Advanced Optogenetic-Based Biosensing and Related Biomaterials
Abstract
:1. Use of Engineered Cells for Cell-Based Sensing Platforms
2. Optogenetics in Sensing
2.1. Key Elements Involved in Cell-Based Biosensing
- (a)
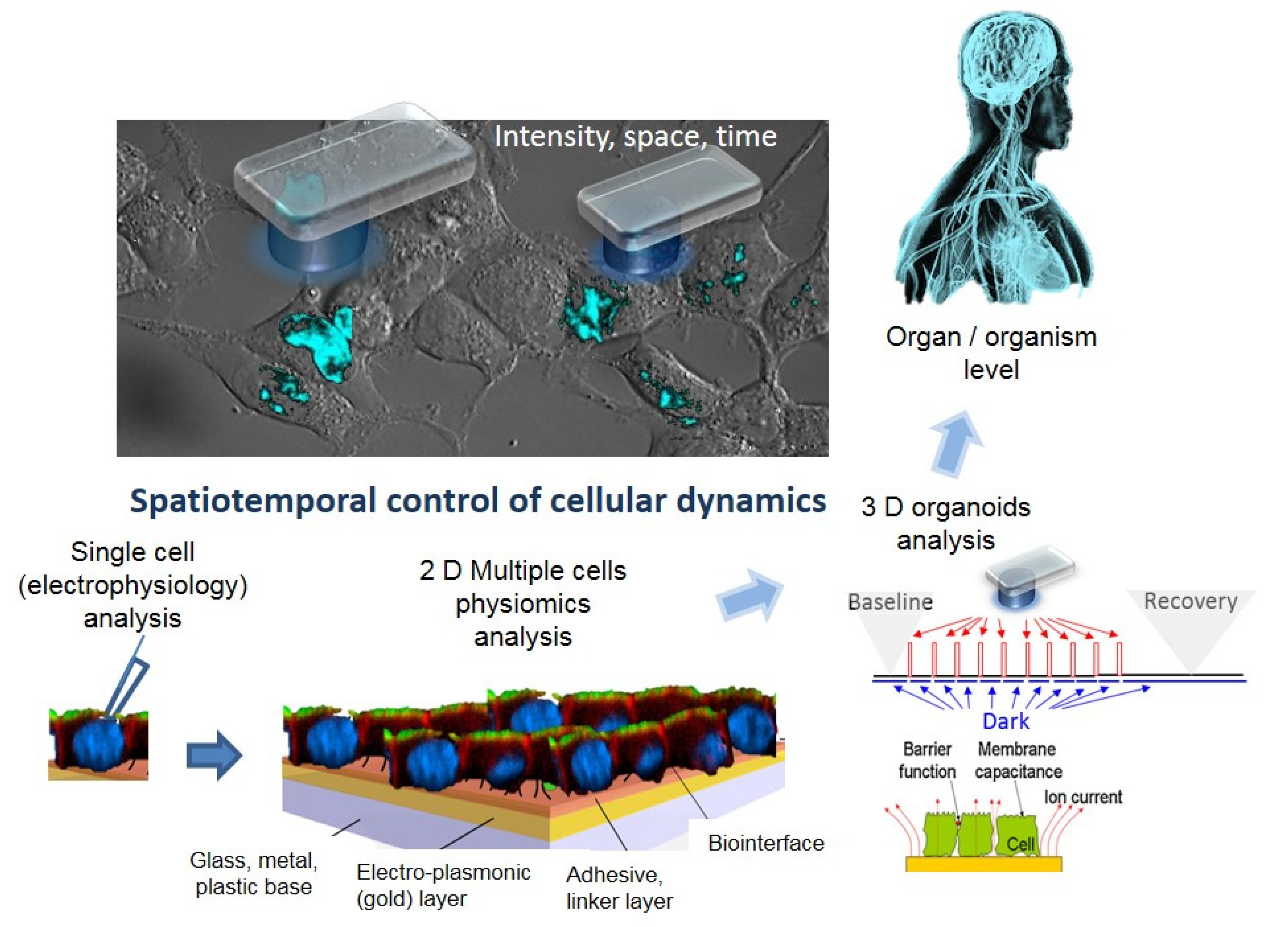
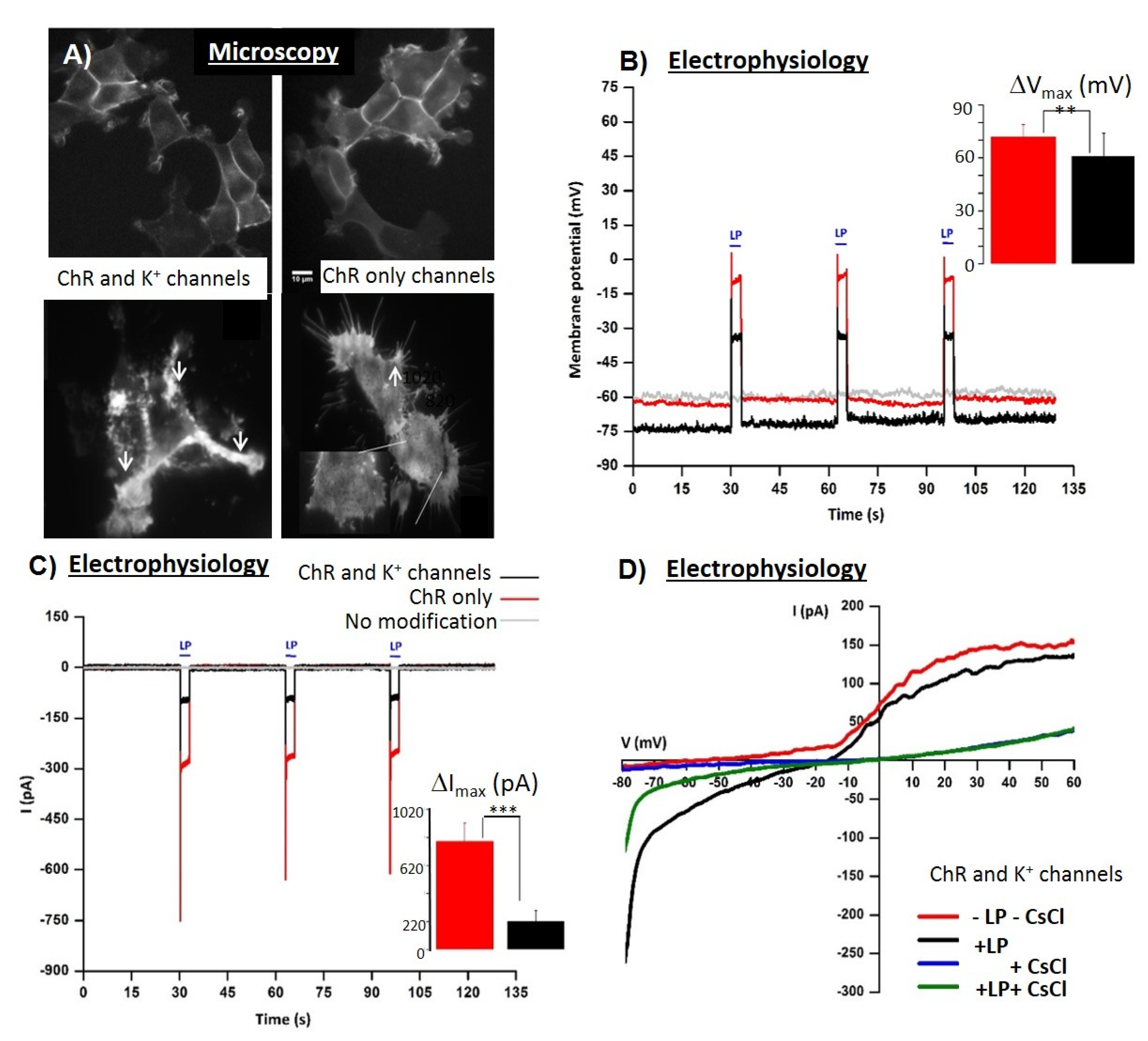
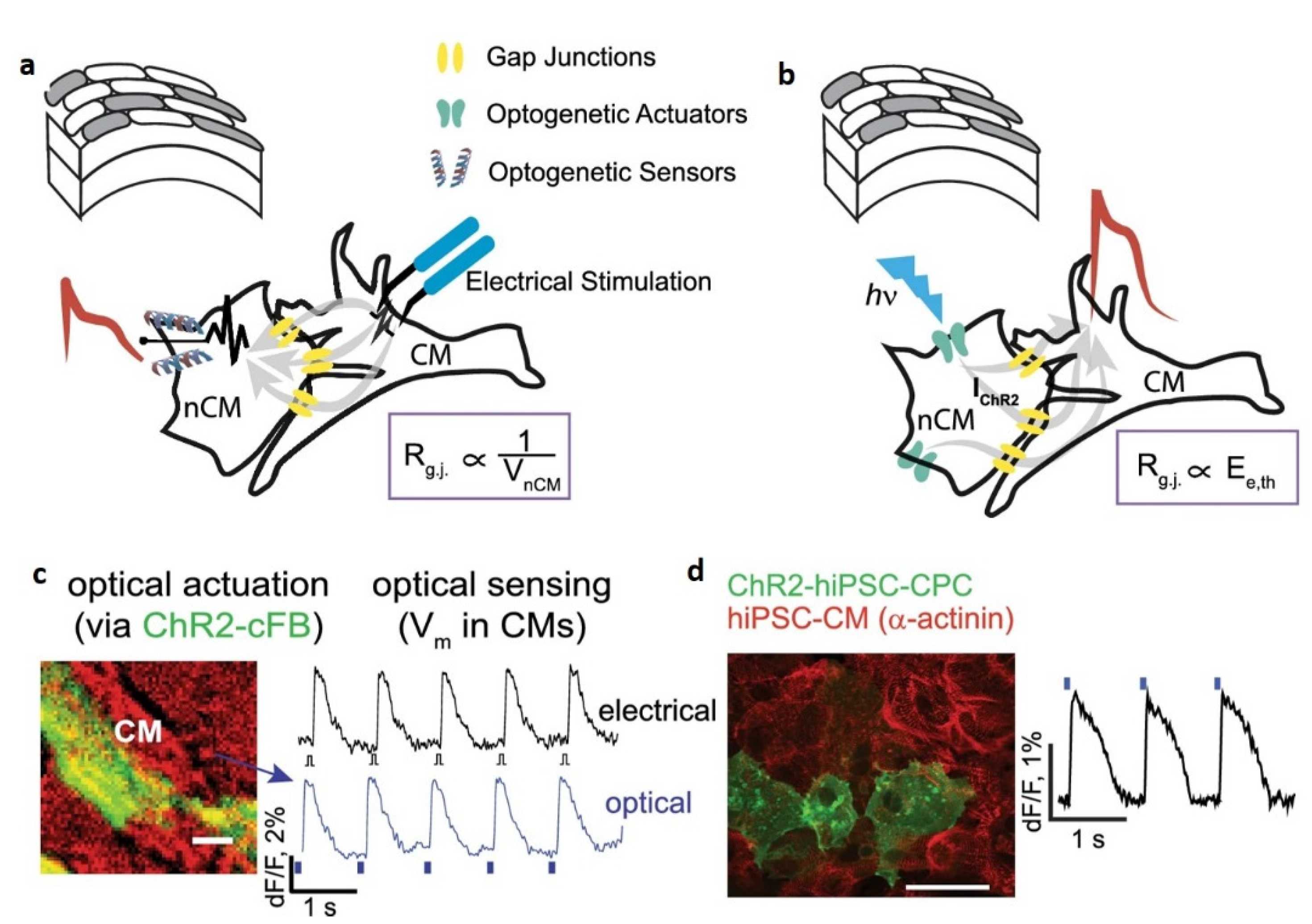
- Active and sensitive control, via light, of cell’s intrinsic dynamic stability, known to play the central role in shaping the response of cells to external perturbation, be it toxic or stimulatory. The selective cell stimulation can be achieved across wide cellular scales and can be combined with electrophysiology or electro-optical assays of cellular status for cell-based sensing platforms, according to Scheme 1. Laser light of specific wavelengths can be used to achieve cellular control spatially restricted to single cells/subcellular volumes, whereas in larger 2D cell sheets, light-emitting diode, LED-based optogenetic stimulation can provide electrical pacing integrable with time-based impedance assays.
- (b)
- Minimally perturbing actuators (as provided by optogenetics toolbox) compatible with live reporters. The reporter cells, i.e., live cells modified with light-sensitive molecules, are the key components of a specialized class of biosensing platforms that highlight the response of cells toward quantitative evaluation of the changes in their microenvironment, including occurrence/presence of bioactive molecules. There is a wide variety of robust excitable/non-excitable cells with tailored light responsiveness and homeostatic control that can be developed and complementary tested (e.g., via electrophysiological tools) and interfaced with electro-analytic assays with integrated controlled microfluidics and optical stimulation.
- (c)
- Fast and affordable electro/optical-analytics amenable for standardization. Indeed, optical methods can be easily implemented and are relatively inexpensive in particular when combined with multiparametric optical readouts of cell physiology within microscopy platforms [29]. Moreover, time-lapse fast impedance assays [25,26] on electrode adherent cells are capable of assessing with exquisite sensitivity the minute changes of cellular state triggered by exposure at bioactive compounds/stimuli and reveal, alone or in combination with optical assays [30,31,32], fast and affordable detection avenues and provide ideal transducers/cell physiomics analysis platforms.
2.2. Advantages and Disadvantages of Cell-Based Biosensors and Optogenetic Approaches
2.3. Envisaged Optogenetics Applications to the Field of Cell-Based Biosensing/Cellular Reporters
- (a)
- Point of Need (PON) quantitative analysis of various stressors’ bio-impact;
- (b)
- Assessment of disease phenotypes and pharmacological modulation for preclinical assays and pharmaceutic industry;
- (c)
- Ingestible/implantable bioelectronic biosensing devices and theranostic platforms;
- (d)
- Development of smart cell sentinels.
3. Generation of Optogenetically Modified Cell Lines and the Complementary Tools for Their Characterization
4. The Optogenetic Toolbox
4.1. The Optogenetic Actuators
4.2. The Optogenetic Reporters
4.3. Optogenetic Control of Intracellular Signals
4.4. (Upcoming) Engineered Opto-Chemogenetic Tools
4.5. Opto-Control of Cell Adhesion and Patterning for Improved Biosensing
5. (Label-Free) Analytical Tools Relevant for Optogenetic Cell Platforms
5.1. Electrical Impedance Sensing (EIS) Platforms
5.2. Combined Electro-Optical Platforms
5.3. Multimodal Functional Imaging for Cell-Based Optogenetic Platforms
6. Wide Biosensing Relevance
- (a)
- Design of novel multifunctionality biosensing probes to allow assessment of stimuli induced, normal or pathological aggregation processes. Many proteins undergo aggregation in vitro and in vivo and, as for amyloid type aggregation, this process is involved in the pathology of many degenerative diseases (e.g., amyloid β42 in Alzheimer’s disease or deposits of amyloid lysozyme fibers on the kidney that are characteristic in patients suffering from familial amyloidosis). It is thus of enormous biosensing/biomedical relevance. Indeed, using a label-free platform [31] integrating improved SPR and impedance assays with cell cultures, we showed continuous, quantitative monitoring of cell monolayer under Amyloidβ42 exposure capable of providing a new perspective on the dynamic processes at various levels within an in vitro cellular system. Kaur. et al. [118] demonstrated the use of optogenetic Amyloidβ to monitor in vivo protein aggregation while fluorescent optogenetic Amyloid-beta was shown to enable discrimination between metabolic and physical damages in neurodegeneration [119] as well in vivo settings.
- (b)
- Development of cell-free systems, as part of the synthetic biology field, to become a critical platform in biological studies [120]. The optogenetic tool has been widely proven as an ideal control switch for protein synthesis due to its nontoxicity and excellent time–space conversion. Zhang et al. [120] used a blue light-regulated two-component system to control cell-free protein synthesis and achieve two-way control: a five-fold dynamic protein expression by blue light repression and three-fold dynamic expression by blue light activation. The cell-free blue light-sensing system was used to perform imaging, light-controlled antibody synthesis and light-triggered artificial cell assembly as a proof of principle expansion of optogenetics tools applications in cell-free synthetic biology.
- (c)
- (d)
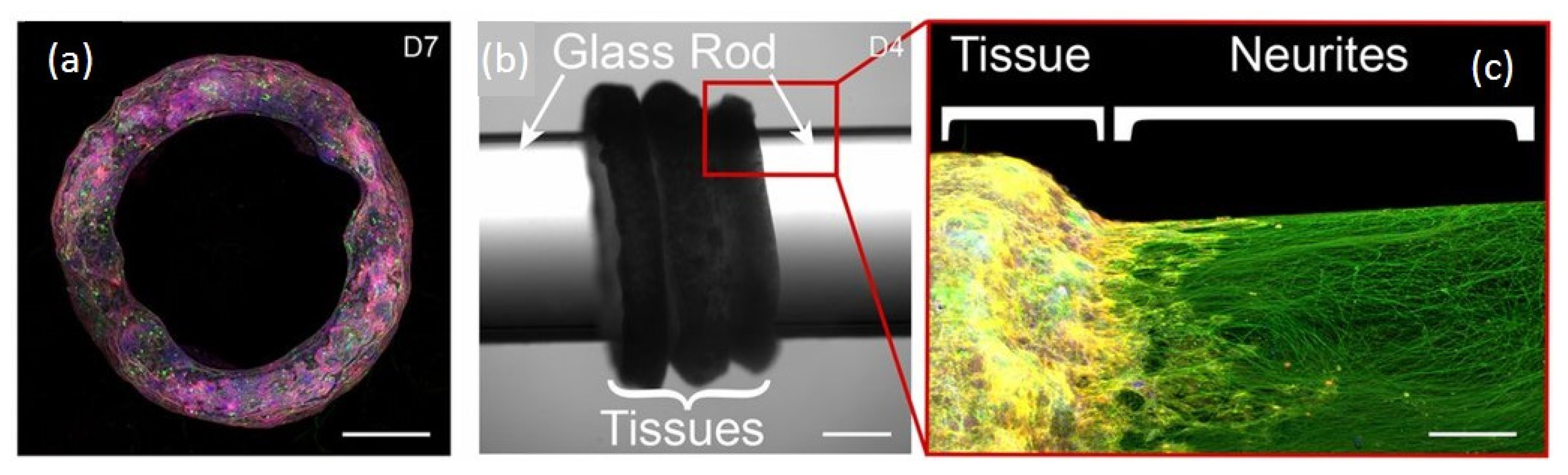
- Optogenetic-inspired tools (optogels) to construct light-responsive extracellular matrix (ECM) mimetic hydrogels better mimicking natural ECM [39] and having light adjustable mechanical properties [38]. Optogels have immediate use in dissecting the cellular response to acute mechanical inputs and are suitable extensions towards 3D cellular biosensing platforms.
- (1)
- Design and engineer synthetic genetically encoded functional nucleic acids FNAs nanostructures and nanodevices [89], extending the traditional biological roles of nucleic acids as catalytic enzymes, intracellular regulatory molecules and carriers of genetic information towards directing the assembly and functionality of materials at the nanoscale. Versatile FNAs-based, light-controlled nanodevices are expected to be broadly used in the near future to probe and program cells and other biological systems (e.g., regulating and compartmentalizing cellular gene expression, imaging, logic operation).
- (2)
- Design of reporter synthetic cells for environmental, nanotechnology, nanomedicine applications. Functionality gains are widespread: from adding targeted mobility of bio-particles [121] and biohybrid swimmers [122] to designer control of optogenetic-enabled biohybrid cellular sentinels [44,45,46,123]. Optogenetics is poised to decode the minimum instruction set required to direct cell behaviors.
- (3)
- Metabolic cybergenetics [47], i.e., use computer interfaces to enable feedback controls over biological processes and engineered metabolic pathways in real-time.
- (4)
- Novel strategies for designing effective and intelligent drug carriers, novel integrated platforms for targeted drug delivery (e.g., photo-responsive polymersomes for drug delivery [48]).
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.E.; Lee, I.C. The Current Trends of Biosensors in Tissue Engineering. Biosensors 2020, 10, 88. [Google Scholar] [CrossRef]
- Lee, J.H.; Luo, J.; Choi, H.K.; Chueng ST, D.; Lee, K.B.; Choi, J.W. Functional nanoarrays for investigating stem cell fate and function. Nanoscale 2020, 12, 9306–9326. [Google Scholar] [CrossRef]
- Gheorghiu, M. A short review on cell-based biosensing: Challenges and breakthroughs in biomedical analysis. J. Biomed Res. 2020, 1–9. [Google Scholar] [CrossRef]
- Gupta, N.; Renugopalakrishnan, V.; Liepmann, D.; Paulmurugan, R.; Malhotra, B.D. Cell-based biosensors: Recent trends, challenges and future perspectives. Biosens. Bioelectron. 2019, 141, 111435. [Google Scholar] [CrossRef]
- Xie, M.; Fussenegger, M. Designing cell function: Assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol. 2018, 19, 507–525. [Google Scholar] [CrossRef]
- Sedlmayer, F.; Aubel, D.; Fussenegger, M. Synthetic gene circuits for the detection, elimination and prevention of disease. Nat. Biomed. Eng. 2018, 2, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Soleymani, J.; Shafiei-Irannejad, V.; Hamblin, M.R.; Hasanzadeh, M.; Somi, M.H.; Jouyban, A. Applications of advanced materials in bio-sensing in live cells: Methods and applications. Mat. Sci. Eng. C-Mater. 2021, 121, 111691. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Umezawa, Y. Cell-Based Biosensor to Visualize Nitric Oxide Release from Living Cells for Toxicity Assessment. Methods Mol. Biol. 2021, 2240, 57–64. [Google Scholar] [PubMed]
- Incaviglia, I.; Frutiger, A.; Blickenstorfer, Y.; Treindl, F.; Ammirati, G.; Lüchtefeld, I.; Reichmuth, A.M.; Dreier, B.; Plückthun, A.; Vörös, J.; et al. An Approach for the Real-Time Quantification of Cytosolic Protein-Protein Interactions in Living Cells. ACS Sens. 2021, 6, 1572–1582. [Google Scholar] [CrossRef]
- Guo, H.; Ji, J.; Sun, J.; Zhang, Y.; Sun, X. Development of a living mammalian cell-based biosensor for the monitoring and evaluation of synergetic toxicity of cadmium and deoxynivalenol. Sci. Total Environ. 2021, 771, 144823. [Google Scholar] [CrossRef] [PubMed]
- Hedayatipour, A.; Aslanzadeh, S.; McFarlane, N. CMOS based whole cell impedance sensing: Challenges and future outlook. Biosens. Bioelectron. 2019, 143, 111600. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, K. Cell-impedance-based label-free technology for the identification of new drugs. Expert Opin. Drug Discov. 2017, 12, 335–343. [Google Scholar] [CrossRef]
- Calabretta, M.M.; Lopreside, A.; Montali, L.; Cevenini, L.; Roda, A.; Michelini, E. A Genetically Encoded Bioluminescence Intracellular Nanosensor for Androgen Receptor Activation Monitoring in 3D Cell Models. Sensors 2021, 21, 893. [Google Scholar] [CrossRef]
- Xu, T.; Kirkpatrick, A.; Toperzer, J.; Ripp, S.; Close, D. Improving Estrogenic Compound Screening Efficiency by Using Self-Modulating, Continuously Bioluminescent Human Cell Bioreporters Expressing a Synthetic Luciferase. Toxicol. Sci. 2019, 168, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Lopreside, A.; Calabretta, M.M.; Montali, L.; Ferri, M.; Tassoni, A.; Branchini, B.R.; Southworth, T.; D’Elia, M.; Roda, A.; Michelini, E. Pret-a-porter nanoYESalpha and nanoYESbeta bioluminescent cell biosensors for ultrarapid and sensitive screening of endocrine-disrupting chemicals. Anal. Bioanal. Chem. 2019, 411, 4937–4949. [Google Scholar] [CrossRef]
- Michelini, E.; Cevenini, L.; Calabretta, M.M.; Calabria, D.; Roda, A. Exploiting in vitro and in vivo bioluminescence for the implementation of the three Rs principle (replacement, reduction, and refinement) in drug discovery. Anal. Bioanal. Chem. 2014, 406, 5531–5539. [Google Scholar] [CrossRef] [PubMed]
- Michelini, E.; Cevenini, L.; Calabretta, M.M.; Spinozzi, S.; Camborata, C.; Roda, A. Field-deployable whole-cell bioluminescent biosensors: So near and yet so far. Anal. Bioanal. Chem. 2013, 405, 6155–6163. [Google Scholar] [CrossRef]
- Michelini, E.; Cevenini, L.; Mezzanotte, L.; Coppa, A.; Roda, A. Cell-based assays: Fuelling drug discovery. Anal. Bioanal. Chem. 2010, 398, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Deisseroth, K. Optogenetics. Nat. Methods 2011, 8, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Gui, Q.; Lawson, T.; Shan, S.; Yan, L.; Liu, Y. The Application of Whole Cell-Based Biosensors for Use in Environmental Analysis and in Medical Diagnostics. Sensors 2017, 17, 1623. [Google Scholar] [CrossRef] [Green Version]
- Miesenbock, G. The optogenetic catechism. Science 2009, 326, 395–399. [Google Scholar] [CrossRef]
- Mattis, J.; Tye, K.M.; Ferenczi, E.A.; Ramakrishnan, C.; O’shea, D.J.; Prakash, R.; Gunaydin, L.A.; Fenno, L.E.; Hyun, M.; Gradinaru, V.; et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat. Methods 2011, 9, 159–172. [Google Scholar] [CrossRef] [Green Version]
- Entcheva, E.; Kay, M.W. Cardiac optogenetics: A decade of enlightenment. Nat. Rev. Cardiol. 2021, 18, 349–367. [Google Scholar] [CrossRef]
- Oh, T.-J.; Fan, H.; Skeeters, S.S.; Zhang, K. Steering Molecular Activity with Optogenetics: Recent Advances and Perspectives. Adv. Biol. 2021, 5, 2000180. [Google Scholar] [CrossRef]
- Gheorghiu, M.; Stanica, L.; Polonschii, C.; David, S.; Ruckenstein, A.; Popescu, O.; Badea, T.; Gheorghiu, E. Modulation of Cellular Reactivity for Enhanced Cell-Based Biosensing. Anal. Chem. 2020, 92, 806–814. [Google Scholar] [CrossRef]
- Gheorghiu, M.; Stănică, L.; Tegla, M.G.G.; Polonschii, C.; Bratu, D.; Popescu, O.; Badea, T.; Gheorghiu, E. Cellular sensing platform with enhanced sensitivity based on optogenetic modulation of cell homeostasis. Biosens. Bioelectron. 2020, 154, 112003. [Google Scholar] [CrossRef] [PubMed]
- Kolar, K.; Knobloch, C.; Stork, H.; Znidaric, M.; Weber, W. OptoBase: A Web Platform for Molecular Optogenetics. ACS Synth. Biol. 2018, 7, 1825–1828. Available online: https://www.optobase.org/switches/ (accessed on 18 June 2021). [CrossRef] [PubMed]
- Forster, D.; Kramer, A.; Baier, H.; Kubo, F. Optogenetic precision toolkit to reveal form, function and connectivity of single neurons. Methods 2018, 150, 42–48. [Google Scholar] [CrossRef]
- Polonschii, C.; Gheorghiu, M.; David, S.; Gáspár, S.; Melinte, S.; Majeed, H.; Kandel, M.E.; Popescu, G.; Gheorghiu, E. High-resolution impedance mapping using electrically activated quantitative phase imaging. Light. Sci. Appl. 2021, 10, 20. [Google Scholar] [CrossRef]
- Gáspár, S.; David, S.; Polonschii, C.; Marcu, I.; Gheorghiu, M.; Gheorghiu, E. Simultaneous impedimetric and amperometric interrogation of renal cells exposed to a calculus-forming salt. Anal. Chim. Acta 2012, 713, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Gheorghiu, M.; David, S.; Polonschii, C.; Olaru, A.; Gaspar, S.; Bajenaru, O.; Popescu, B.O.; Gheorghiu, E. Label free sensing platform for amyloid fibrils effect on living cells. Biosens. Bioelectron. 2014, 52, 89–97. [Google Scholar] [CrossRef]
- Polonschii, C.; David, S.; Gáspár, S.; Gheorghiu, M.; Rosu-Hamzescu, M.; Gheorghiu, E. Complementarity of EIS and SPR to reveal specific and nonspecific binding when interrogating a model bioaffinity sensor; perspective offered by plasmonic based EIS. Anal. Chem. 2014, 86, 8553–8562. [Google Scholar] [CrossRef]
- Williams, L.A.; Joshi, V.; Murphy, M.; Ferrante, J.; Werley, C.A.; Brookings, T.; McManus, O.; Grosse, J.; Davies, C.H.; Dempsey, G.T. Scalable Measurements of Intrinsic Excitability in Human iPS Cell-Derived Excitatory Neurons Using All-Optical Electrophysiology. Neurochem. Res. 2019, 44, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Lapp, H.; Bruegmann, T.; Malan, D.; Friedrichs, S.; Kilgus, C.; Heidsieck, A.; Sasse, P. Frequency-dependent drug screening using optogenetic stimulation of human iPSC-derived cardiomyocytes. Sci. Rep. 2017, 7, 9629. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.; Du, Y.; Zhang, Y.; Wang, Z.; Zhang, D.; He, L.; Tan, W.; Qiu, L.; Jiang, J. Aptamer-based optical manipulation of protein subcellular localization in cells. Nat. Commun. 2020, 11, 1347. [Google Scholar] [CrossRef]
- Mansouri, M.; Lichtenstein, S.; Strittmatter, T.; Buchmann, P.; Fussenegger, M. Construction of a Multiwell Light-Induction Platform for Traceless Control of Gene Expression in Mammalian Cells. Methods Mol. Biol. 2020, 2173, 189–199. [Google Scholar] [PubMed]
- Pagan-Diaz, G.J.; Ramos-Cruz, K.P.; Sam, R.; Kandel, M.E.; Aydin, O.; Saif, M.T.A.; Popescu, G.; Bashir, R. Engineering geometrical 3-dimensional untethered in vitro neural tissue mimic. Proc. Natl. Acad. Sci. USA 2019, 116, 25932–25940. [Google Scholar] [CrossRef] [PubMed]
- Horner, M.; Hoess, P.; Emig, R.; Rebmann, B.; Weber, W. Synthesis of a Light-Controlled Phytochrome-Based Extracellular Matrix with Reversibly Adjustable Mechanical Properties. Methods Mol. Biol. 2020, 2173, 217–231. [Google Scholar]
- Hopkins, E.; Valois, E.; Stull, A.; Le, K.; Pitenis, A.A.; Wilson, M.Z. An Optogenetic Platform to Dynamically Control the Stiffness of Collagen Hydrogels. ACS Biomater. Sci. Eng. 2021, 7, 408–414. [Google Scholar] [CrossRef]
- Choi, M.; Choi, J.W.; Kim, S.; Nizamoglu, S.; Hahn, S.K.; Yun, S.H. Light-guiding hydrogels for cell-based sensing and optogenetic synthesis in vivo. Nat. Photonics 2013, 7, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.F.; Broyles, C.N.; Brook, F.A.; Davies, M.J.; Turtle, C.W.; Nagai, T.; Daniels, M.J. Non-invasive phenotyping and drug testing in single cardiomyocytes or beta-cells by calcium imaging and optogenetics. PLoS ONE 2017, 12, e0174181. [Google Scholar] [CrossRef]
- Hicks, M.; Bachmann, T.T.; Wang, B. Synthetic Biology Enables Programmable Cell-Based Biosensors. Chemphyschem 2020, 21, 132–144. [Google Scholar] [CrossRef] [Green Version]
- Pedone, E.; De Cesare, I.; Zamora-Chimal, C.G.; Haener, D.; Postiglione, L.; La Regina, A.; Marucci, L.; Shannon, B.; Savery, N.J.; Grierson, C.S.; et al. Cheetah: A Computational Toolkit for Cybergenetic Control. ACS Synth. Biol. 2021, 10, 979–989. [Google Scholar] [CrossRef]
- McKay, R.; Hauk, P.; Quan, D.; Bentley, W.E. Development of Cell-Based Sentinels for Nitric Oxide: Ensuring Marker Expression and Unimodality. ACS Synth. Biol. 2018, 7, 1694–1701. [Google Scholar] [CrossRef]
- Young, B.P.; Post, K.L.; Chao, J.T.; Meili, F.; Haas, K.; Loewen, C.J.R. Sentinel interaction mapping—a generic approach for the functional analysis of human disease gene variants using yeast. Dis. Model. Mech. 2020, 13, dmm044560. [Google Scholar] [CrossRef]
- Dixon, T.A.; Williams, T.C.; Pretorius, I.S. Sensing the future of bio-informational engineering. Nat. Commun. 2021, 12, 388. [Google Scholar] [CrossRef]
- Carrasco-Lopez, C.; Garcia-Echauri, S.A.; Kichuk, T.; Avalos, J.L. Optogenetics and biosensors set the stage for metabolic cybergenetics. Curr. Opin. Biotechnol. 2020, 65, 296–309. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, R.; Yang, H.; Bao, C.; Fan, J.; Wang, C.; Lin, Q.; Zhu, L. Light-responsive polymersomes with a charge-switch for targeted drug delivery. J. Mat. Chem. B 2020, 8, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.W.; Han, H.W.; Yang, C.S.; Shrestha, L.K.; Ariga, K.; Hsu, S.H. Optogenetic Modulation and Reprogramming of Bacteriorhodopsin-Transfected Human Fibroblasts on Self-Assembled Fullerene C60 Nanosheets. Adv. Biosyst. 2019, 3, e1800254. [Google Scholar] [CrossRef]
- Reyer, A.; Häßler, M.; Scherzer, S.; Huang, S.; Pedersen, J.T.; Al-Rascheid, K.A.; Bamberg, E.; Palmgren, M.; Dreyer, I.; Nagel, G.; et al. Channelrhodopsin-mediated optogenetics highlights a central role of depolarization-dependent plant proton pumps. Proc. Natl. Acad. Sci. USA 2020, 117, 20920–20925. [Google Scholar] [CrossRef]
- Figueroa, D.; Rojas, V.; Romero, A.; Larrondo, L.F.; Salinas, F. The rise and shine of yeast optogenetics. Yeast 2021, 38, 131–146. [Google Scholar] [CrossRef]
- Marsafari, M.; Ma, J.; Koffas, M.; Xu, P. Genetically-encoded biosensors for analyzing and controlling cellular process in yeast. Curr. Opin. Biotech. 2020, 64, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Chait, R.; Ruess, J.; Bergmiller, T.; Tkacik, G.; Guet, C.C. Shaping bacterial population behavior through computer-interfaced control of individual cells. Nat. Commun. 2017, 8, 1535. [Google Scholar] [CrossRef] [Green Version]
- Boyle, P.M.; Yu, J.; Klimas, A.; Williams, J.C.; Trayanova, N.A.; Entcheva, E. OptoGap is an optogenetics-enabled assay for quantification of cell-cell coupling in multicellular cardiac tissue. Sci. Rep. 2021, 11, 9310. [Google Scholar] [CrossRef]
- Bugaj, L.J.; Choksi, A.T.; Mesuda, C.K.; Kane, R.S.; Schaffer, D.V. Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 2013, 10, 249–252. [Google Scholar] [CrossRef]
- Beiert, T.; Bruegmann, T.; Sasse, P. Optogenetic activation of Gq signalling modulates pacemaker activity of cardiomyocytes. Cardiovasc. Res. 2014, 102, 507–516. [Google Scholar] [CrossRef] [Green Version]
- Goglia, A.G.; Toettcher, J.E. A bright future: Optogenetics to dissect the spatiotemporal control of cell behavior. Curr. Opin. Chem. Biol. 2019, 48, 106–113. [Google Scholar] [CrossRef]
- Dugue, G.P.; Akemann, W.; Knopfel, T. A comprehensive concept of optogenetics. Prog. Brain Res. 2012, 196, 1–28. [Google Scholar] [PubMed]
- Kleinlogel, S.; Feldbauer, K.; Dempski, R.E.; Fotis, H.; Wood, P.G.; Bamann, C.; Bamberg, E. Ultra light-sensitive and fast neuronal activation with the Ca2+-permeable channelrhodopsin CatCh. Nat. Neurosci. 2011, 14, 513–518. [Google Scholar] [CrossRef] [Green Version]
- Williams, D.C.; El Bejjani, R.; Ramirez, P.M.; Coakley, S.; Kim, S.A.; Lee, H.; Hammarlund, M.; Wen, Q.; Samuel, A.; Lu, H.; et al. Rapid and permanent neuronal inactivation in vivo via subcellular generation of reactive oxygen with the use of KillerRed. Cell Rep. 2013, 5, 553–563. [Google Scholar] [CrossRef] [Green Version]
- Wegener, J. Cell-Based Microarrays for In Vitro Toxicology. Annu. Rev. Anal. Chem. 2015, 8, 335–358. [Google Scholar] [CrossRef] [Green Version]
- Atienzar, F.A.; Gerets, H.; Tilmant, K.; Toussaint, G.; Dhalluin, S. Evaluation of impedance-based label-free technology as a tool for pharmacology and toxicology investigations. Biosensors 2013, 3, 132–156. [Google Scholar] [CrossRef] [PubMed]
- Novellino, A.; Scelfo, B.; Palosaari, T.; Price, A.; Sobanski, T.; Shafer, T.J.; Whelan, M.; Johnstone, A.F.M.; Gross, J.W.; Gramowski, A.; et al. Development of micro-electrode array based tests for neurotoxicity: Assessment of interlaboratory reproducibility with neuroactive chemicals. Front. Neuroeng. 2011, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- Muller, K.; Weber, W. Optogenetic tools for mammalian systems. Mol. Biosyst. 2013, 9, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Provencio, I.; Jiang, G.; De Grip, W.J.; Hayes, W.P.; Rollag, M.D. Melanopsin: An opsin in melanophores, brain, and eye. Proc. Natl. Acad. Sci. USA 1998, 95, 340–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Shen, Y.; Campbell, R.E. Engineering Photosensory Modules of Non-Opsin-Based Optogenetic Actuators. Int. J. Mol. Sci. 2020, 21, 6522. [Google Scholar] [CrossRef] [PubMed]
- Pudasaini, A.; El-Arab, K.K.; Zoltowski, B.D. LOV-based optogenetic devices: Light-driven modules to impart photoregulated control of cellular signaling. Front. Mol. Biosci. 2015, 2, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Agulhon, C.; Schmidt, E.; Oheim, M.; Ropert, N. New tools for investigating astrocyte-to-neuron communication. Front. Cell. Neurosci. 2013, 7, 193. [Google Scholar] [CrossRef] [Green Version]
- Wagner, T.R.; Rothbauer, U. Nanobodies Right in the Middle: Intrabodies as Toolbox to Visualize and Modulate Antigens in the Living Cell. Biomolecules 2020, 10, 1701. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kang, M.; Choo, Y.W.; Go, S.H.; Kwon, S.P.; Song, S.Y.; Soh, H.S.; Hong, J.; Kim, B.-S. Immunomodulatory Lipocomplex Functionalized with Photosensitizer-Embedded Cancer Cell Membrane Inhibits Tumor Growth and Metastasis. Nano Lett. 2019, 19, 5185–5193. [Google Scholar] [CrossRef]
- Micheletto, M.C.; Guidelli, E.J.; Costa-Filho, A.J. Interaction of Genetically Encoded Photosensitizers with Scintillating Nanoparticles for X-ray Activated Photodynamic Therapy. ACS Appl. Mater. Interfaces 2021, 13, 2289–2302. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, E.C.; Mehta, S.; Zhang, J. Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem. Rev. 2018, 118, 11707–11794. [Google Scholar] [CrossRef]
- Alford, S.C.; Wu, J.; Zhao, Y.; Campbell, R.E.; Knopfel, T. Optogenetic reporters. Biol. Cell 2013, 105, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Herron, T.J.; Lee, P.; Jalife, J. Optical imaging of voltage and calcium in cardiac cells & tissues. Circ. Res. 2012, 110, 609–623. [Google Scholar] [PubMed] [Green Version]
- Sriram, K.; Insel, P.A. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol. Pharmacol. 2018, 93, 251–258. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.H.; Venkatachalam, V.; Cohen, A.E. Temporal dynamics of microbial rhodopsin fluorescence reports absolute membrane voltage. Biophys. J. 2014, 106, 639–648. [Google Scholar] [CrossRef] [Green Version]
- Spudich, J.L. A molecular voltmeter based on fluorescence dynamics. Biophys. J. 2014, 106, 497–499. [Google Scholar] [CrossRef] [Green Version]
- Quinn, T.A.; Camelliti, P.; Rog-Zielinska, E.A.; Siedlecka, U.; Poggioli, T.; O’Toole, E.T.; Knöpfel, T.; Kohl, P. Electrotonic coupling of excitable and nonexcitable cells in the heart revealed by optogenetics. Proc. Natl. Acad. Sci. USA 2016, 113, 14852–14857. [Google Scholar] [CrossRef] [Green Version]
- Klimas, A.; Ambrosi, C.M.; Yu, J.; Williams, J.C.; Bien, H.; Entcheva, E. OptoDyCE as an automated system for high-throughput all-optical dynamic cardiac electrophysiology. Nat. Commun. 2016, 7, 11542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.X.; Chung, H.K.; Lam, A.J.; Lin, M.Z. Optical control of protein activity by fluorescent protein domains. Science 2012, 338, 810–814. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, P.R.; Gautam, N. Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol. Biol. Cell 2014, 25, 2305–2314. [Google Scholar] [CrossRef]
- Deng, W.; Bates, J.A.; Wei, H.; Bartoschek, M.D.; Conradt, B.; Leonhardt, H. Tunable light and drug induced depletion of target proteins. Nat. Commun. 2020, 11, 304. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Lee, H.; Hong, J.; Jung, H.; Jo, Y.; Oh, B.-H.; Park, B.O.; Heo, W.D. Optogenetic activation of intracellular antibodies for direct modulation of endogenous proteins. Nat. Methods 2019, 16, 1095–1100. [Google Scholar] [CrossRef]
- Goto, Y.; Kondo, Y.; Aoki, K. Visualization and Manipulation of Intracellular Signaling. Adv. Exp. Med. Biol. 2021, 1293, 225–234. [Google Scholar]
- He, L.; Jing, J.; Zhu, L.; Tan, P.; Ma, G.; Zhang, Q.; Nguyen, N.T.; Wang, J.; Zhou, Y.; Huang, Y. Optical control of membrane tethering and interorganellar communication at nanoscales. Chem. Sci. 2017, 8, 5275–5281. [Google Scholar] [CrossRef] [Green Version]
- Hansen, J.N.; Kaiser, F.; Klausen, C.; Stüven, B.; Chong, R.; Bönigk, W.; Mick, D.U.; Möglich, A.; Jurisch-Yaksi, N.; Schmidt, F.I.; et al. Nanobody-directed targeting of optogenetic tools to study signaling in the primary cilium. eLife 2020, 9, e57907. [Google Scholar] [CrossRef]
- Kwon, E.; Heo, W.D. Optogenetic tools for dissecting complex intracellular signaling pathways. Biochem. Biophys. Res. Commun. 2020, 527, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; Lee, S.; Yu, J.; Kim, N.; Won, S.S.; Park, H.; Do Heo, W. Optogenetic control of mRNA localization and translation in live cells. Nat. Cell Biol. 2020, 22, 341–352. [Google Scholar] [CrossRef]
- Yu, Q.; Ren, K.; You, M. Genetically encoded RNA nanodevices for cellular imaging and regulation. Nanoscale 2021, 13, 7988–8003. [Google Scholar] [CrossRef]
- Van Geel, O.; Cheung, S.; Gadella TW, J. Combining optogenetics with sensitive FRET imaging to monitor local microtubule manipulations. Sci. Rep. 2020, 10, 6034. [Google Scholar] [CrossRef]
- Beyer, H.M.; Naumann, S.; Weber, W.; Radziwill, G. Optogenetic control of signaling in mammalian cells. Biotechnol. J. 2015, 10, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, L. Optogenetic Tools for Manipulating Protein Subcellular Localization and Intracellular Signaling at Organelle Contact Sites. Curr. Protoc. 2021, 1, e71. [Google Scholar] [CrossRef]
- Menard, C.; Russo, S.J. Non-invasive chemogenetics. Nat. Biomed. Eng. 2018, 2, 467–468. [Google Scholar] [CrossRef]
- Vogt, N. Potent chemogenetics. Nat. Methods 2019, 16, 363. [Google Scholar] [CrossRef]
- Berglund, K.; Clissold, K.; Li, H.E.; Wen, L.; Park, S.Y.; Gleixner, J.; Klein, M.E.; Lu, D.; Barter, J.W.; Rossi, M.A.; et al. Luminopsins integrate opto- and chemogenetics by using physical and biological light sources for opsin activation. Proc. Natl. Acad. Sci. USA 2016, 113, E358–E367. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Cumberbatch, D.; Centanni, S.; Shi, S.Q.; Winder, D.; Webb, D.; Johnson, C.H. Coupling optogenetic stimulation with NanoLuc-based luminescence (BRET) Ca(++) sensing. Nat. Commun. 2016, 7, 13268. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.Z.; Lampson, M.A.; Chenoweth, D.M. Photoactivatable trimethoprim-based probes for spatiotemporal control of biological processes. Method Enzymol. 2020, 638, 273–294. [Google Scholar]
- Asphahani, F.; Thein, M.; Veiseh, O.; Edmondson, D.; Kosai, R.; Veiseh, M.; Xu, J.; Zhang, M. Influence of cell adhesion and spreading on impedance characteristics of cell-based sensors. Biosens. Bioelectron. 2008, 23, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Bondarenko, A.; Cortés-Salazar, F.; Gheorghiu, M.; Gáspár, S.; Momotenko, D.; Stanica, L.; Lesch, A.; Gheorghiu, E.; Girault, H.H. Electrochemical push-pull probe: From scanning electrochemical microscopy to multimodal altering of cell microenvironment. Anal. Chem. 2015, 87, 4479–4486. [Google Scholar] [CrossRef] [Green Version]
- Munteanu, R.E.; Stanica, L.; Gheorghiu, M.; Gaspar, S. Measurement of the Extracellular pH of Adherently Growing Mammalian Cells with High Spatial Resolution Using a Voltammetric pH Microsensor. Anal. Chem. 2018, 90, 6899–6905. [Google Scholar] [CrossRef]
- Rosu-Hamzescu, M.; Polonschii, C.; Oprea, S.; Popescu, D.; David, S.; Bratu, D.; Gheorghiu, E. High speed CMOS acquisition system based on FPGA embedded image processing for electro-optical measurements. Rev. Sci. Instrum. 2018, 89, 065103. [Google Scholar] [CrossRef] [PubMed]
- Munteanu, R.-E.; Ye, R.; Polonschii, C.; Ruff, A.; Gheorghiu, M.; Gheorghiu, E.; Boukherroub, R.; Schuhmann, W.; Melinte, S.; Gáspár, S. High spatial resolution electrochemical biosensing using reflected light microscopy. Sci. Rep. 2019, 9, 15196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gheorghiu, M.; Gersing, E.; Gheorghiu, E. Quantitative analysis of impedance spectra of organs during ischemia. Ann. NY Acad. Sci. 1999, 873, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.J.; Cramer, L.P. Actin-based cell motility and cell locomotion. Cell 1996, 84, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Germain, R.N.; Robey, E.A.; Cahalan, M.D. A decade of imaging cellular motility and interaction dynamics in the immune system. Science 2012, 336, 1676–1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanica, L.; Gheorghiu, M.; Stan, M.; Polonschii, C.; David, S.; Bratu, D.; Dinischiotu, A.; Supuran, C.T.; Gheorghiu, E. Quantitative assessment of specific carbonic anhydrase inhibitors effect on hypoxic cells using electrical impedance assays. J. Enzyme Inhib. Med. Chem. 2017, 32, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Kim, P.H.; Choi, J.-W.; Oh-Joon, K.; Kim, K.; Kim, D.; Yun, C.-O.; Yoo, K.-H. Capacitance-based real time monitoring of receptor-mediated endocytosis. Biosens. Bioelectron. 2010, 25, 1325–1332. [Google Scholar] [CrossRef]
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121. [Google Scholar] [CrossRef] [PubMed]
- Gheorghiu, M.; Enciu, A.M.; Popescu, B.O.; Gheorghiu, E. Functional and molecular characterization of the effect of amyloid-beta42 on an in vitro epithelial barrier model. J. Alzheimer’s Dis. 2014, 38, 787–798. [Google Scholar] [CrossRef]
- Inda, M.E.; Mimee, M.; Lu, T.K. Cell-based biosensors for immunology, inflammation, and allergy. J. Allergy Clin. Immunol. 2019, 144, 645–647. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Guo, H.; Sun, X. Recent progress on cell-based biosensors for analysis of food safety and quality control. Biosens. Bioelectron. 2019, 126, 389–404. [Google Scholar] [CrossRef]
- Klimas, A.; Ortiz, G.; Boggess, S.C.; Miller, E.W.; Entcheva, E. Multimodal on-axis platform for all-optical electrophysiology with near-infrared probes in human stem-cell-derived cardiomyocytes. Prog. Biophys. Mol. Biol. 2020, 154, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Kandel, M.E.; He, Y.R.; Lee, Y.J.; Chen, T.H.-Y.; Sullivan, K.M.; Aydin, O.; Saif, M.T.A.; Kong, H.; Sobh, N.; Popescu, G. Phase imaging with computational specificity (PICS) for measuring dry mass changes in sub-cellular compartments. Nat. Commun. 2020, 11, 6256. [Google Scholar] [CrossRef]
- Kandel, M.E.; Kim, E.; Lee, Y.J.; Tracy, G.; Chung, H.J.; Popescu, G. Multiscale Assay of Unlabeled Neurite Dynamics Using Phase Imaging with Computational Specificity. ACS Sens. 2021, 6, 1864–1874. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chen, X.; Qian, Y.; Shao, J.; Li, X.; Liu, S.; Zhu, L.; Zhao, Y.; Ye, H.; Yang, Y. A synthetic BRET-based optogenetic device for pulsatile transgene expression enabling glucose homeostasis in mice. Nat. Commun. 2021, 12, 615. [Google Scholar] [CrossRef]
- Denelavas, A.; Weibel, F.; Prummer, M.; Imbach, A.; Clerc, R.G.; Apfel, C.M.; Hertel, C. Real-time cellular impedance measurements detect Ca(2+) channel-dependent oscillations of morphology in human H295R adrenoma cells. Biochim. Biophys. Acta (BBA) 2011, 1813, 754–762. [Google Scholar] [CrossRef] [Green Version]
- Venturelli, L.; Kohler, A.; Stupar, P.; Villalba, M.I.; Kalauzi, A.; Radotic, K.; Bertacchi, M.; Dinarelli, S.; Girasole, M.; Pešić, M.; et al. A perspective view on the nanomotion detection of living organisms and its features. J. Mol. Recognit. 2020, 33, e2849. [Google Scholar] [CrossRef]
- Kaur, P.; Kibat, C.; Teo, E.; Gruber, J.; Mathuru, A.; Tolwinski, N.S. Use of Optogenetic Amyloid-beta to Monitor Protein Aggregation in Drosophila melanogaster, Danio rerio and Caenorhabditis elegans. Bio-Protocol. 2020, 10, e3856. [Google Scholar] [CrossRef]
- Lim, C.H.; Kaur, P.; Teo, E.; Lam VY, M.; Zhu, F.; Kibat, C.; Tolwinski, N.S.; Gruber, J.; Mathuru, A.S.; Tolwinski, N.S.; et al. Application of optogenetic Amyloid-beta distinguishes between metabolic and physical damages in neurodegeneration. eLife 2020, 9, e52589. [Google Scholar] [CrossRef]
- Zhang, P.; Yang, J.; Cho, E.; Lu, Y. Bringing Light into Cell-Free Expression. ACS Synth. Biol. 2020, 9, 2144–2153. [Google Scholar] [CrossRef]
- Bunea, A.I.; Pavel, I.A.; David, S.; Gaspar, S. Sensing based on the motion of enzyme-modified nanorods. Biosens. Bioelectron 2015, 67, 42–48. [Google Scholar] [CrossRef]
- Aydin, O.; Zhang, X.; Nuethong, S.; Pagan-Diaz, G.J.; Bashir, R.; Gazzola, M.; Saif, M.T.A. Neuromuscular actuation of biohybrid motile bots. Proc. Natl. Acad. Sci. USA 2019, 116, 19841–19847. [Google Scholar] [CrossRef] [Green Version]
- Kojima, R.; Aubel, D.; Fussenegger, M. Toward a world of theranostic medication: Programming biological sentinel systems for therapeutic intervention. Adv. Drug Deliv. Rev. 2016, 105, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cheng, X.; Zhao, Y.; Yang, Y. Lighting up Live-Cell and In Vivo Central Carbon Metabolism with Genetically Encoded Fluorescent Sensors. Annu. Rev. Anal. Chem. 2020, 13, 293–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hongdusit, A.; Zwart, P.H.; Sankaran, B.; Fox, J.M. Minimally disruptive optical control of protein tyrosine phosphatase 1B. Nat. Commun. 2020, 11, 788. [Google Scholar] [CrossRef] [Green Version]
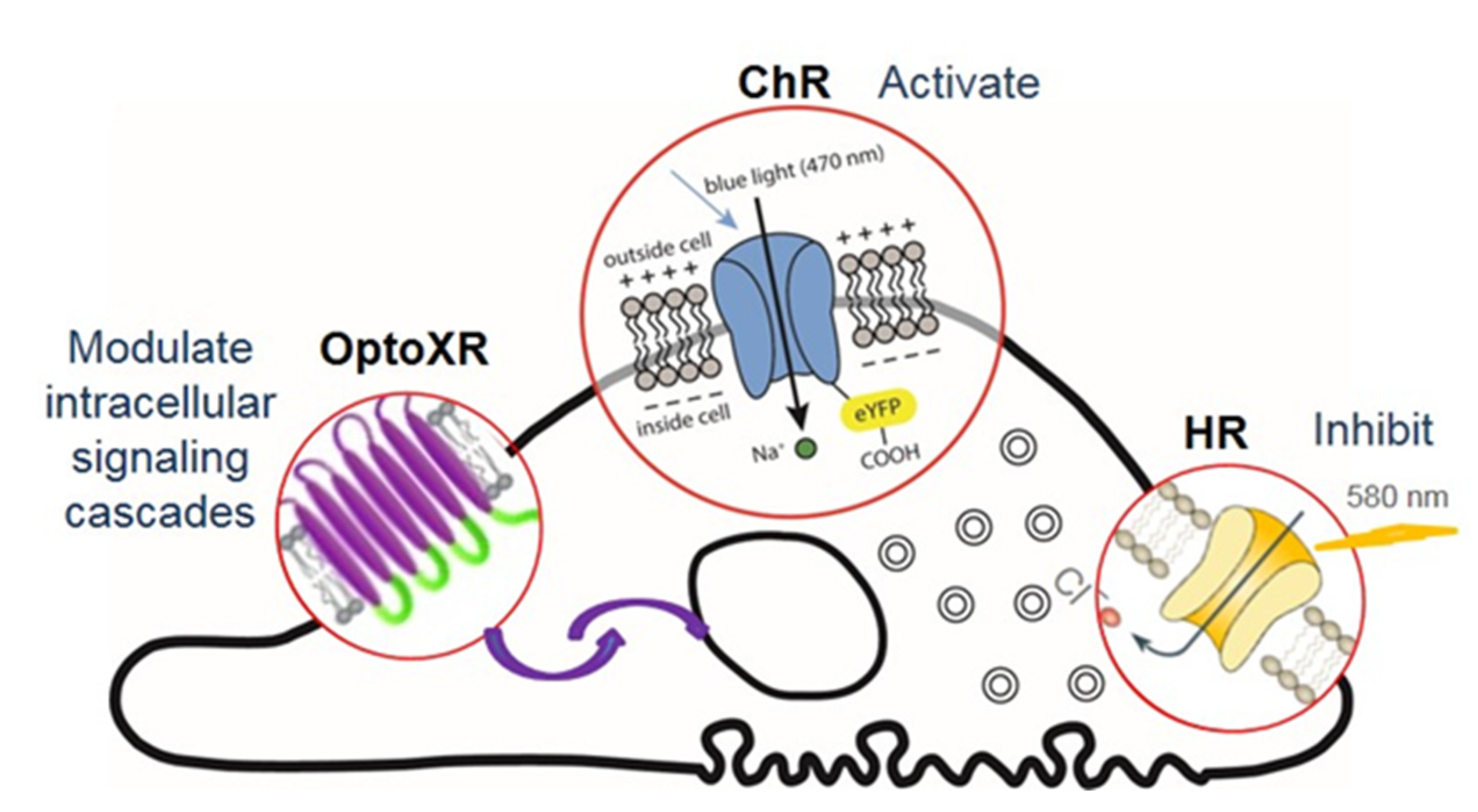
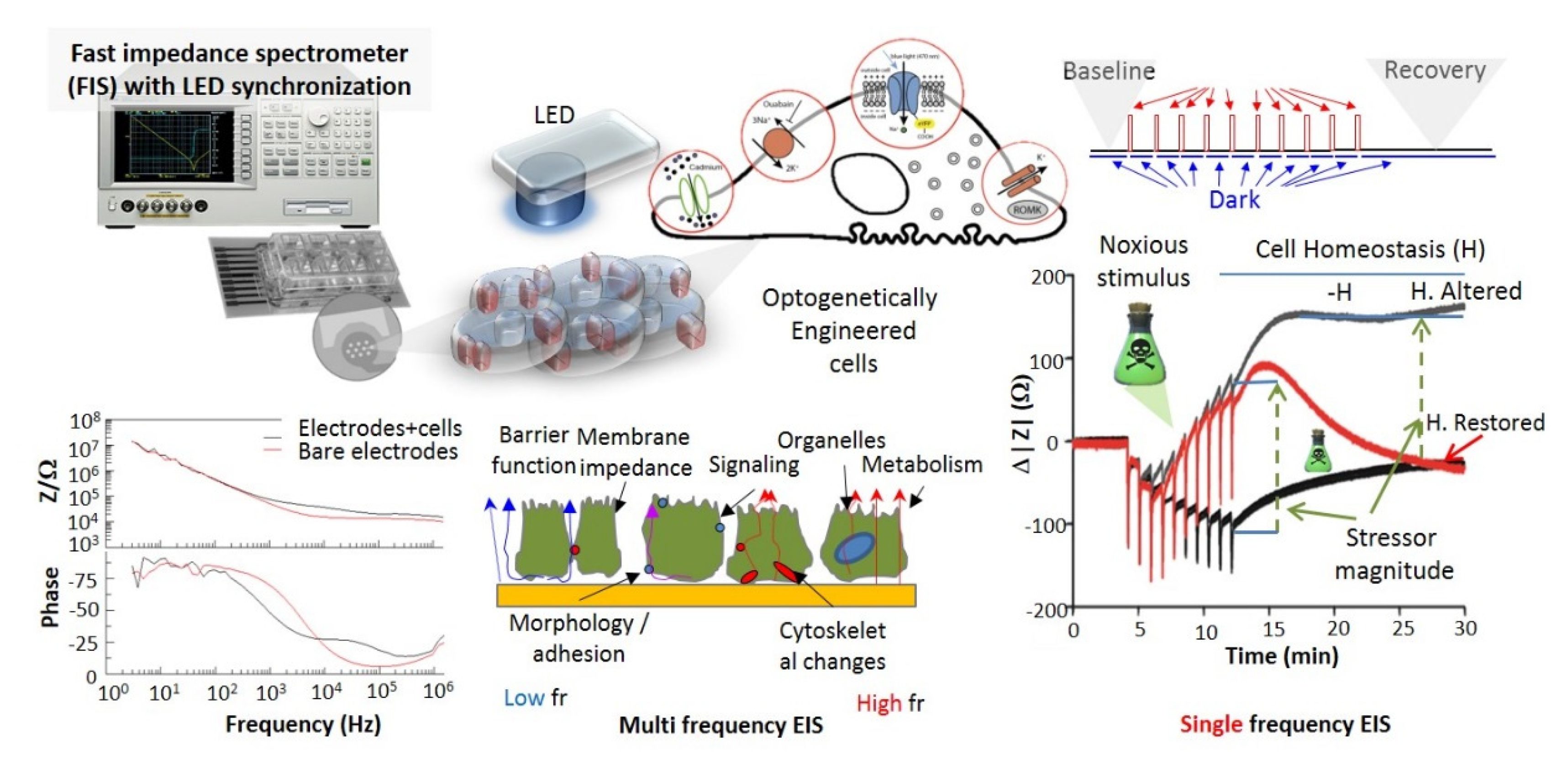
| Field | Status | Reference |
|---|---|---|
| PON stressor’s bioimpact analysis | Demonstrated and validated proof of concept for an environmental toxicant | [25,26] |
| Assessment of disease phenotypes and pharmacological modulation | Demonstrated proof of concepts mostly in cardiac research | [33,34] |
| Design of novel multifunctionality biosensing probes for stimuli induced, normal or pathological aggregation processes | Demonstrated for a fibrillar protein of biomedical relevance | [35] |
| Reference platforms development | Multiwell Light-Induction Platform | [36] |
| Design of better cell matrices and 3D constructs for bioanalysis | 3D tissue mimic reported; bioanalysis not demonstrated | [37] |
| Demonstrated for extracellular matrices | [38] | |
| Demonstrated for hydrogels | [39,40] | |
| Ingestible/implantable bioelectronic biosensing devices | Potential | [41,42,43] |
| Theranostics (sense and respond)-oriented cell sentinels | Potential-relevant (connected) progress | [44,45,46,47] |
| Synthetic cells | Emergent | [48] |
| Target Cells | Optogenetic Molecule | Application | Reference |
|---|---|---|---|
| hIPSC-derived excitatory neurons | CheRiff—voltage actuator QuasAr—voltage reporter | All-Optical Electrophysiology for measurement of intrinsic excitability | [33] |
| hIPSC-derived cardiomyocytes | ChR2—channel | Frequency-dependent drug screening, high content cardiac toxicity screening or personalized medicine for inherited cardiac channelopathies | [34] |
| Human fibroblasts | (HE)bacteriorhodopsin—a pump | Optogenetic Modulation and Reprogramming on fullerene C60 nanosheets | [49] |
| Human embryonic kidney cells HEK * 293 FLPN—2D monolayers | ChR2 alone or with K+ channel | Optogenetic pacing combined with fast electrical impedance assay for quantification of Cd and ouabain | [25,26] |
| HEK 293 T | ChR2 | ‘tandem-cell-unit’ method of optogenetic stimulation | [54] |
| Cryptochrome 2 (Cry2, fused to mCherry (Cry2-mCh) | Tunable control of protein oligomerization and control of intracellular signaling cascades | [55] | |
| Mouse embryonic stem cells in rod and toroidal fibrin scaffolds | ChR2H134R | Untethered and geometrically stable, functional optogenetic Neural tissue mimics | [37] |
| Mouse Embryonic stem cells | Melanopsin | Cardiomyocytes (embryoid bodies) with Optogenetic activation of Gq signaling | [56] |
| Neurons | Various opto tools | Ultra light-sensitive and fast neuronal activation | [57,58] |
| Ca2+-permeable channelrhodopsin CatCh | [59] | ||
| KillerRed | Permanent inactivation of selected neurons—in Caenorhabditis elegans | [60] | |
| Cardiomyocytes | ChETATC /R-GECO | Noninvasive phenotyping Drug testing | [41] |
| Beta cells | ChETATC /R-GECO | Noninvasive phenotyping | [41] |
| Plant cells (mesophyll) | ChR2—channel actuator | Optogenetics stimulation combined with voltage-sensing microelectrodes for assessment of stress-associated physiological responses | [50] |
| Yeasts | Various optogenetic switches (light-controlled on–off gene expression systems) | Optogenetic control of metabolic pathways, heterologous protein production and flocculation | [51] (review) |
| Yeasts | GPCR-based biosensors Optogenetic switches | Directing protein assembly and controlling metabolic fluxes | [52] (review) |
| Bacteria Escherichia coli | Light-switchable (red/far-red and green/red) photo-reversible, two-component signal transduction systems | Hybrid oscillators for detection of sub-inhibitory antibiotic concentrations-generated cell behavior | [53] |
| Optogenetic Toolbox Component | Property | Wavelength | Effect | Reference |
|---|---|---|---|---|
| Actuators | Opsins | 470/570 | Membrane polarisation | [23,56,59,65] |
| Channelrhodopsin Ch | Na+ channel | 470 nm | Membrane depolarization via internal surface potential shift to positive values | [23] |
| Calcium translocating channelrhodopsin CatCh | Ca2+-permeable channelrhodopsin | 473 nm | Membrane depolarization via internal surface potential shift to positive values | [59] |
| Halorhodopsin HR | Cl− ion pump | 570–590 nm | Membrane hyperpolarization | [65] |
| Archaerhodopsin-3 Arch | Cl− ion pump | 570 nm | Membrane hyperpolarization | [23] |
| Melanopsin | Activation of Gq | 480 nm | Membrane depolarization through GPCR signaling cascade | [56] |
| OptoXR Opsin/G protein- coupled receptor chimeric molecules | Activation of Gq | 470 nm | Merge an extracellular light sensitive part and an intracellular G protein-coupled receptor to initiate a signaling cascade upon activation | [56] |
| Actuators | Non-opsin-based | various | various | [66,67] |
| Light-oxygen-voltage-sensing domain (LOV) cryptochrome (CRY2) phytochrome (PhyB and BphP) | various | various | various | [66] |
| fluorescent protein (FP)-based photosensitive domains (Dronpa and PhoCl) | various | various | various | [67] |
| Light activated enzyme—killer red | various | various | ROS generation | [66] |
| Sensors | various | various | various | [41,68,69] |
| Optogenetic Ca2+ indicator probes (e.g., red calcium indicator protein, R-GECO) | various | various | Ca2+ indicator probes of G protein-coupled receptors activation | [41] |
| various | various | Ca2+ indicator probes of functional cell–cell communication | [68] | |
| Traceable intracellular binding molecules—Intrabodies | various | various | Antibody fragments of heavy-chain only antibodies of camelids (nanobodies) to visualize bioactive antigens | [69] |
| Opto-switches | various | Various excitation/reversion wavelengths | The transfer of biochemical information from sensor domain to the actuator domain is mediated by conformational changes and aggregation processes that are able to be switched on and off | [27] and https://www.optobase.org/switches [27] |
| UV receptors | receptors | 300nm/dark | Heterodimerization/homodimerization, dissociation | [27] |
| Cyanobacteriochromes CcaS/CcaR | photoreceptor | 535/670 | Gene expression | [27] |
| BLUF domains bPAC (BlaC) | - | 450/dark | cAMP production | [27] |
| LOV (light, oxygen, voltage) domains | various | various | various | [27,66,67] |
| Cryptochromes | various | various | various | [27,66] |
| Fluorescent proteins (Dronpa, PhoCl, PYP) | various | various | /photocleavage/ | [27,66,67] |
| Phytochromes | PhyB | 660/740 | heterodimerization | [66,67] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gheorghiu, M.; Polonschii, C.; Popescu, O.; Gheorghiu, E. Advanced Optogenetic-Based Biosensing and Related Biomaterials. Materials 2021, 14, 4151. https://doi.org/10.3390/ma14154151
Gheorghiu M, Polonschii C, Popescu O, Gheorghiu E. Advanced Optogenetic-Based Biosensing and Related Biomaterials. Materials. 2021; 14(15):4151. https://doi.org/10.3390/ma14154151
Chicago/Turabian StyleGheorghiu, Mihaela, Cristina Polonschii, Octavian Popescu, and Eugen Gheorghiu. 2021. "Advanced Optogenetic-Based Biosensing and Related Biomaterials" Materials 14, no. 15: 4151. https://doi.org/10.3390/ma14154151









