Interactions of Aminopropyl–Azithromycin Derivatives, Precursors in the Synthesis of Bioactive Macrozones, with E. coli Ribosome: NMR and Docking Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antibacterial Testing
2.2. NMR
2.3. Molecular Docking
3. Results and Discussion
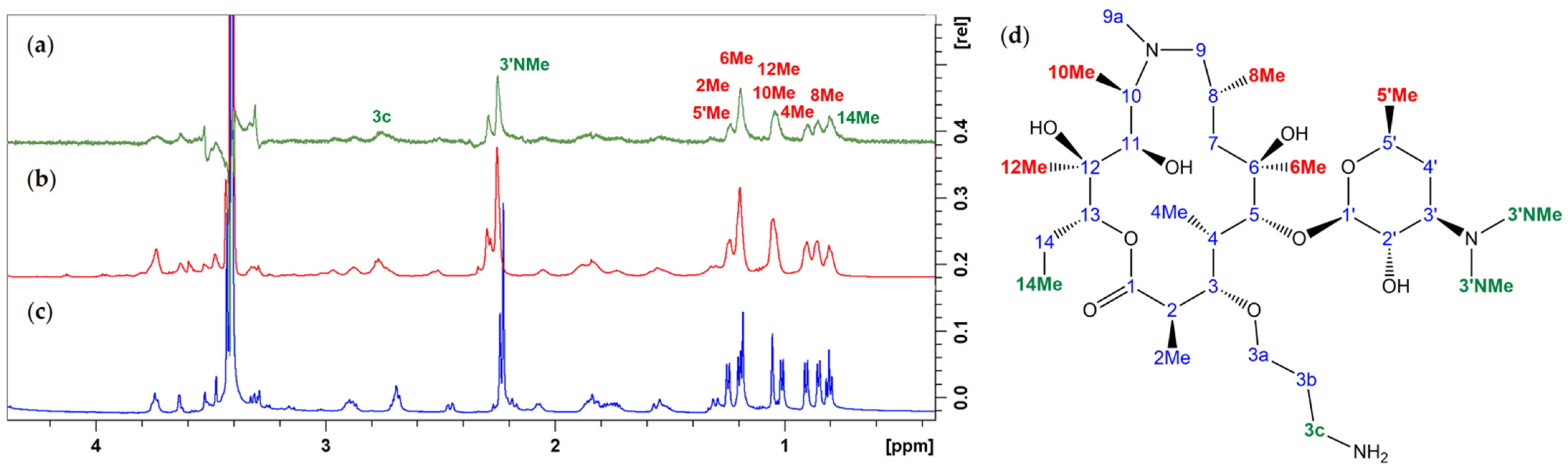
3.1. Structure Elucidation of 1a–c
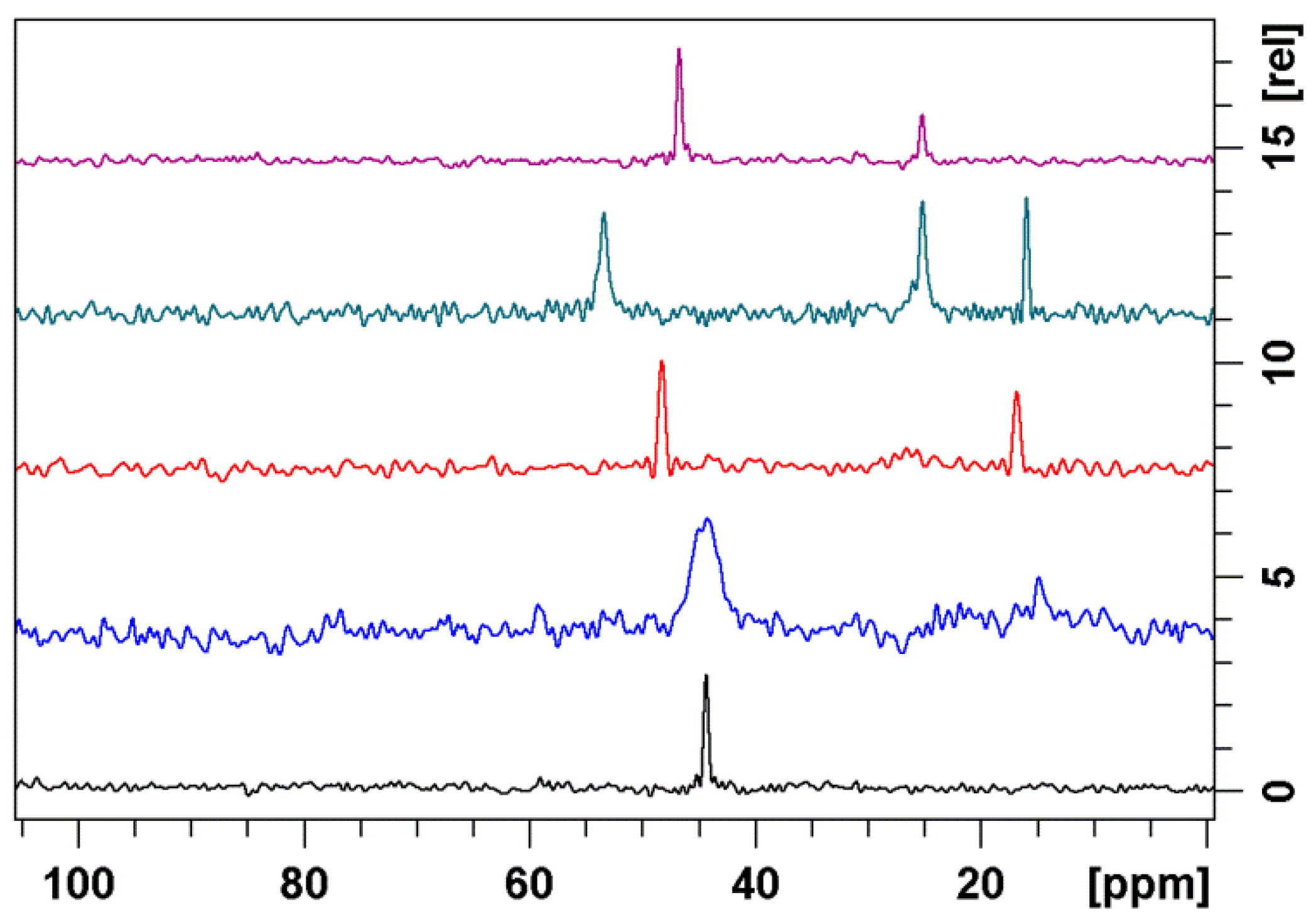
3.2. Bound Structures and Interactions with E. coli Ribosome
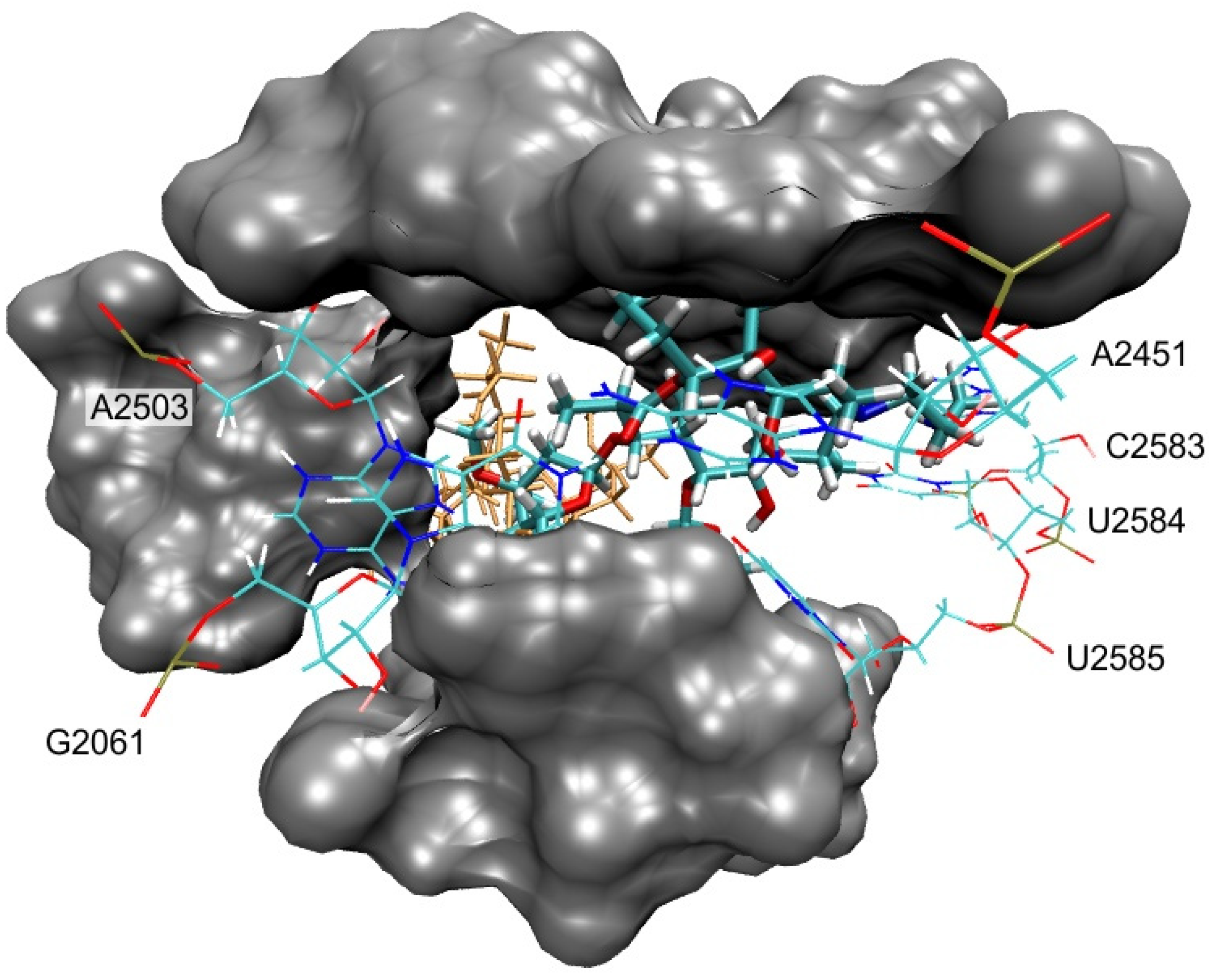
3.3. Molecular Docking
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Grgičević, I.; Mikulandra, I.; Bukvić, M.; Banjanac, M.; Radovanović, V.; Habinovec, I.; Bertoša, B.; Novak, P. Discovery of macrozones, new antimicrobial thiosemicarbazone-based azithromycin conjugates: Design, synthesis and in vitro biological activity. Int. J. Antimicrob. Agents 2020, 56, 106147. [Google Scholar] [CrossRef] [PubMed]
- Jednačak, T.; Mikulandra, I.; Novak, P. Advanced methods for studying structure and interactions of macrolide antibiotics. Int. J. Mol. Sci. 2020, 21, 7799. [Google Scholar] [CrossRef]
- Arsić, B.; Barber, J.; Novak, P. The macrolide antibiotics and their semi-synthetic derivatives. In Macrolides: Properties, Synthesis and Applications; Arsić, B., Ed.; Walter de Gruyter GmbH: Berlin, Germany; Boston, MA, USA, 2018; pp. 1–30. [Google Scholar] [CrossRef]
- Chancey, S.T.; Zhou, X.; Zähner, D.; Stephens, D.S. Induction of efflux-mediated macrolide resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2011, 55, 3413–3422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlünzen, F.; Harms, J.M.; Franceschi, F.; Hansen, H.A.S.; Bartels, H.; Zarivach, R.; Yonath, A. Structural basis for the antibiotic activity of ketolides and azalides. Structure 2003, 11, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Tu, D.; Blaha, G.; Moore, P.B.; Steitz, T.A. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 2005, 121, 257–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunkle, J.A.; Xiong, L.; Mankin, A.S.; Cate, J.H.D. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. USA 2010, 107, 17152–17157. [Google Scholar] [CrossRef] [Green Version]
- Novak, P.; Banić Tomišić, Z.; Tepeš, P.; Lazarevski, G.; Plavec, J.; Turkalj, G. Conformational analysis of oleandomycin and its 8-methylene-9-oxime derivative by NMR and molecular modeling. Org. Biomol. Chem. 2005, 3, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Tatić, I.; Tepeš, P.; Koštrun, S.; Barber, J. Systematic approach to understanding macrolide-ribosome interactions: NMR and modeling studies of oleandomycin and its derivatives. J. Phys. Chem. A 2006, 110, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Barber, J.; Čikoš, A.; Arsić, B.; Plavec, J.; Lazarevski, G.; Tepeš, P.; Košutić-Hulita, N. Free and bound state structures of 6-O-methyl homoerythromycins and epitope mapping of their interactions with ribosomes. Bioorg. Med. Chem. 2009, 17, 5857–5867. [Google Scholar] [CrossRef] [PubMed]
- Kosol, S.; Schrank, E.; Krajačić, M.B.; Wagner, G.E.; Meyer, N.H.; Göbl, C.; Rechberger, G.N.; Zangger, K.; Novak, P. Probing the interactions of macrolide antibiotics with membrane-mimetics by NMR spectroscopy. J. Med. Chem. 2012, 55, 5632–5636. [Google Scholar] [CrossRef]
- Glanzer, S.; Pulido, S.A.; Tutz, S.; Wagner, G.E.; Kriechbaum, M.; Gubensäk, N.; Trifunović, J.; Dorn, M.; Fabian, W.M.F.; Novak, P.; et al. Structural and functional implications of the interaction between macrolide antibiotics and bile acids. Chem. Eur. J. 2015, 21, 4350–4358. [Google Scholar] [CrossRef] [Green Version]
- Novak, P.; Tepeš, P.; Lazić, V. Epitope mapping of macrolide antibiotics to bovine serum albumin by saturation transfer difference NMR spectroscopy. Croat. Chem. Acta 2007, 80, 211–216. [Google Scholar]
- Novak, P. Interactions of macrolides with their biological targets. In Macrolides: Properties, Synthesis and Applications; Arsić, B., Ed.; Walter de Gruyter GmbH: Berlin, Germany; Boston, MA, USA, 2018; pp. 63–77. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]






| Compound | S. pneumoniae B0652 | S. pneumoniae B0326 | S. pyogenes B0542 | S. pyogenes B0545 | S. aureus ATCC 29213 | E. faecalis ATCC 29212 | M. catarrhalis ATCC 23246 | E. coli ATCC 25922 |
|---|---|---|---|---|---|---|---|---|
| eryS | M | eryS | M | eryS | ||||
| MIC (μg/mL) a | ||||||||
| 1a | 1 | 64 | 0.5 | 64 | 8 | 64 | 4 | 32 |
| 1b | 16 | 64 | 32 | 64 | >64 | >64 | 2 | 64 |
| 1c | 0.5 | 16 | 2 | 32 | 16 | 64 | 2 | 32 |
| azithromycin | ≤0.125 | 16 | ≤0.125 | 8 | 1 | 8 | ≤0.125 | >64 |
| Compound | 3JH2,H3/Hz | |
|---|---|---|
| Acetonitrile | Tris Buffer | |
| 1a | 6.60 | 6.43 |
| 1b | 10.14 | 9.88 |
| 1c | 5.39 | 5.11 |
| NOE Contact | Compound | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 1c | |||||||
| acet. a | Tris b | rib. c | acet. a | Tris b | rib. c | acet. a | Tris b | rib. c | |
| H3-H11 | − | − | − | − | − | − | − | − | − |
| H4-H11 | + | + | + | + | + | + | + | + | + |
| H2-H4 | + | + | + | + | + | + | + | + | + |
| H4-H6Me | − | − | − | − | − | − | − | − | − |
| H3-H8 | − | − | − | − | − | − | − | − | − |
| H5-H6Me | + | + | + | + | + | + | + | + | + |
| H8-H11 | − | − | − | − | − | − | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikulandra, I.; Jednačak, T.; Bertoša, B.; Parlov Vuković, J.; Kušec, I.; Novak, P. Interactions of Aminopropyl–Azithromycin Derivatives, Precursors in the Synthesis of Bioactive Macrozones, with E. coli Ribosome: NMR and Docking Studies. Materials 2021, 14, 5561. https://doi.org/10.3390/ma14195561
Mikulandra I, Jednačak T, Bertoša B, Parlov Vuković J, Kušec I, Novak P. Interactions of Aminopropyl–Azithromycin Derivatives, Precursors in the Synthesis of Bioactive Macrozones, with E. coli Ribosome: NMR and Docking Studies. Materials. 2021; 14(19):5561. https://doi.org/10.3390/ma14195561
Chicago/Turabian StyleMikulandra, Ivana, Tomislav Jednačak, Branimir Bertoša, Jelena Parlov Vuković, Iva Kušec, and Predrag Novak. 2021. "Interactions of Aminopropyl–Azithromycin Derivatives, Precursors in the Synthesis of Bioactive Macrozones, with E. coli Ribosome: NMR and Docking Studies" Materials 14, no. 19: 5561. https://doi.org/10.3390/ma14195561






