Femtosecond Laser-Based Integration of Nano-Membranes into Organ-on-a-Chip Systems
Abstract
:1. Introduction
2. Materials and Methods
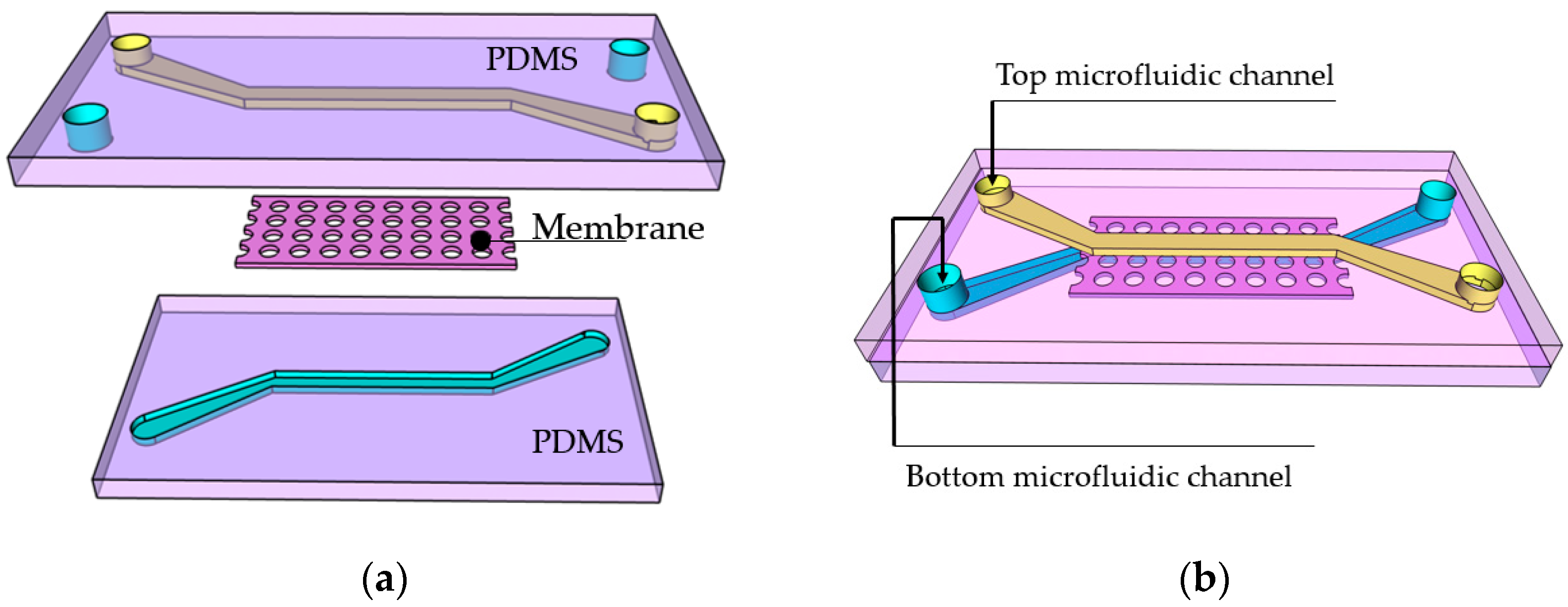
2.1. Step 1—PDMS Chip Preparation
2.2. Steps 2 and 3—Introducing and Drying Photoresist
2.3. Step 4—In Situ Laser Writing
2.4. Steps 5 and 6—Photoresist Development
2.5. Sample Characterization
3. Results and Discussion
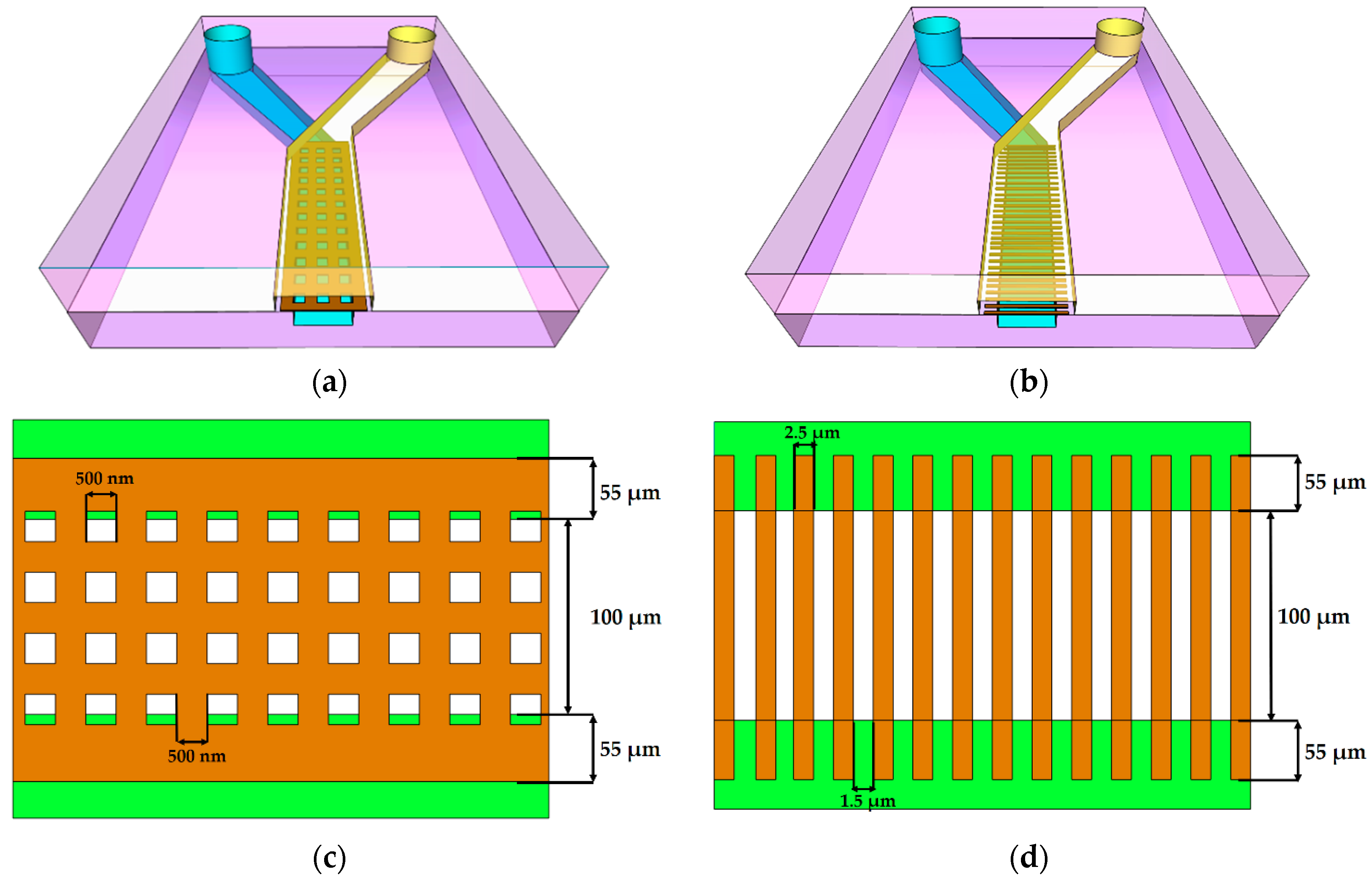
3.1. Membrane Structure Geometry
3.2. Preparation for Printing
3.3. In Situ Direct Laser Writing
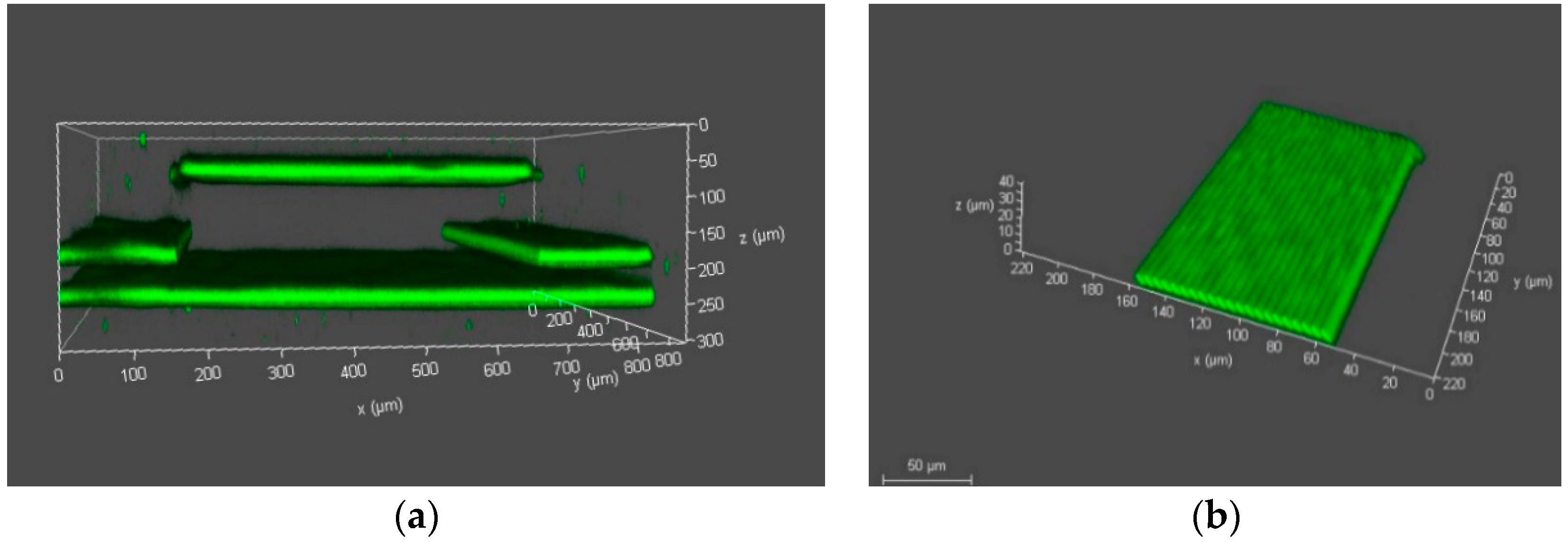
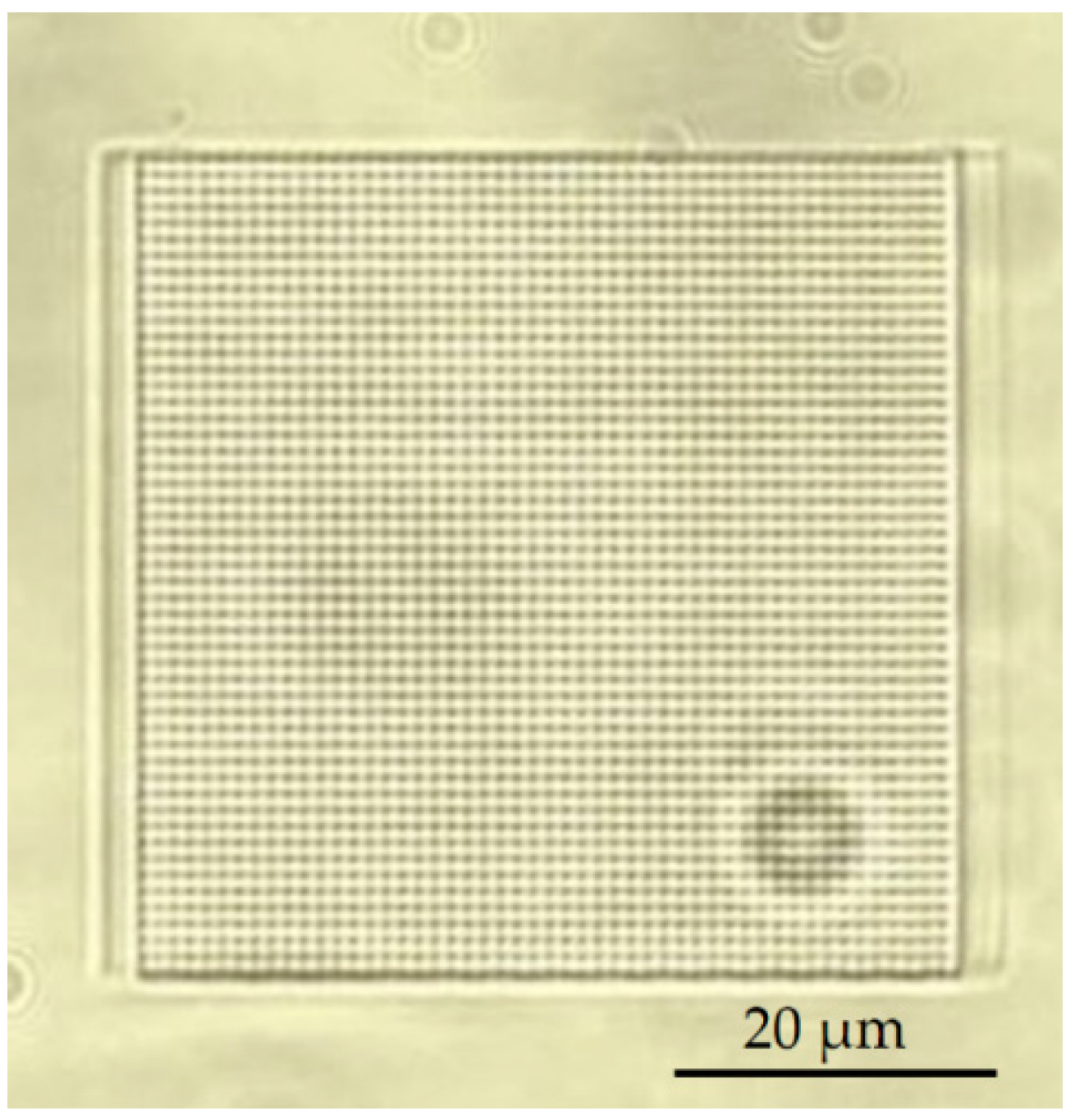
3.4. Experimental Verification of Membrane
3.5. Development in a Closed Microfluidic Channel
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lohasz, C.; Frey, O.; Bonanini, F.; Renggli, K.; Hierlemann, A. Tubing-Free Microfluidic Microtissue Culture System Featuring Gradual, in vivo-Like Substance Exposure Profiles. Front. Bioeng. Biotechnol. 2019, 7, 72. [Google Scholar] [CrossRef]
- Moshksayan, K.; Kashaninejad, N.; Warkiani, M.E.; Lock, J.G.; Moghadas, H.; Firoozabadi, B.; Saidi, M.S.; Nguyen, N.-T. Spheroids-on-a-chip: Recent advances and design considerations in microfluidic platforms for spheroid formation and culture. Sens. Actuators B Chem. 2018, 263, 151–176. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Sun, Q.; Pei, J. Microfluidic-Based 3D Engineered Microvascular Networks and Their Applications in Vascularized Microtumor Models. Micromachines 2018, 9, 493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagamine, K.; Sato, H.; Kai, H.; Kaji, H.; Kanzaki, M.; Nishizawa, M. Contractile Skeletal Muscle Cells Cultured with a Conducting Soft Wire for Effective, Selective Stimulation. Sci. Rep. 2018, 8, 2253. [Google Scholar] [CrossRef] [Green Version]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Kaisar, M.A.; Sajja, R.K.; Prasad, S.; Abhyankar, V.V.; Liles, T.; Cucullo, L. New experimental models of the blood-brain barrier for CNS drug discovery. Expert Opin. Drug Dis. 2017, 12, 89–103. [Google Scholar] [CrossRef] [Green Version]
- Herland, A.; Van der Meer, A.D.; FitzGerald, E.A.; Park, T.E.; Sleeboom, J.J.; Ingber, D.E. Distinct Contributions of Astrocytes and Pericytes to Neuroinflammation Identified in a 3D Human Blood-Brain Barrier on a Chip. PLoS ONE 2016, 3, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; Zhang, X.; Wen, X.; Wu, T.; Wang, W.; Yang, M.; Wang, J.; Fang, M.; Lin, B.; Lin, H. Development of a Functional Glomerulus at the Organ Level on a Chip to Mimic Hypertensive Nephropathy. Sci. Rep. 2016, 6, 31771. [Google Scholar] [CrossRef] [Green Version]
- Jang, K.-J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.-Y.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef]
- Arık, Y.B.; Van der Helm, M.W.; Odijk, M.; Segerink, L.I.; Passier, R.; van den Berg, A.; Van der Meer, A.D. Barriers-on-chips: Measurement of barrier function of tissues in organs-on-chips. Biomicrofluidics 2018, 12, 42218. [Google Scholar] [CrossRef] [Green Version]
- Pasman, T.; Grijpma, D.; Stamatialis, D.; Poot, A. Flat and microstructured polymeric membranes in organs-on-chips. J. R. Soc. Interface 2018, 15, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, A.; Barrile, R.; Van der Meer, A.D.; Mammoto, A.; Mammoto, T.; Ceunynck, K.; Aisiku, O.; Otieno, M.A.; Louden, C.S.; Hamilton, G.A.; et al. Primary Human Lung Alveolus-on-a-chip Model of Intravascular Thrombosis for Assessment of Therapeutics. Clin. Pharmacol. Ther. 2018, 103, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.-H.; Lee, S.-K.; Kim, J.-H.; Park, J.-H. PDMS membrane filter with nano-slit array fabricated using three-dimensional silicon mold for the concentration of particles with bacterial size range. Microelectron. Eng. 2019, 215, 111008. [Google Scholar] [CrossRef]
- Chen, Q.; Wu, J.; Zhuang, Q.; Lin, X.; Zhang, J.; Lin, J.-M. Microfluidic isolation of highly pure embryonic stem cells using feeder-separated co-culture system. Sci. Rep. 2013, 3, 2433. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.I.; Abaci, H.E.; Shuler, M.L. Microfluidic blood-brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol. Bioeng. 2017, 114, 184–194. [Google Scholar] [CrossRef]
- Shim, K.-Y.; Lee, D.; Han, J.; Nguyen, N.-T.; Park, S.; Sung, J.H. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed. Microdevices 2017, 19, 37. [Google Scholar] [CrossRef] [Green Version]
- Jonušauskas, L.; Mackevičiūtė, D.; Kontenis, G.; Purlys, V. Femtosecond lasers: The ultimate tool for high-precision 3D manufacturing. Adv. Opt. Technol. 2019, 8, 241–251. [Google Scholar] [CrossRef]
- Zandrini, T.; Shan, O.; Parodi, V.; Cerullo, G.; Raimondi, M.T.; Osellame, R. Multi-foci laser microfabrication of 3D polymeric scaffolds for stem cell expansion in regenerative medicine. Sci. Rep. 2019, 9, 11761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberale, C.; Cojoc, G.; Bragheri, F.; Minzioni, P.; Perozziello, G.; La Rocca, R.; Ferrara, L.; Rajamanickam, V.; Fabrizio, D.E.; Cristiani, I. Integrated microfluidic device for single-cell trapping and spectroscopy. Sci. Rep. 2013, 3, 1258. [Google Scholar] [CrossRef] [Green Version]
- Maigyte, L.; Purlys, V.; Trull, J.; Peckus, M.; Cojocaru, C.; Gailevičius, D.; Malinauskas, M.; Staliunas, K. Flat lensing in the visible frequency range by woodpile photonic crystals. Opt. Lett 2013, 38, 2376–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danilevičius, P.; Žukauskas, A.; Bičkauskaitė, G.; Purlys, V.; Rutkauskas, M.; Gertus, T.; Paipulas, D.; Matukaitė, J.; Baltriukienė, D.; Malinauskas, M. Laser-micro/nanofabricated 3D polymers for tissue engineering applications. Latv. J. Phys. Tech. Sci. 2011, 48, 32–43. [Google Scholar] [CrossRef]
- Jonušauskas, L.; Baravykas, T.; Andrijec, D.; Gadišauskas, T.; Purlys, V. Stitchless support-free 3D printing of free-form micromechanical structures with feature size on-demand. Sci. Rep. 2019, 9, 17533. [Google Scholar] [CrossRef] [Green Version]
- Hahn, V.; Kalt, S.; Sridharan, G.M.; Wegener, M.; Bhattacharya, S. Polarizing beam splitter integrated onto an optical fiber facet. Opt. Express 2018, 26, 33148–33157. [Google Scholar] [CrossRef] [PubMed]
- Alsharhan, A.T.; Acevedo, R.; Warren, R.; Sochol, R.D. 3D microfluidics via cyclic olefin polymer-based in situ direct laser writing. Lab Chip 2019, 19, 2799–2810. [Google Scholar] [CrossRef]
- Farsari, M.; Vamvakaki, M.; Chichkov, B.N. Multiphoton polymerization of hybrid materials. J. Opt. UK 2010, 12, 124001. [Google Scholar] [CrossRef]
- Barner-Kowollik, C.; Bastmeyer, M.; Blasco, E.; Delaittre, G.; Müller, P.; Richter, B.; Wegener, M. 3D Laser Micro- and Nanoprinting: Challenges for Chemistry. Chem. Int. Ed. 2017, 56, 15828. [Google Scholar] [CrossRef] [PubMed]
- Merkininkaitė, G.; Gailevičius, D.; Šakirzanovas, S.; Jonušauskas, L. Polymers for Regenerative Medicine Structures Made via Multiphoton 3D Lithography. Int. J. Polym. Sci. 2019. [Google Scholar] [CrossRef]
- Jonušauskas, L.; Rekštyte, S.; Buividas, R.; Butkus, S.; Gadonas, R.; Juodkazis, S.; Malinauskas, M. Hybrid subtractive-additive-welding microfabrication for lab-on-chip applications via single amplified femtosecond laser source. Opt. Eng. 2017, 56, 094108. [Google Scholar] [CrossRef]
- Wu, D.; Wu, S.-Z.; Xu, J.; Niu, L.-G.; Midorikawa, K.; Sugioka, K. Hybrid femtosecond laser microfabrication to achieve true 3D glass/polymer composite biochips with multiscale features and high performance: The concept of ship-in-a-bottle biochip. Laser Photonics Rev. 2014, 8, 458–467. [Google Scholar] [CrossRef]
- Tang, L.; Lee, N.Y. A facile route for irreversible bonding of plastic-PDMS hybrid microdevices at room temperature. Lab Chip 2010, 10, 1274–1280. [Google Scholar] [CrossRef]
- Ovsianikov, A.; Viertl, J.; Chichkov, B.; Oubaha, M.; MacCraith, B.; Sakellari, I.; Giakoumaki, A.; Gray, D.; Vamvakaki, M.; Farsari, M.; et al. Ultra-Low Shrinkage Hybrid Photosensitive Material for Two-Photon Polymerization Microfabrication. ACS Nano 2008, 2, 2257–2262. [Google Scholar] [CrossRef] [PubMed]
- Jonušauskas, L.; Gailevičius, D.; Mikoliūnaitė, L.; Sakalauskas, D.; Šakirzanovas, S.; Juodkazis, S.; Malinauskas, M. Optically Clear and Resilient Free-Form µ-Optics 3D-Printed via Ultrafast Laser Lithography. Materials 2017, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Ricci, D.; Nava, M.M.; Zandrini, T.; Cerullo, G.; Raimondi, M.T.; Osellame, R. Scaling-Up Techniques for the Nanofabrication of Cell Culture Substrates via Two-Photon Polymerization for Industrial-Scale Expansion of Stem Cells. Materials 2017, 10, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piruska, A.; Nikcevic, I.; Lee, S.H.; Ahn, C.; Heineman, W.R.; Limbacha, P.A.; Seliskar, C.J. The autofluorescence of plastic materials and chips measured under laser irradiation. Lab Chip 2005, 5, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Gittard, S.D.; Nguyen, A.; Obata, K.; Koroleva, A.; Narayan, R.J.; Chichkov, B.N. Fabrication of microscale medical devices by two-photon polymerization with multiple foci via a spatial light modulator. Biomed. Opt. Express 2011, 2, 3167–3178. [Google Scholar] [CrossRef]
- Römera, G.R.B.E.; Bechtold, P. Electro-optic and Acousto-optic Laser Beam Scanners. Phys. Procedia 2014, 56, 29–39. [Google Scholar] [CrossRef] [Green Version]
- Malinauskas, M.; Bickauskait, G.; Rutkauskas, M.; Paipulas, D.; Purlys, V.; Gadonas, R. Self-polymerization of nano-fibres and nano-membranes induced by two-photon absorption. Lith. J. Phys. 2010, 50, 135–140. [Google Scholar] [CrossRef]
- Honda, T.; Miyazaki, M.; Nakamura, H.; Maeda, H. Controllable polymerization of N-carboxy anhydrides in a microreaction system. Supplementary files. Lab Chip 2005, 5, 812. [Google Scholar] [CrossRef]
- Zeng, H.; Martella, D.; Wasylczyk, P.; Cerretti, G.; Lavocat, J.-C.G.; Ho, C.-H.; Parmeggiani, C.; Wiersma, D.S. High-Resolution 3D Direct Laser Writing for Liquid-Crystalline Elastomer Microstructures. Adv. Mater. 2014, 26, 2319–2322. [Google Scholar] [CrossRef]
- Pashneh-Tala, S.; Owen, R.; Bahmaee, H.; Rekštytė, S.; Malinauskas, M.; Claeyssens, F. Synthesis, Characterization and 3D Micro-Structuring via 2-Photon Polymerization of Poly (glycerol sebacate)-Methacrylate–An Elastomeric Degradable Polymer. Front. Phys. 2018, 6, 41. [Google Scholar] [CrossRef] [Green Version]
- Obata, K.; Slobin, S.; Schonewille, A.; Hohnholz, A.; Unger, C.; Koch, J.; Suttmann, O.; Overmeyer, L. UV laser direct writing of 2D/3D structures using photo-curable polydimethylsiloxane (PDMS). Appl. Phys. A 2017, 123, 495. [Google Scholar] [CrossRef]
- Lamont, A.C.; Alsharhan, A.T.; Sochol, R.D. Geometric Determinants of In-Situ Direct Laser Writing. Sci. Rep. 2019, 9, 394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schizas, C.; Melissinaki, V.; Gaidukeviciute, A.; Reinhardt, C.; Ohrt, C.; Dedoussis, V.; Chichkov, B.N.; Fotakis, C.; Farsari, M.; Karalekas, D. On the design and fabrication by two-photon polymerization of a readily assembled micro-valve. Int. J. Adv. Manuf. Technol. 2010, 48, 435–441. [Google Scholar] [CrossRef]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakhchova, L.; Jonušauskas, L.; Andrijec, D.; Kurachkina, M.; Baravykas, T.; Eremin, A.; Steinmann, U. Femtosecond Laser-Based Integration of Nano-Membranes into Organ-on-a-Chip Systems. Materials 2020, 13, 3076. https://doi.org/10.3390/ma13143076
Bakhchova L, Jonušauskas L, Andrijec D, Kurachkina M, Baravykas T, Eremin A, Steinmann U. Femtosecond Laser-Based Integration of Nano-Membranes into Organ-on-a-Chip Systems. Materials. 2020; 13(14):3076. https://doi.org/10.3390/ma13143076
Chicago/Turabian StyleBakhchova, Liubov, Linas Jonušauskas, Dovilė Andrijec, Marharyta Kurachkina, Tomas Baravykas, Alexey Eremin, and Ulrike Steinmann. 2020. "Femtosecond Laser-Based Integration of Nano-Membranes into Organ-on-a-Chip Systems" Materials 13, no. 14: 3076. https://doi.org/10.3390/ma13143076





