Fluorescent Protein-Based Autophagy Biosensors
Abstract
:1. Introduction
2. Autophagy and Neurodegenerative Diseases
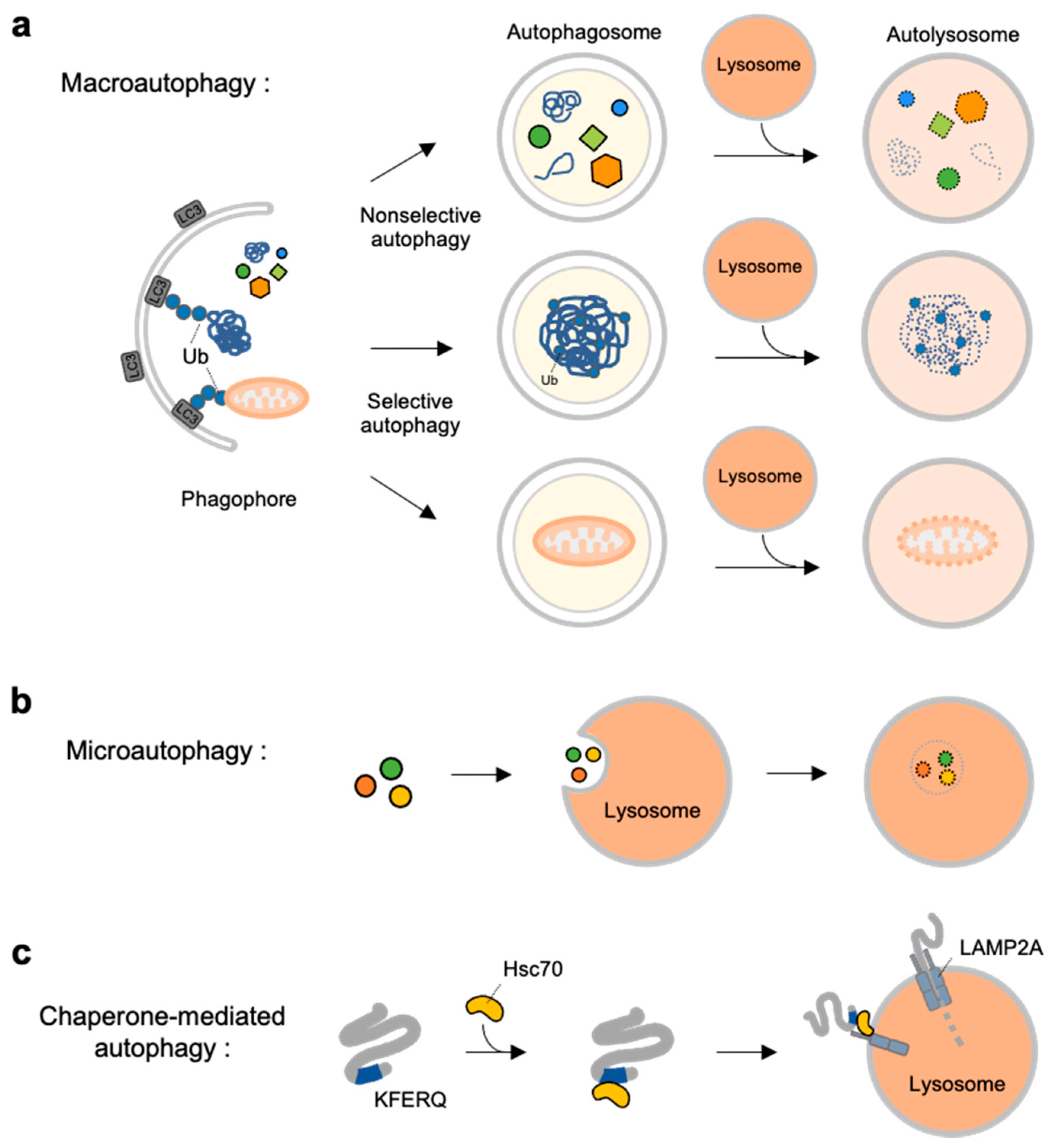
2.1. Autophagy Types
2.2. Molecular Mechanisms at Different Stages of Macroautophagy
2.3. Autophagy Dysfunctions in Neurodegenerative Diseases
3. Methods for the Detection of Autophagy
3.1. Electron Microscopy
3.2. Autophagic Protein Degradation Assay
3.3. Biochemical Assay
3.4. Chemical Probes
3.5. Fluorescent Protein (FP)-Based Biosensors
4. FP-Based Biosensors for the Monitoring of Autophagy Progression
4.1. GFP-LC3
4.2. HyD-LIR-GFP
4.3. RFP-GFP-LC3
4.4. Red, Green and Blue (RGB)-LC3
4.5. GFP-LC3-RFP-LC3∆G
4.6. Fluorescence Resonance Energy Transfer (FRET)-Based Autophagy Sensor
4.7. Mitophagy Sensor
4.8. Chaperone-Mediated Autophagy Sensor
5. Summary and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Aβ | amyloid-beta |
| AD | Alzheimer’s disease |
| AMBRA1 | activating molecule in Beclin 1-regulated autophagy protein 1 |
| AMPK | AMP-activated protein kinase |
| Atg | autophagy-related protein |
| BFP | blue fluorescent protein |
| CMA | chaperone-mediated autophagy |
| ECFP | enhanced cyan fluorescent proteins |
| EYFP | enhanced yellow fluorescent protein |
| FIP200 | focal adhesion kinase family interacting protein of 200-kDa |
| FP | fluorescent protein |
| FRET | fluorescence resonance energy transfer |
| GFP | green fluorescent protein |
| HD | Huntington’s Disease |
| Hsc70 | heat shock cognate protein of 70 kDa |
| Htt | huntingtin |
| HyD | hydrophobic domain |
| LAMP2A | lysosomal-associated membrane protein 2A |
| LC3 | microtubule-associated protein light chain 3 |
| LDH | lactate dehydrogenase |
| LD | lipid droplet |
| LID | light-induced dimer |
| LIR | LC3 interacting region |
| mTOR | mechanistic target of rapamycin |
| NBR1 | neighbor of BRCA1 gene 1 |
| NDP52 | nuclear dot protein 52 kDa |
| PD | Parkinson’s disease |
| PE | phosphatidylethanolamine |
| PINK1 | PTEN-induced kinase 1 |
| PI3P | phosphatidylinositol 3-phosphate |
| PolyQ | polyglutamine |
| PS-CFP2 | photoconvertible cyan FP |
| PS-1 | presenilin 1 |
| RFP | red fluorescent protein |
| SEP | superecliptic pHluourin |
| SF44 | Seoul-Fluor 44 |
| SILAC | stable isotope labeling by amino acid in cell culture |
| SNARE | soluble NSF (N-Ethylmaleimide-Sensitive Factor) Attachment Protein Receptor |
| SRAI | signal-retaining autophagy indicator |
| TEM | transmission electron microscope |
| TOLLES | TOLerance of Lysosomal EnvironmentS |
| ULK1 | Unc-51 like kinase 1 |
| Vps34 | vacuolar protein sorting 34 |
| WIPI | WD-repeat protein interacting with phosphoinositide |
References
- Djajadikerta, A.; Keshri, S.; Pavel, M.; Prestil, R.; Ryan, L.; Rubinsztein, D.C. Autophagy Induction as a Therapeutic Strategy for Neurodegenerative Diseases. J. Mol. Biol. 2020, 432, 2799–2821. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Vicente, M. Autophagy in neurodegenerative diseases: From pathogenic dysfunction to therapeutic modulation. Semin. Cell Dev. Biol. 2015, 40, 115–126. [Google Scholar] [CrossRef]
- Nixon, R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef]
- Kim, D.; Hwang, H.Y.; Kwon, H.J. Targeting Autophagy In Disease: Recent Advances In Drug Discovery. Expert Opin. Drug Discov. 2020, 15, 1045–1064. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B. Autophagy in Human Diseases. N. Engl. J. Med. 2020, 383, 15641576. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Murphy, L.O. Autophagy Assays for Biological Discovery and Therapeutic Development. Trends Biochem. Sci. 2020, 45, 1080–1093. [Google Scholar] [CrossRef]
- Vigneshvar, S.; Sudhakumari, C.C.; Senthilkumaran, B.; Prakash, H. Recent Advances in Biosensor Technology for Potential Applications—An Overview. Front. Bioeng. Biotechnol. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Klionsky, D.J. Autophagy and disease: Unanswered questions. Cell Death Differ. 2020, 27, 858–871. [Google Scholar] [CrossRef]
- Ding, S.; Hong, Y. The fluorescence toolbox for visualizing autophagy. Chem. Soc. Rev. 2020, 49, 8354–8389. [Google Scholar] [CrossRef]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green Fluorescent Protein as a Marker for Gene Expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eason, M.G.; Damry, A.M.; Chica, R.A. Structure-guided rational design of red fluorescent proteins: Towards designer genetically-encoded fluorophores. Curr. Opin. Struct. Biol. 2017, 45, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Goedhart, J.; von Stetten, D.; Noirclerc-Savoye, M.; Lelimousin, M.; Joosen, L.; Hink, M.A.; van Weeren, L.; Gadella, T.W., Jr.; Royant, A. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun 2012, 3, 751. [Google Scholar] [CrossRef] [PubMed]
- Kamiyama, D.; Sekine, S.; Barsi-Rhyne, B.; Hu, J.; Chen, B.; Gilbert, L.A.; Ishikawa, H.; Leonetti, M.D.; Marshall, W.F.; Weissman, J.S.; et al. Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 2016, 7, 11046. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tsien, R.W. pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat. Neurosci. 2012, 15, 1047–1053. [Google Scholar] [CrossRef] [Green Version]
- Sankaranarayanan, S.; De Angelis, D.; Rothman, J.E.; Ryan, T.A. The Use of pHluorins for Optical Measurements of Presynaptic Activity. Biophys. J. 2000, 79, 2199–2208. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Rosendale, M.; Campbell, R.E.; Perrais, D. pHuji, a pH-sensitive red fluorescent protein for imaging of exo- and endocytosis. J. Cell Biol. 2014, 207, 419–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chudakov, D.M.; Matz, M.V.; Lukyanov, S.; Lukyanov, K.A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010, 90, 1103–1163. [Google Scholar] [CrossRef]
- Newman, R.H.; Zhang, J. The design and application of genetically encodable biosensors based on fluorescent proteins. Methods Mol. Biol. 2014, 1071, 1–16. [Google Scholar]
- Nienhaus, K.; Nienhaus, G.U. Fluorescent proteins for live-cell imaging with super-resolution. Chem. Soc. Rev. 2014, 43, 1088–1106. [Google Scholar] [CrossRef]
- Kim, H.; Ju, J.; Lee, H.N.; Chun, H.; Seong, J. Genetically Encoded Biosensors Based on Fluorescent Proteins. Sensors 2021, 21, 3. [Google Scholar]
- Bindels, D.S.; Haarbosch, L.; van Weeren, L.; Postma, M.; Wiese, K.E.; Mastop, M.; Aumonier, S.; Gotthard, G.; Royant, A.; Hink, M.A.; et al. mScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 2017, 14, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Ai, H.W.; Wong, P.; Young, J.D.; Campbell, R.E.; Casey, J.R. Red fluorescent protein pH biosensor to detect concentrative nucleoside transport. J. Biol. Chem. 2009, 284, 20499–20511. [Google Scholar] [CrossRef] [Green Version]
- Kogure, T.; Karasawa, S.; Araki, T.; Saito, K.; Kinjo, M.; Miyawaki, A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat. Biotechnol. 2006, 24, 577–581. [Google Scholar] [CrossRef]
- Lambert, T.J. FPbase: A community-editable fluorescent protein database. Nat. Methods. 2019, 16, 277–278. [Google Scholar] [CrossRef] [PubMed]
- Gross, L.A.; Baird, G.S.; Hoffman, R.C.; Baldridge, K.K.; Tsien, R.Y. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 2000, 97, 11990–11995. [Google Scholar] [CrossRef] [Green Version]
- Liu, A.; Huang, X.; He, W.; Xue, F.; Yang, Y.; Liu, J.; Chen, L.; Yuan, L.; Xu, P. pHmScarlet is a pH-sensitive red fluorescent protein to monitor exocytosis docking and fusion steps. Nat. Commun. 2021, 12, 1413. [Google Scholar] [CrossRef]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Lambert, G.G.; Chammas, A.; Ni, Y.; Cranfill, P.J.; Baird, M.A.; Sell, B.R.; Allen, J.R.; Day, R.N.; Israelsson, M.; et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 2013, 10, 407–409. [Google Scholar] [CrossRef]
- Shcherbo, D.; Merzlyak, E.M.; Chepurnykh, T.V.; Fradkov, A.F.; Ermakova, G.V.; Solovieva, E.A.; Lukyanov, K.A.; Bogdanova, E.A.; Zaraisky, A.G.; Lukyanov, S.; et al. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods 2007, 4, 741–746. [Google Scholar] [CrossRef]
- Shcherbo, D.; Murphy, C.S.; Ermakova, G.V.; Solovieva, E.A.; Chepurnykh, T.V.; Shcheglov, A.S.; Verkhusha, V.V.; Pletnev, V.Z.; Hazelwood, K.L.; Roche, P.M.; et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009, 418, 567–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subach, F.V.; Patterson, G.H.; Manley, S.; Gillette, J.M.; Lippincott-Schwartz, J.; Verkhusha, V.V. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 2009, 6, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Subach, O.M.; Cranfill, P.J.; Davidson, M.W.; Verkhusha, V.V. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS ONE 2011, 6, e28674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tantama, M.; Hung, Y.P.; Yellen, G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J. Am. Chem Soc. 2011, 133, 10034–10037. [Google Scholar] [CrossRef] [Green Version]
- Vrzheshch, E.P.; Dmitrienko, D.V.; Rudanov, G.S.; Zagidullin, V.E.; Paschenko, V.Z.; Razzhivin, A.P.; Saletsky, A.M.; Vrzheshch, P.V. Optical properties of the monomeric red fluorescent protein mRFP1. Mosc. Univ. Biol. Sci. Bull. 2008, 63, 109–112. [Google Scholar] [CrossRef]
- Bar-Yosef, T.; Damri, O.; Agam, G. Dual Role of Autophagy in Diseases of the Central Nervous System. Front. Cell Neurosci. 2019, 13, 196. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy Levy, J.M.; Thorburn, A. Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 2020, 27, 843–857. [Google Scholar] [CrossRef]
- Yim, W.W.; Mizushima, N. Lysosome biology in autophagy. Cell Discov. 2020, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Rubinsztein, D.C.; Shpilka, T.; Elazar, Z. Mechanisms of autophagosome biogenesis. Curr. Biol. 2012, 22, R29–R34. [Google Scholar] [CrossRef] [Green Version]
- Reggiori, F.; Komatsu, M.; Finley, K.; Simonsen, A. Autophagy: More than a nonselective pathway. Int J. Cell Biol. 2012, 2012, 219625. [Google Scholar] [CrossRef] [PubMed]
- Palikaras, K.; Lionaki, E.; Tavernarakis, N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 2018, 20, 1013–1022. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Cuervo, A.M.; Dunn, W.A., Jr.; Levine, B.; van der Klei, I.; Seglen, P.O. How shall I eat thee? Autophagy 2007, 3, 413–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuck, S. Microautophagy—Distinct molecular mechanisms handle cargoes of many sizes. J. Cell Sci. 2020, 133, 17. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol. 2012, 22, 407–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaushik, S.; Bandyopadhyay, U.; Sridhar, S.; Kiffin, R.; Martinez-Vicente, M.; Kon, M.; Orenstein, S.J.; Wong, E.; Cuervo, A.M. Chaperone-mediated autophagy at a glance. J. Cell Sci. 2011, 124, 495–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrpour, M.; Esclatine, A.; Beau, I.; Codogno, P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010, 20, 748–762. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. AMPK and autophagy get connected. EMBO J. 2011, 30, 634–635. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.C.; Guan, K.L. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Invest. 2015, 125, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Zachari, M.; Ganley, I.G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017, 61, 585–596. [Google Scholar]
- Feng, Y.; Klionsky, D.J. Autophagic membrane delivery through ATG9. Cell Res. 2017, 27, 161–162. [Google Scholar] [CrossRef] [PubMed]
- Mack, H.I.; Zheng, B.; Asara, J.M.; Thomas, S.M. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy 2012, 8, 1197–1214. [Google Scholar] [CrossRef] [Green Version]
- Burman, C.; Ktistakis, N.T. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010, 584, 1302–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proikas-Cezanne, T.; Takacs, Z.; Donnes, P.; Kohlbacher, O. WIPI proteins: Essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci. 2015, 128, 207–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walczak, M.; Martens, S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy 2013, 9, 424–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrotis, A.; von Chamier, L.; Oliver, H.; Kiso, K.; Singh, T.; Ketteler, R. Human ATG4 autophagy proteases counteract attachment of ubiquitin-like LC3/GABARAP proteins to other cellular proteins. J. Biol. Chem. 2019, 294, 12610–12621. [Google Scholar] [CrossRef] [Green Version]
- Khaminets, A.; Behl, C.; Dikic, I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016, 26, 6–16. [Google Scholar] [CrossRef]
- Kirkin, V.; Lamark, T.; Sou, Y.S.; Bjorkoy, G.; Nunn, J.L.; Bruun, J.A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P.; et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 2009, 33, 505–516. [Google Scholar] [CrossRef] [Green Version]
- Kirkin, V.; Rogov, V.V. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol. Cell 2019, 76, 268–285. [Google Scholar] [CrossRef] [PubMed]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Overvatn, A.; Bjorkoy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, T.; Noda, N.N. Autophagy-regulating protease Atg4: Structure, function, regulation and inhibition. J. Antibiot. (Tokyo) 2018, 71, 72–78. [Google Scholar] [CrossRef] [Green Version]
- Ao, X.; Zou, L.; Wu, Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014, 21, 348–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, L.; Hou, C.; Lai, Y.; Long, J.; Liu, J.; Zhong, Q.; Diao, J. SNARE-mediated membrane fusion in autophagy. Semin Cell Dev. Biol. 2016, 60, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 conjugation system in mammalian autophagy. Int J. Biochem. Cell Biol. 2004, 36, 2503–2518. [Google Scholar] [CrossRef]
- Chung, C.G.; Lee, H.; Lee, S.B. Mechanisms of protein toxicity in neurodegenerative diseases. Cell Mol. Life Sci. 2018, 75, 3159–3180. [Google Scholar] [CrossRef] [Green Version]
- Harjes, P.; Wanker, E.E. The hunt for huntingtin function: Interaction partners tell many different stories. Trends Biochem. Sci. 2003, 28, 425–433. [Google Scholar] [CrossRef]
- Martinez-Vicente, M.; Talloczy, Z.; Wong, E.; Tang, G.; Koga, H.; Kaushik, S.; de Vries, R.; Arias, E.; Harris, S.; Sulzer, D.; et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat. Neurosci. 2010, 13, 567–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, Y.C.; Holzbaur, E.L. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci. 2014, 34, 1293–1305. [Google Scholar] [CrossRef] [Green Version]
- Koga, H.; Martinez-Vicente, M.; Arias, E.; Kaushik, S.; Sulzer, D.; Cuervo, A.M. Constitutive upregulation of chaperone-mediated autophagy in Huntington’s disease. J. Neurosci. 2011, 31, 18492–18505. [Google Scholar] [CrossRef]
- Nixon, R.A. Autophagy, amyloidogenesis and Alzheimer disease. J. Cell Sci. 2007, 120, 4081–4091. [Google Scholar] [CrossRef] [Green Version]
- Boland, B.; Kumar, A.; Lee, S.; Platt, F.M.; Wegiel, J.; Yu, W.H.; Nixon, R.A. Autophagy induction and autophagosome clearance in neurons: Relationship to autophagic pathology in Alzheimer’s disease. J. Neurosci. 2008, 28, 6926–6937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neely, K.M.; Green, K.N.; LaFerla, F.M. Presenilin is necessary for efficient proteolysis through the autophagy-lysosome system in a gamma-secretase-independent manner. J. Neurosci 2011, 31, 2781–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guglielmotto, M.; Monteleone, D.; Piras, A.; Valsecchi, V.; Tropiano, M.; Ariano, S.; Fornaro, M.; Vercelli, A.; Puyal, J.; Arancio, O.; et al. Abeta1-42 monomers or oligomers have different effects on autophagy and apoptosis. Autophagy 2014, 10, 1827–1843. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, K. Tau in Alzheimer Disease and Related Tauopathies. Curr Alzheimer Res. 2010, 7, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Bhaskar, K. Degradation and Transmission of Tau by Autophagic-Endolysosomal Networks and Potential Therapeutic Targets for Tauopathy. Front. Mol. Neurosci. 2020, 13, 586731. [Google Scholar] [CrossRef]
- Menzies, F.M.; Fleming, A.; Caricasole, A.; Bento, C.F.; Andrews, S.P.; Ashkenazi, A.; Fullgrabe, J.; Jackson, A.; Jimenez Sanchez, M.; Karabiyik, C.; et al. Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron 2017, 93, 1015–1034. [Google Scholar] [CrossRef] [Green Version]
- Stefanis, L. alpha-Synuclein in Parkinson’s disease. Cold Spring Harb Perspect Med. 2012, 2, a009399. [Google Scholar] [CrossRef] [Green Version]
- Cuervo, A.M. Impaired Degradation of mutant alpha synuclein of chaperone-mediated autophagy. Science 2004, 305, 1292–1295. [Google Scholar] [CrossRef]
- Vogiatzi, T.; Xilouri, M.; Vekrellis, K.; Stefanis, L. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J. Biol Chem. 2008, 283, 23542–23556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winslow, A.R.; Chen, C.W.; Corrochano, S.; Acevedo-Arozena, A.; Gordon, D.E.; Peden, A.A.; Lichtenberg, M.; Menzies, F.M.; Ravikumar, B.; Imarisio, S.; et al. alpha-Synuclein impairs macroautophagy: Implications for Parkinson’s disease. J. Cell Biol. 2010, 190, 1023–1037. [Google Scholar] [CrossRef] [Green Version]
- Coune, P.G.; Bensadoun, J.C.; Aebischer, P.; Schneider, B.L. Rab1A over-expression prevents Golgi apparatus fragmentation and partially corrects motor deficits in an alpha-synuclein based rat model of Parkinson’s disease. J. Parkinsons Dis. 2011, 1, 373–387. [Google Scholar] [CrossRef]
- Tanik, S.A.; Schultheiss, C.E.; Volpicelli-Daley, L.A.; Brunden, K.R.; Lee, V.M. Lewy body-like alpha-synuclein aggregates resist degradation and impair macroautophagy. J. Biol. Chem. 2013, 288, 15194–15210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volpicelli-Daley, L.A.; Gamble, K.L.; Schultheiss, C.E.; Riddle, D.M.; West, A.B.; Lee, V.M. Formation of alpha-synuclein Lewy neurite-like aggregates in axons impedes the transport of distinct endosomes. Mol. Biol. Cell 2014, 25, 4010–4023. [Google Scholar] [CrossRef] [PubMed]
- Malpartida, A.B.; Williamson, M.; Narendra, D.P.; Wade-Martins, R.; Ryan, B.J. Mitochondrial Dysfunction and Mitophagy in Parkinson’s Disease: From Mechanism to Therapy. Trends Biochem. Sci. 2021, 46, 329–343. [Google Scholar] [CrossRef]
- Pickrell, A.M.; Youle, R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 2015, 85, 257–273. [Google Scholar] [CrossRef] [Green Version]
- Ge, P.; Dawson, V.L.; Dawson, T.M. PINK1 and Parkin mitochondrial quality control: A source of regional vulnerability in Parkinson’s disease. Mol. Neurodegener 2020, 15, 20. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in mammalian autophagy research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N. Methods for monitoring autophagy. Int J. Biochem. Cell Biol. 2004, 36, 2491–2502. [Google Scholar] [CrossRef]
- du Toit, A.; Hofmeyr, J.S.; Gniadek, T.J.; Loos, B. Measuring autophagosome flux. Autophagy 2018, 14, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Rhodin, J. Correlation of Ultrastructural Organization and Function in Normal and Experimentally Changed Proximal Convoluted Tubule Cells of the Mouse Kidney: An Electron Microscopic Study Including an Experimental Analysis of the Conditions For Fixation of the Renal Tissue For High Reso-Lution Electron Microscopy. Ph.D. Thesis, Karolinska Institutet, Stockholm, Sweden, 1954. [Google Scholar]
- Eskelinen, E.L.; Reggiori, F.; Baba, M.; Kovacs, A.L.; Seglen, P.O. Seeing is believing: The impact of electron microscopy on autophagy research. Autophagy 2011, 7, 935–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barth, S.; Glick, D.; Macleod, K.F. Autophagy: Assays and artifacts. J. Pathol. 2010, 221, 117–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauvy, C.; Meijer, A.J.; Codogno, P. Chapter 4 Assaying of Autophagic Protein Degradation. Autophagy Mamm. Syst. Part B 2009, 452, 47–61. [Google Scholar]
- Zhang, T.; Wolfe, C.; Pierle, A.; Welle, K.A.; Hryhorenko, J.R.; Ghaemmaghami, S. Proteome-wide modulation of degradation dynamics in response to growth arrest. Proc. Natl. Acad. Sci. USA 2017, 114, E10329–E10338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Price, J.C.; Nouri-Nigjeh, E.; Li, J.; Hellerstein, M.K.; Qu, J.; Ghaemmaghami, S. Kinetics of precursor labeling in stable isotope labeling in cell cultures (SILAC) experiments. Anal. Chem. 2014, 86, 11334–11341. [Google Scholar] [CrossRef]
- Luhr, M.; Szalai, P.; Saetre, F.; Gerner, L.; Seglen, P.O.; Engedal, N. A Simple Cargo Sequestration Assay for Quantitative Measurement of Nonselective Autophagy in Cultured Cells. Methods Enzymol. 2017, 587, 351–364. [Google Scholar]
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays Biochem. 2017, 61, 609–624. [Google Scholar]
- Kirisako, T.; Ichimura, Y.; Okada, H.; Kabeya, Y.; Mizushima, N.; Yoshimori, T.; Ohsumi, M.; Takao, T.; Noda, T.; Ohsumi, Y. The Reversible Modification Regulates the Membrane-Binding State of Apg8 Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000, 151, 263–275. [Google Scholar] [CrossRef]
- Wild, P.; McEwan, D.G.; Dikic, I. The LC3 interactome at a glance. J. Cell Sci. 2014, 127, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T. How to interpret LC3 immunoblotting. Autophagy 2007, 3, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Cui, D.; Sun, D.; Wang, X.; Yi, L.; Kulikowicz, E.; Reyes, M.; Zhu, J.; Yang, Z.J.; Jiang, W.; Koehler, R.C. Impaired autophagosome clearance contributes to neuronal death in a piglet model of neonatal hypoxic-ischemic encephalopathy. Cell Death Dis. 2017, 8, e2919. [Google Scholar] [CrossRef] [Green Version]
- Mauthe, M.; Orhon, I.; Rocchi, C.; Zhou, X.; Luhr, M.; Hijlkema, K.J.; Coppes, R.P.; Engedal, N.; Mari, M.; Reggiori, F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018, 14, 1435–1455. [Google Scholar] [CrossRef]
- Mauvezin, C.; Neufeld, T.P. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy 2015, 11, 1437–1438. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Koh, M.; Lim, B.J.; Park, S.B. Emission wavelength prediction of a full-color-tunable fluorescent core skeleton, 9-aryl-1,2-dihydropyrrolo[3,4-b]indolizin-3-one. J. Am. Chem Soc. 2011, 133, 6642–6649. [Google Scholar] [CrossRef]
- Lee, S.; Kim, E.; Park, S.B. Discovery of autophagy modulators through the construction of a high-content screening platform via monitoring of lipid droplets. Chem. Sci. 2013, 4, 8. [Google Scholar] [CrossRef]
- Li, S.S.; Zhang, M.; Wang, J.H.; Yang, F.; Kang, B.; Xu, J.J.; Chen, H.Y. Monitoring the Changes of pH in Lysosomes during Autophagy and Apoptosis by Plasmon Enhanced Raman Imaging. Anal. Chem. 2019, 91, 8398–8405. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Patterson, G.H.; Davidson, M.W. Advances in fluorescent protein technology. J. Cell Sci 2007, 120, 4247–4260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaizuka, T.; Morishita, H.; Hama, Y.; Tsukamoto, S.; Matsui, T.; Toyota, Y.; Kodama, A.; Ishihara, T.; Mizushima, T.; Mizushima, N. An Autophagic Flux Probe that Releases an Internal Control. Mol. Cell 2016, 64, 835–849. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Kim, H.; Choi, J.; Inn, K.S.; Seong, J. Visualization of Autophagy Progression by a Red-Green-Blue Autophagy Sensor. ACS Sens. 2020, 5, 3850–3861. [Google Scholar] [CrossRef]
- Kimura, S.; Noda, T.; Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007, 3, 452–460. [Google Scholar] [CrossRef] [Green Version]
- Maulucci, G.; Chiarpotto, M.; Papi, M.; Samengo, D.; Pani, G.; De Spirito, M. Quantitative analysis of autophagic flux by confocal pH-imaging of autophagic intermediates. Autophagy 2015, 11, 1905–1916. [Google Scholar] [CrossRef] [Green Version]
- Tanida, I.; Ueno, T.; Uchiyama, Y. A super-ecliptic, pHluorin-mKate2, tandem fluorescent protein-tagged human LC3 for the monitoring of mammalian autophagy. PLoS ONE 2014, 9, e110600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Zhong, W.; Zhou, J.; Sheng, F.; Fang, Z.; Wei, Y.; Chen, Y.; Deng, X.; Xia, B.; Lin, J. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy 2012, 8, 1215–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koga, H.; Martinez-Vicente, M.; Macian, F.; Verkhusha, V.V.; Cuervo, A.M. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat. Commun. 2011, 2, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Acunzo, P.; Strappazzon, F.; Caruana, I.; Meneghetti, G.; Di Rita, A.; Simula, L.; Weber, G.; Del Bufalo, F.; Dalla Valle, L.; Campello, S.; et al. Reversible induction of mitophagy by an optogenetic bimodular system. Nat. Commun. 2019, 10, 1533. [Google Scholar] [CrossRef]
- Miesenbock, G.; De Angelis, D.A.; Rothman, J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998, 394, 192–195. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Yamamoto, A.; Oshitani-Okamoto, S.; Ohsumi, Y.; Yoshimori, T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004, 117, 2805–2812. [Google Scholar] [CrossRef] [Green Version]
- Leeman, D.S.; Hebestreit, K.; Ruetz, T.; Webb, A.E.; McKay, A.; Pollina, E.A.; Dulken, B.W.; Zhao, X.; Yeo, R.W.; Ho, T.T.; et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 2018, 359, 1277–1283. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.H.; Cao, L.; Mostoslavsky, R.; Lombard, D.B.; Liu, J.; Bruns, N.E.; Tsokos, M.; Alt, F.W.; Finkel, T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA 2008, 105, 3374–3379. [Google Scholar] [CrossRef] [Green Version]
- Bains, M.; Heidenreich, K.A. Chapter 7 Live-Cell Imaging of Autophagy Induction and Autophagosome-Lysosome Fusion in Primary Cultured Neurons. Autophagy Dis. Clin. Appl. Part C 2009, 453, 145–158. [Google Scholar]
- Demarchi, F.; Bertoli, C.; Copetti, T.; Tanida, I.; Brancolini, C.; Eskelinen, E.L.; Schneider, C. Calpain is required for macroautophagy in mammalian cells. J. Cell Biol. 2006, 175, 595–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flavin, W.P.; Bousset, L.; Green, Z.C.; Chu, Y.; Skarpathiotis, S.; Chaney, M.J.; Kordower, J.H.; Melki, R.; Campbell, E.M. Endocytic vesicle rupture is a conserved mechanism of cellular invasion by amyloid proteins. Acta Neuropathol. 2017, 134, 629–653. [Google Scholar] [CrossRef] [PubMed]
- Jahreiss, L.; Menzies, F.M.; Rubinsztein, D.C. The itinerary of autophagosomes: From peripheral formation to kiss-and-run fusion with lysosomes. Traffic 2008, 9, 574–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N.; Yamamoto, A.; Hatano, M.; Kobayashi, Y.; Kabeya, Y.; Suzuki, K.; Tokuhisa, T.; Ohsumi, Y.; Yoshimori, T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic. J. Cell Biol. 2001, 152, 657–667. [Google Scholar] [CrossRef] [Green Version]
- Tanida, I.; Yamaji, T.; Ueno, T.; Ishiura, S.; Kominami, E.; Hanada, K. Consideration about negative controls for LC3 and expression vectors for four colored fluorescent protein-LC3 negative controls. Autophagy 2008, 4, 131–134. [Google Scholar] [CrossRef] [Green Version]
- Erin, M.; Buckingham, J.E.C. Wallen Jackson Charles Grose, Nuclear LC3-positive puncta in stressed cells do not represent autophagosomes. Biotechniques 2018, 57, 241–244. [Google Scholar]
- Lee, Y.K.; Jun, Y.W.; Choi, H.E.; Huh, Y.H.; Kaang, B.K.; Jang, D.J.; Lee, J.A. Development of LC3/GABARAP sensors containing a LIR and a hydrophobic domain to monitor autophagy. EMBO J. 2017, 36, 1100–1116. [Google Scholar] [CrossRef] [Green Version]
- Rosado, C.J.; Mijaljica, D.; Hatzinisiriou, I.; Prescott, M.; Devenish, R.J. Rosella: A fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy 2008, 4, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Patterson, G.H.; Knobel, S.M.; Sharif, W.D.; Kain, S.R.; Piston, D.W. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 1997, 73, 2782–2790. [Google Scholar] [CrossRef] [Green Version]
- Ai, H.W.; Olenych, S.G.; Wong, P.; Davidson, M.W.; Campbell, R.E. Hue-shifted monomeric variants of Clavularia cyan fluorescent protein: Identification of the molecular determinants of color and applications in fluorescence imaging. BMC Biol. 2008, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Merzlyak, E.M.; Goedhart, J.; Shcherbo, D.; Bulina, M.E.; Shcheglov, A.S.; Fradkov, A.F.; Gaintzeva, A.; Lukyanov, K.A.; Lukyanov, S.; Gadella, T.W.; et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 2007, 4, 555–557. [Google Scholar] [CrossRef]
- Shaner, N.C.; Lin, M.Z.; McKeown, M.R.; Steinbach, P.A.; Hazelwood, K.L.; Davidson, M.W.; Tsien, R.Y. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 2008, 5, 545–551. [Google Scholar] [CrossRef] [Green Version]
- Esposito, A.; Gralle, M.; Dani, M.A.C.; Lange, D.; Wouters, F.S. pHlameleons: A Family of FRET-Based Protein Sensors for Quantitative pH Imaging. Biochemistry 2008, 47, 13115–13126. [Google Scholar] [CrossRef]
- Kremers, G.; Goedhart, J.; van Munster, E.B.; Gadella, T.W.J. Cyan and Yellow Super Fluorescent Proteins with Improved Brightness, Protein Folding, and FRET Förster Radius. Biochemistry 2006, 45, 6570–6580. [Google Scholar] [CrossRef]
- Elsliger, M.; Wachter, R.M.; Hanson, G.T.; Kallio, K.; Remington, S.J. Structural and Spectral Response of Green Fluorescent Protein Variants to Changes in pH. Biochemistry 1999, 38, 5296–5301. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Shimozono, S.; Hosoi, H.; Mizuno, H.; Fukano, T.; Tahara, T.; Miyawaki, A. Concatenation of Cyan and Yellow Fluorescent Proteins for Efficient Resonance Energy Transfer. Biochemistry 2006, 45, 6267–6271. [Google Scholar] [CrossRef]
- Burgstaller, S.; Bischof, H.; Gensch, T.; Stryeck, S.; Gottschalk, B.; Ramadani-Muja, J.; Eroglu, E.; Rost, R.; Balfanz, S.; Baumann, A.; et al. pH-Lemon, a Fluorescent Protein-Based pH Reporter for Acidic Compartments. ACS Sens. 2019, 4, 883–891. [Google Scholar] [CrossRef] [Green Version]
- Katayama, H.; Hama, H.; Nagasawa, K.; Kurokawa, H.; Sugiyama, M.; Ando, R.; Funata, M.; Yoshida, N.; Homma, M.; Nishimura, T.; et al. Visualizing and Modulating Mitophagy for Therapeutic Studies of Neurodegeneration. Cell 2020, 181, 1176–1187.e16. [Google Scholar] [CrossRef]
- Karasawa, S.; Araki, T.; Yamamoto-Hino, M.; Miyawaki, A. A green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J. Biol. Chem. 2003, 278, 34167–34171. [Google Scholar] [CrossRef] [Green Version]
- Rudolf, R.; Mongillo, M.; Magalhaes, P.J.; Pozzan, T. In vivo monitoring of Ca(2+) uptake into mitochondria of mouse skeletal muscle during contraction. J. Cell Biol. 2004, 166, 527–536. [Google Scholar] [CrossRef] [Green Version]
- Gilon, T.; Chomsky, O.; Kulka, R.G. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998, 17, 2759–2766. [Google Scholar] [CrossRef]
- Ghoda, L.; Sidney, D.; Macrae, M.; Coffino, P. Structural elements of Ornithine decarboxylase required for intracellular degradation and polyamine-dependent regulation. Mol. Cell. Biol. 1992, 12, 2178–2185. [Google Scholar] [CrossRef] [Green Version]
- Chudakov, D.M.; Verkhusha, V.V.; Staroverov, D.B.; Souslova, E.A.; Lukyanov, S.; Lukyanov, K.A. Photoswitchable cyan fluorescent protein for protein tracking. Nat. Biotechnol. 2004, 22, 1435–1439. [Google Scholar] [CrossRef]
- Heckmann, B.L.; Teubner, B.J.W.; Tummers, B.; Boada-Romero, E.; Harris, L.; Yang, M.; Guy, C.S.; Zakharenko, S.S.; Green, D.R. LC3-Associated Endocytosis Facilitates beta-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer’s Disease. Cell 2019, 178, 536–551.e14. [Google Scholar] [CrossRef]
- Kim, Y.D.; Jeong, E.I.; Nah, J.; Yoo, S.M.; Lee, W.J.; Kim, Y.; Moon, S.; Hong, S.H.; Jung, Y.K. Pimozide reduces toxic forms of tau in TauC3 mice via 5’ adenosine monophosphate-activated protein kinase-mediated autophagy. J. Neurochem. 2017, 142, 734–746. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Lu, J.; Xu, Y.; Wang, J.; Qiu, X.; Fan, L.; Li, B.; Liu, W.; Mao, F.; Zhu, J.; et al. Discovery of nitazoxanide-based derivatives as autophagy activators for the treatment of Alzheimer’s disease. Acta Pharm. Sin. B 2020, 10, 646–666. [Google Scholar] [CrossRef]
- Tien, N.T.; Karaca, I.; Tamboli, I.Y.; Walter, J. Trehalose Alters Subcellular Trafficking and the Metabolism of the Alzheimer-associated Amyloid Precursor Protein. J. Biol. Chem. 2016, 291, 10528–10540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, I.; Zhang, Y.; Seegobin, S.P.; Pruvost, M.; Wang, Q.; Purtell, K.; Zhang, B.; Yue, Z. Microglia Clear Neuron-released α-Synuclein via Selective Autophagy and Prevent Neurodegeneration. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehay, B.; Bove, J.; Rodriguez-Muela, N.; Perier, C.; Recasens, A.; Boya, P.; Vila, M. Pathogenic lysosomal depletion in Parkinson’s disease. J. Neurosci. 2010, 30, 12535–12544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xilouri, M.; Vogiatzi, T.; Vekrellis, K.; Park, D.; Stefanis, L. Abberant alpha-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS ONE 2009, 4, e5515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, X.X.; Wang, S.F.; Tan, Y.; Song, J.X.; Zhu, Z.; Wang, Z.Y.; Wu, M.Y.; Cai, C.Z.; Huang, Z.J.; Tan, J.Q.; et al. Pharmacological enhancement of TFEB-mediated autophagy alleviated neuronal death in oxidative stress-induced Parkinson’s disease models. Cell Death Dis. 2020, 11, 128. [Google Scholar] [CrossRef] [Green Version]
- Ashkenazi, A.; Bento, C.F.; Ricketts, T.; Vicinanza, M.; Siddiqi, F.; Pavel, M.; Squitieri, F.; Hardenberg, M.C.; Imarisio, S.; Menzies, F.M.; et al. Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 2017, 545, 108–111. [Google Scholar] [CrossRef] [Green Version]
- Filimonenko, M.; Stuffers, S.; Raiborg, C.; Yamamoto, A.; Malerod, L.; Fisher, E.M.; Isaacs, A.; Brech, A.; Stenmark, H.; Simonsen, A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007, 179, 485–500. [Google Scholar] [CrossRef]
- Proenca, C.C.; Stoehr, N.; Bernhard, M.; Seger, S.; Genoud, C.; Roscic, A.; Paganetti, P.; Liu, S.; Murphy, L.O.; Kuhn, R.; et al. Atg4b-dependent autophagic flux alleviates Huntington’s disease progression. PLoS ONE 2013, 8, e68357. [Google Scholar] [CrossRef]
- Feng, Q.; Luo, Y.; Zhang, X.N.; Yang, X.F.; Hong, X.Y.; Sun, D.S.; Li, X.C.; Hu, Y.; Li, X.G.; Zhang, J.F.; et al. MAPT/Tau accumulation represses autophagy flux by disrupting IST1-regulated ESCRT-III complex formation: A vicious cycle in Alzheimer neurodegeneration. Autophagy 2020, 16, 641–658. [Google Scholar] [CrossRef]
- Gao, J.; Chen, X.; Ma, T.; He, B.; Li, P.; Zhao, Y.; Ma, Y.; Zhuang, J.; Yin, Y. PEG-Ceramide Nanomicelles Induce Autophagy and Degrade Tau Proteins in N2a Cells. Int. J. Nanomed. 2020, 15, 6779–6789. [Google Scholar] [CrossRef]
- Guo, Y.L.; Duan, W.J.; Lu, D.H.; Ma, X.H.; Li, X.X.; Li, Z.; Bi, W.; Kurihara, H.; Liu, H.Z.; Li, Y.F.; et al. Autophagy-dependent removal of alpha-synuclein: A novel mechanism of GM1 ganglioside neuroprotection against Parkinson’s disease. Acta Pharmacol. Sin. 2021, 42, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Pal, R.; Bajaj, L.; Sharma, J.; Palmieri, M.; Di Ronza, A.; Lotfi, P.; Chaudhury, A.; Neilson, J.; Sardiello, M.; Rodney, G.G. NADPH oxidase promotes Parkinsonian phenotypes by impairing autophagic flux in an mTORC1-independent fashion in a cellular model of Parkinson’s disease. Sci Rep. 2016, 6, 22866. [Google Scholar] [CrossRef] [Green Version]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef] [PubMed]
- Kunze, M.; Berger, J. The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance. Front. Physiol. 2015, 6, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guntas, G.; Hallett, R.A.; Zimmerman, S.P.; Williams, T.; Yumerefendi, H.; Bear, J.E.; Kuhlman, B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. USA 2015, 112, 112–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seong, J.; Lin, M.Z. Optobiochemistry: Genetically Encoded Control of Protein Activity by Light. Annu. Rev. Biochem. 2021, 90. [Google Scholar] [CrossRef] [PubMed]




| Fluorescent Protein | λex | λem | pKa | EC | QY | Brightness | Fluorescence Fold Change (pH 5.5–7.5) | Reference |
|---|---|---|---|---|---|---|---|---|
| mTagBFP2 | 399 | 454 | 2.7 | 50,600 | 0.64 | 32.38 | 1 | [32], This work |
| mTurquoise2 | 434 | 474 | 3.1 | 30,000 | 0.93 | 27.9 | - | [12,24] |
| CFP | 456 | 480 | - | - | - | - | - | [24] |
| PS-CFP2(cyan) | 400 | 468 | - | 43,000 | 0.2 | 8.6 | - | [24] |
| pHRed | 440 | 610 | 6.6 | - | - | - | - | [33] |
| mKeima | 440 | 620 | 6.5 | 14,400 | 0.24 | 3.46 | - | [23,24] |
| EGFP | 488 | 507 | 6 | 55,900 | 0.6 | 33.54 | 1 | [24], This work |
| PS-CFP2(green) | 490 | 511 | - | 47,000 | 0.23 | 10.81 | - | [24] |
| mWasabi | 493 | 509 | 6.5 | 70,000 | 0.8 | 56 | - | [24,28] |
| SE-pHluorin | 495 | 512 | 7.2 | - | - | - | 50 | [16] |
| pHoran1 | 547 | 564 | 6.7 | - | - | - | 10 | [16] |
| pHoran4 | 547 | 561 | 7.5 | - | - | - | 17 | [16] |
| mOrange | 548 | 562 | 6.5 | 71,000 | 0.69 | 48.99 | 5 | [16,24] |
| mApple | 548 | 592 | 6.5 | 75,000 | 0.49 | 36.75 | 4 | [16,24] |
| pHTomato | 550 | 580 | 7.8 | - | - | - | - | [14] |
| tdTomato | 554 | 581 | 4.7 | 138,000 | 0.69 | 95.22 | - | [24] |
| mNectarine | 558 | 578 | 6.9 | 58,000 | 0.45 | 30 | 6 | [22,24] |
| DsRed | 558 | 583 | - | 72,500 | 0.68 | 49.3 | - | [24,25] |
| pHmScarlet | 562 | 585 | 7.4 | 85,000 | 0.47 | 39.73 | - | [26] |
| PAmCherry1 | 564 | 595 | 6.3 | 18,000 | 0.46 | 8.28 | - | [24,31] |
| pHuji | 566 | 598 | 7.7 | 31,000 | 0.22 | 6.82 | 22 | [16,24] |
| mScarlet | 569 | 594 | 5.3 | 100,000 | 0.7 | 70 | 1.3 | [24], This work |
| mRFP1 | 584 | 607 | 4.5 | 50,000 | 0.25 | 12.5 | - | [14,21] |
| mCherry | 587 | 610 | 4.5 | 72,000 | 0.22 | 15.84 | 1 | [25], This work |
| mKate2 | 588 | 633 | 5.4 | 62,500 | 0.4 | 25 | 1.3 | [23], This work |
| mKate | 588 | 635 | 6.2 | 45,000 | 0.33 | 14.85 | - | [24,29] |
| HcRed | 592 | 645 | - | 20,000 | 0.015 | 0.3 | - | [24] |
| FP-Based Autophagy Biosensor type | Molecular Design | Detected Autophagy Types (and Stages) | Biosensor Applications in Neurodegenerative Disease |
|---|---|---|---|
| FP-LC3 | EGFP-LC3 RFP-LC3 mCherry-LC3 YFP-LC3 HcRed-LC3 CFP-LC3 | Macroautophagy (autophagosome) | Huntingtin disease [68] Alzheimer’s disease [71,146,147,148,149] Parkinson’s disease [150,151,152,153] |
| HyD-LIR-GFP | HyD-LIR(Fy)-GFP HyD-LIR(TP)-GFP | Macroautophagy (autophagosome) | - |
| RFP-GFP-LC3 | mRFP-GFP-LC3 mCherry-EGFP-LC3 mTagRFP-mWasabi-LC3 mKate-SEP-LC3 | Macroautophagy (autophagosome, autolysosome) | Huntingtin disease [154,155,156] Alzheimer’s disease [73,157,158] Parkinson’s disease [159,160] |
| GFP-LC3-RFP-LC3ΔG | GFP-LC3-RFP-LC3ΔG | Macroautophagy (autophagosome, autolysosome) | - |
| BFP-RFP-GFP-LC3 | mTagBFP2-mApple-SEP-LC3 | Macroautophagy (phagophore, autophagosome, fusion, autolysosome) | Alzheimer’s disease [73] |
| pH-Lemon-LC3 | mTurquioise2- EYFP-LC3 | Macroautophagy (autophagosome, autolysosome) | - |
| mito-SRAI | mito-TOLLES-Ypet | Mitophagy (autophagosome, autolysosome) | - |
| CMA sensor | KFERQ-PS-CFP2 KFERQ-PA-mCherry | Chaperon-mediated autophagy | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Seong, J. Fluorescent Protein-Based Autophagy Biosensors. Materials 2021, 14, 3019. https://doi.org/10.3390/ma14113019
Kim H, Seong J. Fluorescent Protein-Based Autophagy Biosensors. Materials. 2021; 14(11):3019. https://doi.org/10.3390/ma14113019
Chicago/Turabian StyleKim, Heejung, and Jihye Seong. 2021. "Fluorescent Protein-Based Autophagy Biosensors" Materials 14, no. 11: 3019. https://doi.org/10.3390/ma14113019
APA StyleKim, H., & Seong, J. (2021). Fluorescent Protein-Based Autophagy Biosensors. Materials, 14(11), 3019. https://doi.org/10.3390/ma14113019






