Scaling Scientific Cellular Automata Microstructure Evolution Model of Static Recrystallization toward Practical Industrial Calculations
Abstract
:1. Introduction
2. SRX Model
3. Parallelization Modes
4. Communication Mechanism
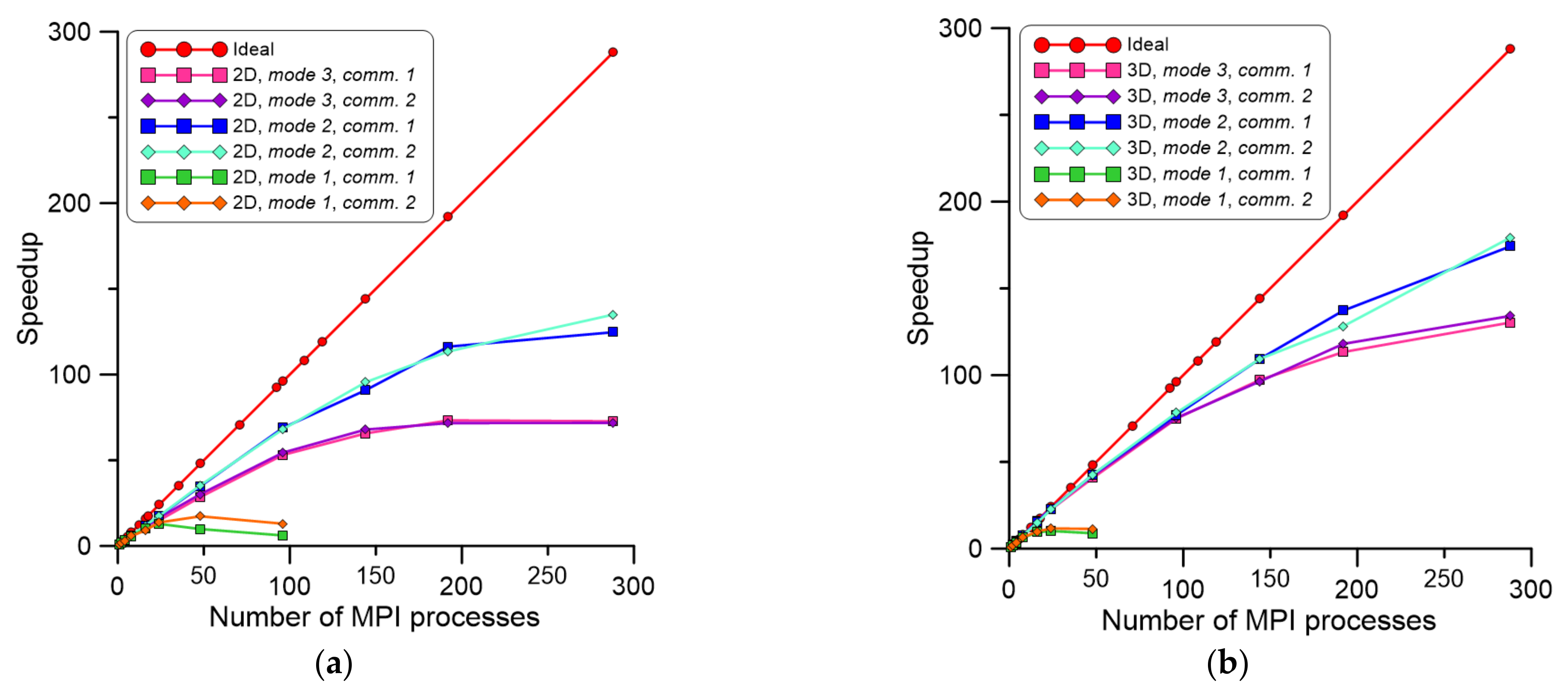
5. Practical Case Studies
6. Conclusions
- The cellular automata method has great potential for algorithm adaptation to the distributed environment via parallelization.
- The parallelization mode 1 is clearly affected by communication overheads.
- The parallelization mode 2 is the best solution for large CA spaces but requires an additional data preparation stage.
- Parallelization is more effective with an increasing number of CA cells processed during the entire simulation; this is clearly visible for 3D scenarios.
- Communication mechanisms used during CA cell interaction in each time step directly influence model execution times.
- The speedup decreases for more than 96 and 144 MPI processes in 2D and 3D, respectively, which is related to sequential parts of the code.
- The developed model provides a possibility to obtain scaled speedup over 700 times for a 2D scenario and over 800 times for a 3D scenario, which is crucial for practical applications in industrial practice.
- With the developed solution, the computations of more than one billion CA cells are possible with reasonable time, which provides a possibility to simulate 3D microstructure models with a large number of grains with varying sizes.
- Simulations based on experimental data from the 3D reconstruction algorithm can also be undertaken with a reasonable time.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pokharel, R.; Lind, J.; Li, S.F.; Kenesei, P.; Lebensohn, R.A.; Suter, R.M.; Rollett, A.D. In-situ observation of bulk 3D grain evolution during plastic deformation in polycrystalline Cu. Int. J. Plast. 2015, 67, 217–234. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Beladi, H.; Pan, L.; Fang, F.; Hu, J.; Kong, L.; Hodgson, P.D.; Timokhina, I. Interphase precipitation hardening of a TiMo microalloyed dual-phase steel produced by continuous cooling. Mater. Sci. Eng. A 2021, 804, 140518. [Google Scholar] [CrossRef]
- Villaret, F.; Hary, B.; de Carlan, Y.; Baudin, T.; Logé, R.; Maire, L.; Bernacki, M. Probabilistic and deterministic full field approaches to simulate recrystallization in ODS steels. Comput. Mater. Sci. 2020, 179, 109646. [Google Scholar] [CrossRef]
- Roters, F.; Diehl, M.; Shanthraj, P.; Eisenlohr, P.; Reuber, C.; Wong, S.L.; Maiti, T.; Ebrahimi, A.; Hochrainer, T.; Fabritius, H.O.; et al. DAMASK—The Düsseldorf Advanced Material Simulation Kit for modeling multi-physics crystal plasticity, thermal, and damage phenomena from the single crystal up to the component scale. Comput. Mater. Sci. 2019, 158, 420–478. [Google Scholar] [CrossRef]
- Szeliga, D.; Bzowski, K.; Rauch, L.; Kuziak, R.; Pietrzyk, M. Mean field and full field modelling of microstructure evolution and phase transformations during hot forming and cooling of low carbon steels. Comp. Methods Mater. Sci. 2020, 20, 121–132. [Google Scholar] [CrossRef]
- Després, A.; Greenwood, M.; Sinclair, C.W. A mean-field model of static recrystallization considering orientation spreads and their time-evolution. Acta Mater. 2020, 199, 116–128. [Google Scholar] [CrossRef]
- Allwood, J.M.; Duncan, S.R.; Cao, J.; Groche, P.; Hirt, G.; Kinsey, B.; Kuboki, T.; Liewald, M.; Sterzing, A.; Tekkaya, A.E. Closed-loop control of product properties in metal forming. CIRP Ann. Manuf. Technol. 2016, 65, 573–596. [Google Scholar] [CrossRef] [Green Version]
- Rowenhorst, D.J.; Nguyen, L.; Murphy-Leonard, A.D.; Fonda, R.W. Characterization of Microstructure in Additively Manufactured 316L using Automated Serial Sectioning. Curr. Opin. Solid State Mater. Sci. 2020, 24, 100819. [Google Scholar] [CrossRef]
- Poulsen, H.F.; Jensen, D.J.; Vaughan, G.B.M. Three-Dimensional X-Ray Diffraction Microscopy Using High-Energy X-Rays. MRS Bull. 2004, 29, 166–169. [Google Scholar] [CrossRef]
- Hefferan, C.M.; Lind, J.; Li, S.F.; Lienert, U.; Rollett, A.D.; Suter, R.M. Observation of recovery and recrystallization in high-purity aluminum measured with forward modeling analysis of high-energy diffraction microscopy. Acta Mater. 2012, 60, 4311–4318. [Google Scholar] [CrossRef]
- Lookman, T.; Eidenbenz, S.; Alexander, F.; Barnes, C. Materials Discovery and Design. by Means of Data Science and Optimal Learning; Springer: Berlin/Heidelberg, Germany, 2018; Available online: http://link.springer.com/10.1007/978-3-319-99465-9 (accessed on 20 July 2021).
- Min, K.M.; Jeong, W.; Hong, S.H.; Lee, C.A.; Cha, P.R.; Han, H.N.; Lee, M.G. Integrated crystal plasticity and phase field model for prediction of recrystallization texture and anisotropic mechanical properties of cold-rolled ultra-low carbon steels. Int. J. Plast. 2020, 127, 102644. [Google Scholar] [CrossRef]
- Mellbin, Y.; Hallberg, H.; Ristinmaa, M. Recrystallization and texture evolution during hot rolling of copper, studied by a multiscale model combining crystal plasticity and vertex models. Model. Simul. Mater. Sci. Eng. 2016, 24. [Google Scholar] [CrossRef]
- Vafaeenezhad, H.; Seyedein, S.H.; Aboutalebi, M.R.; Eivani, A.R. Hybrid Monte Carlo—Finite element simulation of microstructural evolution during annealing of severely deformed Sn-5Sb alloy. Comput. Mater. Sci. 2019, 163, 196–208. [Google Scholar] [CrossRef]
- Serajzadeh, P.A.S. Microstructural Changes During Static Recrystallization of Austenitic Stainless Steel 304L: Cellular Automata Simulation. Metallogr. Microstruct. Anal. 2020. [Google Scholar] [CrossRef]
- Kugler, G.; Turk, R. Study of the influence of initial microstructure topology on the kinetics of static recrystallization using a cellular automata model. Comput. Mater. Sci. 2006, 37, 284–291. [Google Scholar] [CrossRef]
- Madej, L.; Sitko, M.; Legwand, A.; Perzynski, K.; Michalik, K. Development and evaluation of data transfer protocols in the fully coupled random cellular automata finite element model of dynamic recrystallization. J. Comput. Sci. 2018, 26, 66–77. [Google Scholar] [CrossRef]
- Salehi, M.S.; Serajzadeh, S. Simulation of static recrystallization in non-isothermal annealing using a coupled cellular automata and finite element model. Comput. Mater. Sci. 2012, 53, 145–152. [Google Scholar] [CrossRef]
- Svyetlichnyy, D.S. Modelling of the microstructure: From classical cellular automata approach to the frontal one. Comput. Mater. Sci. 2010, 50, 92–97. [Google Scholar] [CrossRef]
- Lin, M.; Prahl, U. A parallelized model for coupled phase field and crystal plasticity simulation. Comput. Methods Mater. Sci. 2016, 16, 156–162. [Google Scholar]
- Millán, E.N.; Bederian, C.S.; Piccoli, M.F.; Garino, C.G.; Bringa, E.M. Performance analysis of Cellular Automata HPC implementations. Comput. Electr. Eng. 2015, 48, 12–24. [Google Scholar] [CrossRef]
- Kolnoochenko, A.; Menshutina, N. CUDA-optimized cellular automata for diffusion limited processes. Comput. Aided Chem. Eng. 2015, 37, 551–556. [Google Scholar] [CrossRef]
- Bernabé, G.; Fernández, R.; García, J.M.; Acacio, M.E.; González, J. An efficient implementation of a 3D wavelet transform based encoder on hyper-threading technology. Parallel Comput. 2007, 33, 54–72. [Google Scholar] [CrossRef] [Green Version]
- Gibson, M.J.; Keedwell, E.C.; Savić, D.A. An investigation of the efficient implementation of cellular automata on multi-core CPU and GPU hardware. J. Parallel Distrib. Comput. 2015, 77, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Innocenti, E.; Silvani, X.; Muzy, A.; Hill, D.R.C. A software framework for fine grain parallelization of cellular models with OpenMP: Application to fire spread. Environ. Model. Softw. 2009, 24, 819–831. [Google Scholar] [CrossRef]
- Millán, E.N.; Martínez, P.; Costa, G.V.G.; Piccoli, M.F.; Printista, A.M.; Bederian, C.; Garino, C.G.; Bringa, E.M. Parallel implementation of a cellular automata in a hybrid CPU/GPU environment. In Proceedings of the XVIII Congreso Argentino de Ciencias de la Computación, Universidad Nacional de La Plata, La Plata, Argentina, 21–25 October 2013; pp. 184–193. [Google Scholar]
- Bandini, S.; Magagnini, M. Parallel Processing Simulation of Dynamic Properties of Filled Rubber Compounds Based On Cellular Automata. Parallel Comput. 2001, 27, 643–661. [Google Scholar] [CrossRef]
- Shterenlikht, A.; Margetts, L.; Cebamanos, L. Modelling fracture in heterogeneous materials on HPC systems using a hybrid MPI/Fortran coarray multi-scale CAFE framework. Adv. Eng. Softw. 2018, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Shterenlikht, A.; Cebamanos, L. MPI vs Fortran coarrays beyond 100k cores: 3D cellular automata. Parallel Comput. 2019, 84, 37–49. [Google Scholar] [CrossRef]
- Kühbach, M.; Barrales-Mora, L.A.; Gottstein, G. A massively parallel cellular automaton for the simulation of recrystallization. Model. Simul. Mater. Sci. Eng. 2014, 22, 075016. [Google Scholar] [CrossRef]
- Sitko, M.; Chao, Q.; Wang, J.; Perzynski, K.; Muszka, K.; Madej, L. A parallel version of the cellular automata static recrystallization model dedicated for high performance computing platforms—Development and verification. Comput. Mater. Sci. 2020, 172, 109283. [Google Scholar] [CrossRef]
- Sitko, M.; Madej, L.; Perzynski, K.; Kwiaton, N.; Kuziak, R.; Radwański, K. Numerical investigation of the static recrystallization inhomogeneities across the plate thickness during continuous annealing. Comput. Methods Mater. Sci. 2015, 15, 371–379. [Google Scholar]
- Zheng, C.; Xiao, N.; Li, D.; Li, Y. Microstructure prediction of the austenite recrystallization during multi-pass steel strip hot rolling: A cellular automaton modeling. Comput. Mater. Sci. 2008, 44, 507–514. [Google Scholar] [CrossRef]
- Han, F.; Tang, B.; Kou, H.; Cheng, L.; Li, J.; Feng, Y. Static recrystallization simulations by coupling cellular automata and crystal plasticity finite element method using a physically based model for nucleation. J. Mater. Sci. 2014, 49, 3253–3267. [Google Scholar] [CrossRef]
- Zhu, G.; Mao, W.; Yu, Y. Calculation of misorientation distribution between recrystallized grains and deformed matrix. Scr. Mater. 1999, 42, 37–41. [Google Scholar] [CrossRef]
- Madej, L.; Sitko, M.; Radwanski, K.; Kuziak, R. Validation and predictions of coupled finite element and cellular automata model: Influence of the degree of deformation on static recrystallization kinetics case study. Mater. Chem. Phys. 2016, 179, 282–294. [Google Scholar] [CrossRef]
- Sieradzki, L.; Madej, L. A perceptive comparison of the cellular automata and Monte Carlo techniques in application to static recrystallization modeling in polycrystalline materials. Comput. Mater. Sci. 2013, 67, 156–173. [Google Scholar] [CrossRef]
- Babu, K.A.; Prithiv, T.S.; Gupta, A.; Mandal, S. Modeling and simulation of dynamic recrystallization in super austenitic stainless steel employing combined cellular automaton, artificial neural network and finite element method. Comput. Mater. Sci. 2021, 195, 110482. [Google Scholar] [CrossRef]
- Cyfronet Krakow: Prometheus Supercomputer. 2021. Available online: https://www.cyfronet.krakow.pl/computers/15226,artykul,prometheus.html (accessed on 20 July 2021).
- Sitko, M.; Mojzeszko, M.; Rychlowski, L.; Cios, G.; Bala, P.; Muszka, K.; Madej, L. Numerical procedure of three-dimensional reconstruction of ferrite-pearlite microstructure data from SEM/EBSD serial sectioning. Procedia Manuf. 2020, 47, 1217–1222. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sitko, M.; Banaś, K.; Madej, L. Scaling Scientific Cellular Automata Microstructure Evolution Model of Static Recrystallization toward Practical Industrial Calculations. Materials 2021, 14, 4082. https://doi.org/10.3390/ma14154082
Sitko M, Banaś K, Madej L. Scaling Scientific Cellular Automata Microstructure Evolution Model of Static Recrystallization toward Practical Industrial Calculations. Materials. 2021; 14(15):4082. https://doi.org/10.3390/ma14154082
Chicago/Turabian StyleSitko, Mateusz, Krzysztof Banaś, and Lukasz Madej. 2021. "Scaling Scientific Cellular Automata Microstructure Evolution Model of Static Recrystallization toward Practical Industrial Calculations" Materials 14, no. 15: 4082. https://doi.org/10.3390/ma14154082





