR). Bioresour. Technol. 2013, 136, 73–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersson, A.; Thomsen, M.H.; Hauggaard-Nielsen, H.; Thomsen, A.B. Potential Bioethanol and Biogas Production Using Lignocellulosic Biomass from Winter Rye, Oilseed Rape and Faba Bean. Biomass Bioenergy 2007, 31, 812–819. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Feng, L.; Zhang, R.; He, Y.; Liu, X.; Xiao, X.; Ma, X.; Chen, C.; Liu, G. Influence of Inoculum Source and Pre-Incubation on Bio-Methane Potential of Chicken Manure and Corn Stover. Appl. Biochem. Biotechnol. 2013, 171, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Dhamodharan, K.; Kumar, V.; Kalamdhad, A.S. Effect of Different Livestock Dungs as Inoculum on Food Waste Anaerobic Digestion and Its Kinetics. Bioresour. Technol. 2015, 180, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Zwain, H.M.; Dahlan, I. Biological Treatment of Recycled Paper Mill Wastewater Using Modified Anaerobic Inclining-Baffled Bioreactor (MAIB-R). In Wastewater Engineering: Advanced Wastewater Treatment Systems; International Journal of Scientific Research (IJSR) Publications: Penang, Malaysia, 2014; pp. 72–86. [Google Scholar]
- Zaier, H.; Chmingui, W.; Rajhi, H.; Bouzidi, D.; Roussos, S.; Rhouma, A. Physico-Chemical and Microbiological Characterization of Olive Mill Wastewater (OMW) of Different Regions of Tunisia (North, Sahel, South). J. New Sci. Agric. Biotechnol. 2017, 48, 2897–2906. [Google Scholar]
- Huang, X.; Yun, S.; Zhu, J.; Du, T.; Zhang, C.; Li, X. Mesophilic Anaerobic Co-Digestion of Aloe Peel Waste with Dairy Manure in the Batch Digester: Focusing on Mixing Ratios and Digestate Stability. Bioresour. Technol. 2016, 218, 62–68. [Google Scholar] [CrossRef]
- Wang, Z.; Yun, S.; Xu, H.; Wang, C.; Zhang, Y.; Chen, J.; Jia, B. Mesophilic Anaerobic Co-Digestion of Acorn Slag Waste with Dairy Manure in a Batch Digester: Focusing on Mixing Ratios and Bio-Based Carbon Accelerants. Bioresour. Technol. 2019, 286, 121394. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, J.; Krooneman, J.; Euverink, G.J.W. Strategies to Boost Anaerobic Digestion Performance of Cow Manure: Laboratory Achievements and Their Full-Scale Application Potential. Sci. Total Environ. 2021, 755, 142940. [Google Scholar] [CrossRef]
- Jiang, Y.; Heaven, S.; Banks, C.J. Strategies for Stable Anaerobic Digestion of Vegetable Waste. Renew. Energy 2012, 44, 206–214. [Google Scholar] [CrossRef] [Green Version]
- Rouez, M. Dégradation Anaérobie de Déchets Solides: Caractérisation, Facteurs d’Influence et Modélisations. Ph.D. Thesis, Civil Engineering and Environmental Engineering Laboratory (LGCIE), INSA Lyon—National Institute of Applied Sciences of Lyon, Lyon, France, 2008. ISBN 3865376657. [Google Scholar]
- Berle, E. Methane Production Through Anaerobic Digestion of Various Organic Substrates. Bachelor’s Thesis, Novia University of Applied Sciences, Vaasa, Finland, 2016; p. 82. [Google Scholar]
- Xiao, Y.; Zan, F.; Zhang, W.; Hao, T. Alleviating Nutrient Imbalance of Low Carbon-to-Nitrogen Ratio Food Waste in Anaerobic Digestion by Controlling the Inoculum-to-Substrate Ratio. Bioresour. Technol. 2021, 346, 126342. [Google Scholar] [CrossRef]
- Abbas, Y.; Yun, S.; Wang, K.; Ali Shah, F.; Xing, T.; Li, B. Static-Magnetic-Field Coupled with Fly-Ash Accelerant: A Powerful Strategy to Significantly Enhance the Mesophilic Anaerobic-Co-Digestion. Bioresour. Technol. 2021, 327, 124793. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yun, S.; Xing, T.; Wang, K.; Ke, T.; An, J. A Strategy for Understanding the Enhanced Anaerobic Co-Digestion via Dual-Heteroatom Doped Bio-Based Carbon and Its Functional Groups. Chem. Eng. J. 2021, 425, 130473. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Zhang, S.; Luo, G. Reactor Performances and Microbial Communities of Biogas Reactors: Effects of Inoculum Sources. Appl. Microbiol. Biotechnol. 2016, 100, 987–995. [Google Scholar] [CrossRef]
- Pramanik, S.K.; Suja, F.B.; Zain, S.M.; Pramanik, B.K. The Anaerobic Digestion Process of Biogas Production from Food Waste: Prospects and Constraints. Bioresour. Technol. Rep. 2019, 8, 100310. [Google Scholar] [CrossRef]
- Leung, D.Y.C.; Wang, J. An Overview on Biogas Generation from Anaerobic Digestion of Food Waste. Int. J. Green Energy 2016, 13, 119–131. [Google Scholar] [CrossRef]
- Yadvika; Santosh; Sreekrishnan, T.R.; Kohli, S.; Rana, V. Enhancement of Biogas Production from Solid Substrates Using Different Techniques—A Review. Bioresour. Technol. 2004, 95, 1–10. [Google Scholar] [CrossRef]
- Edwiges, T.; Frare, L.; Mayer, B.; Lins, L.; Mi Triolo, J.; Flotats, X.; de Mendonça Costa, M.S.S. Influence of Chemical Composition on Biochemical Methane Potential of Fruit and Vegetable Waste. Waste Manag. 2018, 71, 618–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Li, Y.; Ge, X.; Yang, L.; Li, Y. Anaerobic Digestion of Food Waste—Challenges and Opportunities. Bioresour. Technol. 2018, 247, 1047–1058. [Google Scholar] [CrossRef]
- Neves, L.; Oliveira, R.; Alves, M.M. Influence of Inoculum Activity on the Bio-Methanization of a Kitchen Waste under Different Waste/Inoculum Ratios. Process Biochem. 2004, 39, 2019–2024. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Gu, J.; Wang, X.; Zhang, K.; Yin, Y.; Zhang, R.; Zhang, S. Effects of Tylosin, Ciprofloxacin, and Sulfadimidine on McrA Gene Abundance and the Methanogen Community during Anaerobic Digestion of Cattle Manure. Chemosphere 2019, 221, 81–88. [Google Scholar] [CrossRef] [PubMed]
- El Gnaoui, Y.; Sounni, F.; Bakraoui, M.; Karouach, F.; Benlemlih, M.; Barz, M.; El Bari, H. Anaerobic Co-Digestion Assessment of Olive Mill Wastewater and Food Waste: Effect of Mixture Ratio on Methane Production and Process Stability. J. Environ. Chem. Eng. 2020, 8, 103874. [Google Scholar] [CrossRef]
- Yavini, T.D.; Chia, A.I.; John, A. Evaluation of the Effect of Total Solids Concentration on Biogas Yields of Agricultural Wastes. Int. Res. J. Environ. Sci. 2014, 3, 70–75. [Google Scholar]
- Yang, Y.; Yu, K.; Xia, Y.; Lau, F.T.K.; Tang, D.T.W.; Fung, W.C.; Fang, H.H.P.; Zhang, T. Metagenomic Analysis of Sludge from Full-Scale Anaerobic Digesters Operated in Municipal Wastewater Treatment Plants. Appl. Microbiol. Biotechnol. 2014, 98, 5709–5718. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, B.; Wright, A.D.G. Comparative Metagenomic Analysis of Bacterial Populations in Three Full-Scale Mesophilic Anaerobic Manure Digesters. Appl. Microbiol. Biotechnol. 2014, 98, 2709–2717. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.L.; Jangid, K.; Whitman, W.B.; Das, K.C. Transition of Microbial Communities during the Adaption to Anaerobic Digestion of Carrot Waste. Bioresour. Technol. 2011, 102, 7249–7256. [Google Scholar] [CrossRef]
- Schwarz, W.H. The Cellulosome and Cellulose Degradation by Anaerobic Bacteria. Appl. Microbiol. Biotechnol. 2001, 56, 634–649. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; Van Zyl, W.H.; Isak, S. Microbial Cellulose Utilization: Fundamentals and Biotechnology Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Yun, S.; Shi, J.; Han, F.; Liu, B.; Wang, R.; Li, X. Critical Evidence for Direct Interspecies Electron Transfer with Tungsten-Based Accelerants: An Experimental and Theoretical Investigation. Bioresour. Technol. 2020, 311, 123519. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.; Xing, T.; Han, F.; Shi, J.; Wang, Z.; Fan, Q.; Xu, H. Enhanced Direct Interspecies Electron Transfer with Transition Metal Oxide Accelerants in Anaerobic Digestion. Bioresour. Technol. 2021, 320, 124294. [Google Scholar] [CrossRef] [PubMed]
- Martí-Herrero, J.; Alvarez, R.; Flores, T. Evaluation of the Low Technology Tubular Digesters in the Production of Biogas from Slaughterhouse Wastewater Treatment. J. Clean. Prod. 2018, 199, 633–642. [Google Scholar] [CrossRef]
- Traversi, D.; Villa, S.; Lorenzi, E.; Degan, R.; Gilli, G. Application of a Real-Time QPCR Method to Measure the Methanogen Concentration during Anaerobic Digestion as an Indicator of Biogas Production Capacity. J. Environ. Manag. 2012, 111, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, K.; Zhang, X.; Gong, H.; Xia, Y.; Holmes, D.E. Application of In-Situ H2-Assisted Biogas Upgrading in High-Rate Anaerobic Wastewater Treatment. Bioresour. Technol. 2020, 299, 122598. [Google Scholar] [CrossRef] [PubMed]



| Inoc | Cellulosic Inoculum1 (C.I1) | Cellulosic Inoculum2 (C.I2) | Lipidic Inoculum (Li.I) | Lactic Inoculum1 (La.I1) | Lactic Inoculum2 (La.I2) | Saccharidicinoculum (Sacc.I) |
|---|---|---|---|---|---|---|
| Composition (in % w/w) | 30% Leaf litter 40% Wheat straw 10% Milk 10% Glucose 10% Water | 40% Wheat straw 40% rice straw 20% corn stover | 10%vegetabl peels 11% Coffee grounds 5% Yoghurt 30% Leaves and branches of plants 44% Olive pomace | 10% Glucose 90% Lactoserum | 5% Yeast 25% Flour 70% natural yeast | 100% molasses |
| Parameter | C.I1 | C.I2 | Li.I | La.I1 | La.I2 | Sacc.I | Substrate (CM) |
|---|---|---|---|---|---|---|---|
| pH | 6.15 ± 0.03 d | 6.52 ± 0.02 c | 7.08 ± 0.06 a | 7.12 ± 0.02 a | 4.62 ± 0.11 e | 6.81 ± 0.06 b | 7.19 ± 0.01 a |
| Total Solids (%wet basis) | 41.70 ± 0.2 cd | 26.16 ± 0.97 ef | 19.33 ± 2.28 f | 76.12 ± 3.30 b | 33.89 ± 1.93 de | 44.33 ± 4.6 c | 94.87 ± 5.25 a |
| Volatiles Solids (%wet basis) | 36.85 ± 1.53 c | 22.27 ± 1.73 de | 14.25 ± 1.08 e | 62.25 ± 4.58 b | 24.85 ± 3.66 d | 26.38 ± 1.16 d | 89.43 ± 5.17 a |
| VS/TS (% TS) | 88.36 | 85.12 | 73.71 | 81.77 | 73.32 | 59.5 | 94.26 |
| Total lipid % | 20.26 ± 1.39 bc | 13.66 ± 1.49 d | 35.05 ± 2.93 a | 24.08 ± 0.57 b | 12.9 ± 0.98 | 5.04 ± 0.6 e | 16.11 ± 1.78 cd |
| Total sugar % | 7.12 ± 0.61 ab | 4.06 ± 0.5 cd | 2.21 ± 0.68 d | 5.75 ± 1.16 bc | 7.27 ± 0.56 ab | 8.15 ± 0.66 a | 4.16 ± 0.01 cd |
| Total protein % | 4.44 ± 1.34 c | 14 ± 1.81 a | 6.08 ± 1.6 bc | 5.17 ± 1.37 c | 4.59 ± 0.84 c | 2.33 ± 0.62 c | 10.01 ± 1.49 ab |
| P (mg·g−1) | 12.72 ± 1.05 b | 15.70 ± 0.16 a | 8.75 ± 1.28 c | 6.75 ± 0.27 c | 7.75 ± 0.23 c | 11.1 ± 0.73 b | 11.13 ± 0.42 c |
| Ca (mg·g−1) | 0.4 ± 0.00 c | 0.4 ± 0.19 c | 5.48 ± 0.20 a | 0.84 ± 0.15 c | 4.38 ± 0.32 b | 0.51 ± 0.14 c | 4.63 ± 0.39 b |
| Particle size (mm) | 2 | ≤2 | ≤3 | 1–1.5 | - | - | 1–1.5 |
| Parameter | C.I1 | C.I2 | Li.I | La.I1 | La.I2 | Sacc.I | Substrate (CM) |
|---|---|---|---|---|---|---|---|
| Initial pH | 6.87 ± 0.31 b | 7.12 ± 0.08 ab | 6.13 ± 0.45 c | 6.81 ± 0.00 b | 7.23 ± 0.04 ab | 7.48 ± 0.04 a | 7.11 ± 0.18 ab |
| Final pH | 6.50 ± 0.00 a | 6.16 ± 0.01 bcd | 6 ± 0.00 d | 5.17 ± 0.05 e | 6.32 ± 0.11 b | 6.12 ± 0.00 cd | 6.18 ± 0.03 bc |
| Total Carbon (TC) % | 52.18 ± 2.02 c | 67.04 ± 1.92 b | 18.58 ± 0.66 e | 36.32 ± 3.2 d | 66.37 ± 1.92 b | 22.62 ± 1.78 e | 76.95 ± 2.57 a |
| Total Nitrogen (TN) % | 1.537 ± 0.04 cd | 3.199 ± 0.2 a | 2.444 ± 0.11 b | 1.609 ± 0.1 cd | 2.106 ± 0.32 bc | 1.160 ± 0.27 d | 1.941 ± 0.38 bc |
| C/N | 33.94 | 20.95 | 7.60 | 22.57 | 31.51 | 19.5 | 39.64 |
| COD efficiency (%) | 77.28 | 39.23 | 15.07 | 68.23 | 64.81 | 14.97 | 43.05 |
| BOD5 efficiency | 96.45 | 4 | 26 | 33.33 | 51.85 | 33.33 | 41.17 |
| Initial COD/BOD5 | 1.144 | 1.933 | 1.05 | 2.368 | 2.517 | 2.412 | 1.09 |
| Final COD/BOD5 | 1.35 | 1.182 | 1.213 | 1.128 | 1.839 | 4.16 | 2.64 |
| Temperature °C | 37 | 37 | 37 | 37 | 37 | 37 | 37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niya, B.; Yaakoubi, K.; Azaroual, S.E.; Beraich, F.Z.; Arouch, M.; Meftah Kadmiri, I. Anaerobic Digestion of Agricultural Waste Using Microbial Inocula: Performance and Characterization of Bacterial Communities Using 16S rRNA Sequencing Approach. Energies 2023, 16, 3300. https://doi.org/10.3390/en16083300
Niya B, Yaakoubi K, Azaroual SE, Beraich FZ, Arouch M, Meftah Kadmiri I. Anaerobic Digestion of Agricultural Waste Using Microbial Inocula: Performance and Characterization of Bacterial Communities Using 16S rRNA Sequencing Approach. Energies. 2023; 16(8):3300. https://doi.org/10.3390/en16083300
Chicago/Turabian StyleNiya, Btissam, Kaoutar Yaakoubi, Salah Eddine Azaroual, Fatima Zahra Beraich, Moha Arouch, and Issam Meftah Kadmiri. 2023. "Anaerobic Digestion of Agricultural Waste Using Microbial Inocula: Performance and Characterization of Bacterial Communities Using 16S rRNA Sequencing Approach" Energies 16, no. 8: 3300. https://doi.org/10.3390/en16083300
APA StyleNiya, B., Yaakoubi, K., Azaroual, S. E., Beraich, F. Z., Arouch, M., & Meftah Kadmiri, I. (2023). Anaerobic Digestion of Agricultural Waste Using Microbial Inocula: Performance and Characterization of Bacterial Communities Using 16S rRNA Sequencing Approach. Energies, 16(8), 3300. https://doi.org/10.3390/en16083300







