Antibacterial Polymers Based on Poly(2-hydroxyethyl methacrylate) and Thiazolium Groups with Hydrolytically Labile Linkages Leading to Inactive and Low Cytotoxic Compounds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
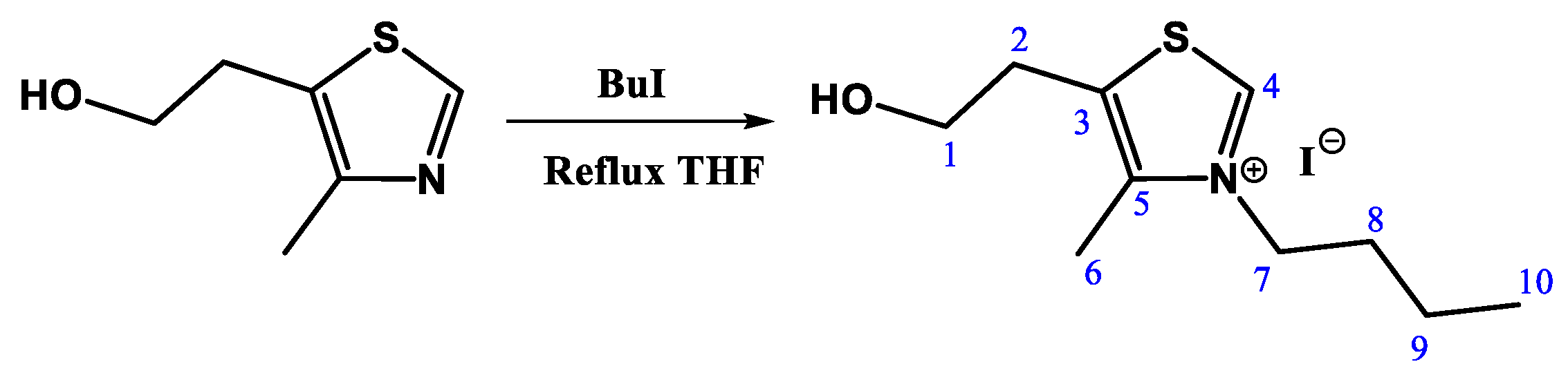
2.2. N-Alkylation Reaction of 5-(2-hydroxyethyl)-4-methylthiazole (TZ-Bu)
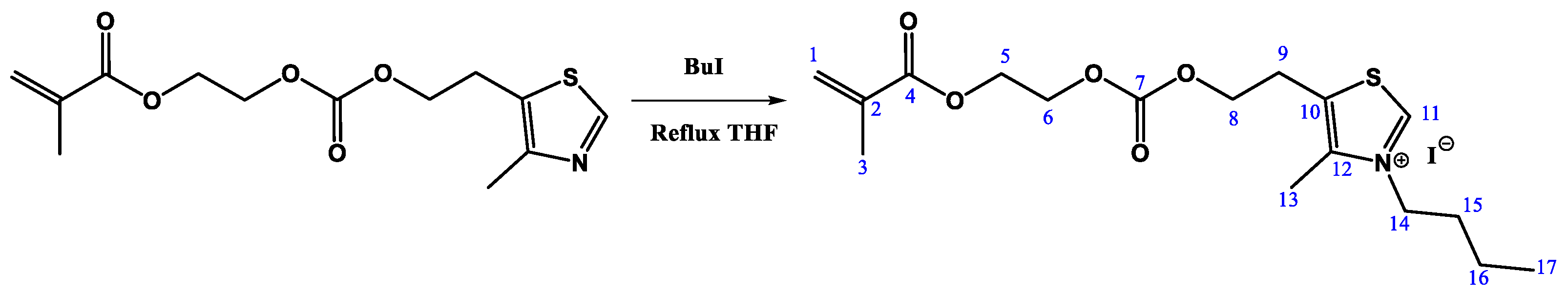
2.3. N-Alkylation Reaction of 2-(((2-(4-Methylthiazol-5-yl)ethoxy)carbonyl)oxy)ethyl Methacrylate. Synthesis of MTZ-Bu
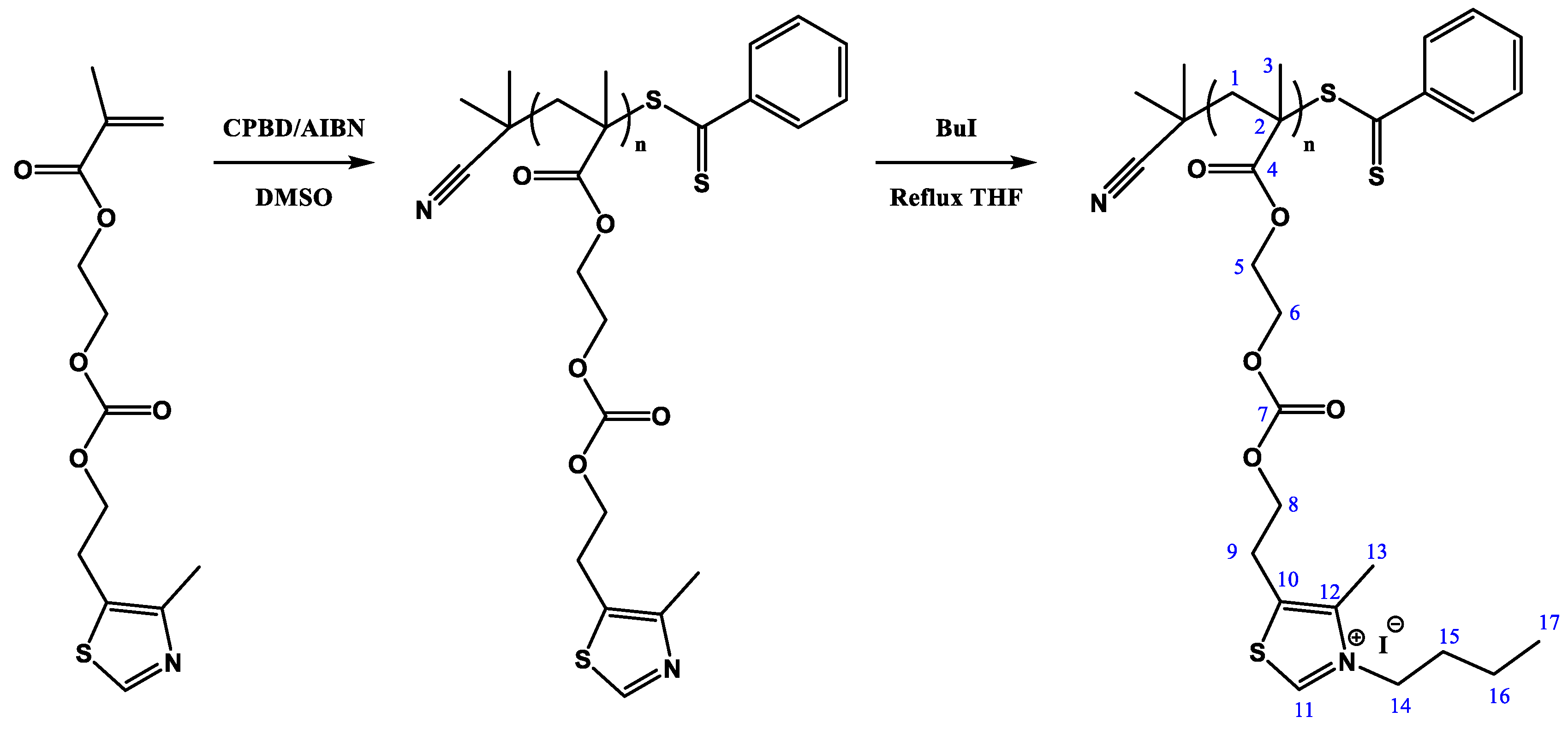
2.4. Polymerization via RAFT of MTZ. Synthesis of the Polymer PMTZ-Bu
2.5. Characterization
2.6. Evaluation of Monomer and Polymer Hydrolysis
2.7. Antibacterial Activity Measurements
2.8. Hemolysis Assay
3. Results and Discussion
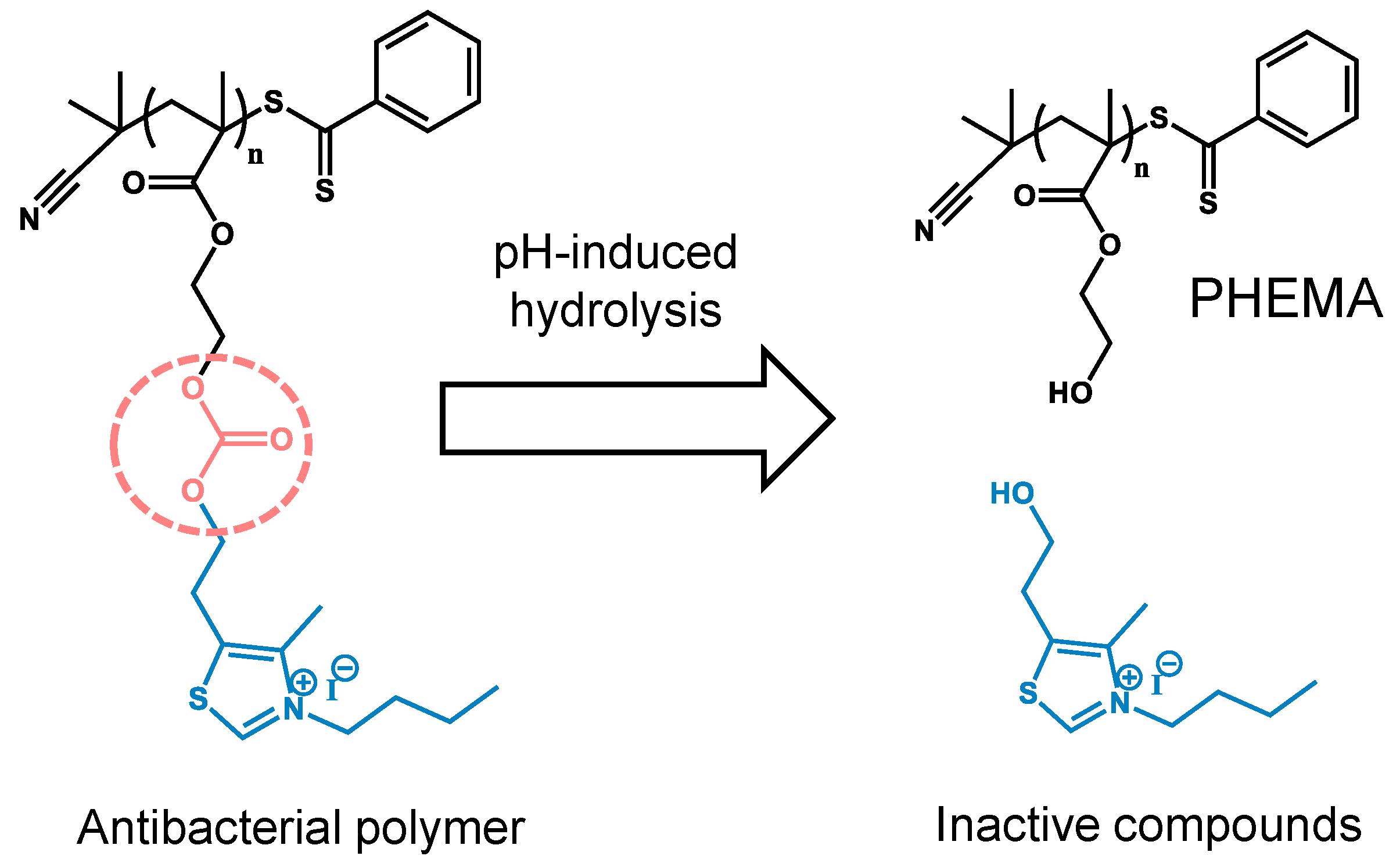
3.1. Material Design and Synthesis
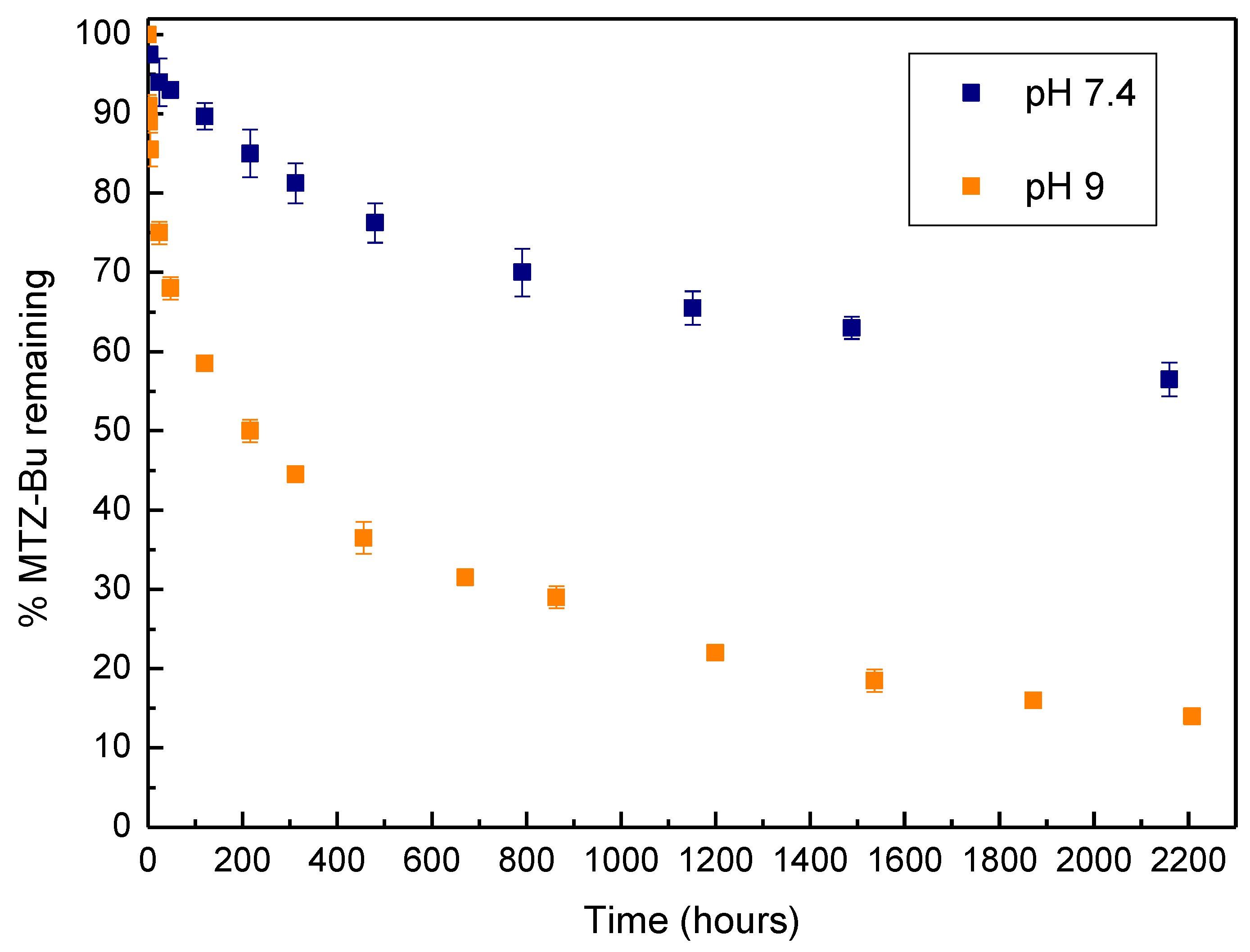
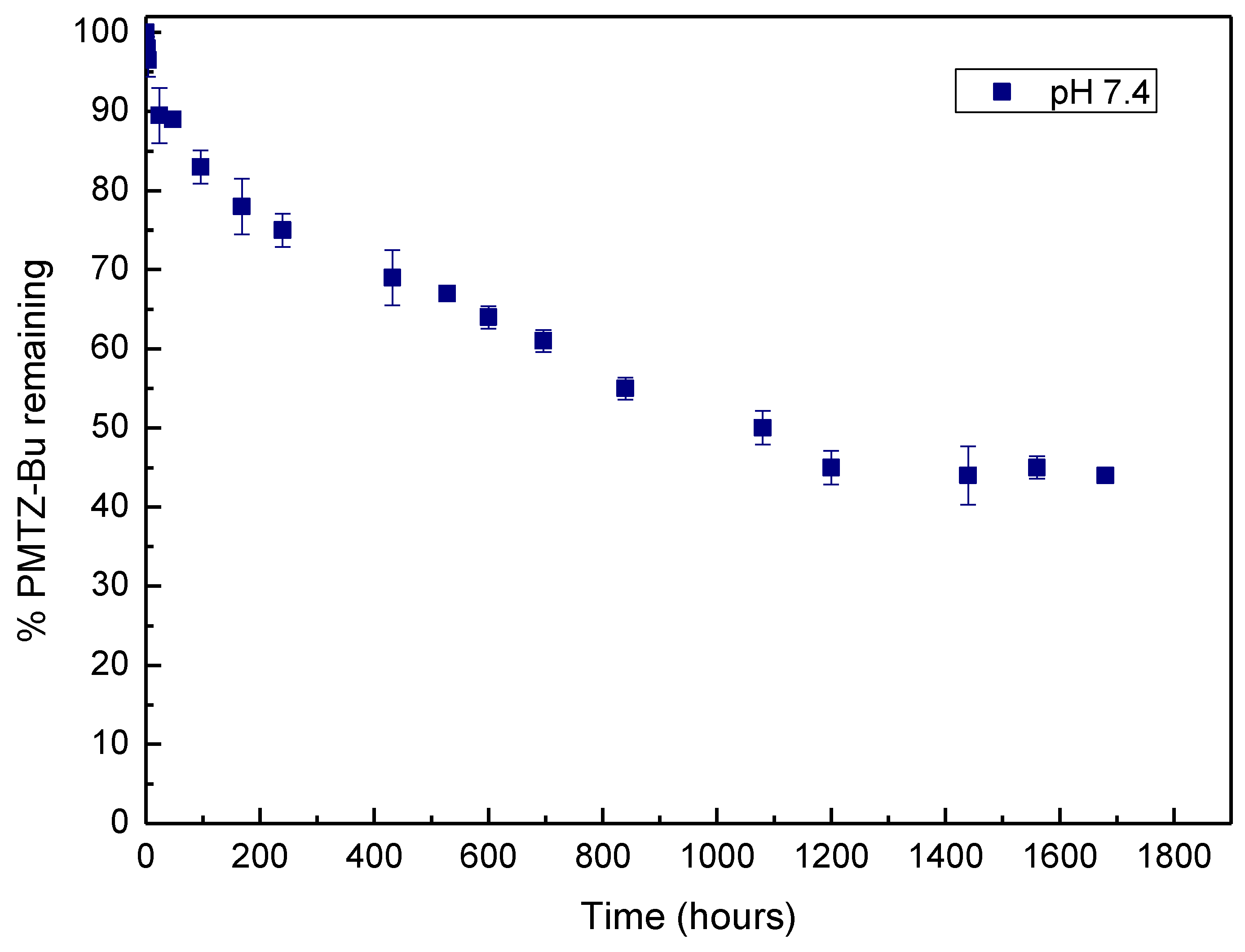
3.2. pH-Induced Hydrolysis Profiles
3.3. Antibacterial and Hemolytic Activity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sugden, R.; Kelly, R.; Davies, S. Combatting antimicrobial resistance globally. Nat. Microbiol. 2016, 1, 16187. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, J.L. Antibiotics and Antibiotic Resistance Genes in Natural Environments. Science 2008, 321, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Cycon, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment-Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef]
- Khatoon, U.T.; Rao, G.N.; Mohan, K.M.; Ramanaviciene, A.; Ramanavicius, A. Antibacterial and antifungal activity of silver nanospheres synthesized by tri-sodium citrate assisted chemical approach. Vacuum 2017, 146, 259–265. [Google Scholar] [CrossRef]
- Dai, X.; Chen, X.; Zhao, J.; Zhao, Y.; Guo, Q.; Zhang, T.; Chu, C.; Zhang, X.; Li, C. Structure–Activity Relationship of Membrane-Targeting Cationic Ligands on a Silver Nanoparticle Surface in an Antibiotic-Resistant Antibacterial and Antibiofilm Activity Assay. ACS Appl. Mater. Interfaces 2017, 9, 13837–13848. [Google Scholar] [CrossRef]
- Mohammed, H.; Kumar, A.; Bekyarova, E.; Al-Hadeethi, Y.; Zhang, X.; Chen, M.; Ansari, M.S.; Cochis, A.; Rimondini, L. Antimicrobial Mechanisms and Effectiveness of Graphene and Graphene-Functionalized Biomaterials. A Scope Review. Front. Bioeng. Biotechnol. 2020, 8, 465. [Google Scholar] [CrossRef]
- Sun, B.; Slomberg, D.L.; Chudasama, S.L.; Lu, Y.; Schoenfisch, M.H. Nitric Oxide-Releasing Dendrimers as Antibacterial Agents. Biomacromolecules 2012, 13, 3343–3354. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Bonilla, A.; Fernández-García, M. Polymeric materials with antimicrobial activity. Prog. Polym. Sci. 2012, 37, 281–339. [Google Scholar] [CrossRef]
- Ergene, C.; Yasuhara, K.; Palermo, E.F. Biomimetic antimicrobial polymers: Recent advances in molecular design. Polym. Chem. 2018, 9, 2407–2427. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.; Duvvuri, L.S.; Farah, S.; Beyth, N.; Domb, A.J.; Khan, W. Antimicrobial polymers. Adv. Healthc. Mater. 2014, 3, 1969–1985. [Google Scholar] [CrossRef]
- Konai, M.M.; Bhattacharjee, B.; Ghosh, S.; Haldar, J. Recent Progress in Polymer Research to Tackle Infections and Antimicrobial Resistance. Biomacromolecules 2018, 19, 1888–1917. [Google Scholar] [CrossRef]
- Qiu, H.; Si, Z.; Luo, Y.; Feng, P.; Wu, X.; Hou, W.; Zhu, Y.; Chan-Park, M.B.; Xu, L.; Huang, D. The Mechanisms and the Applications of Antibacterial Polymers in Surface Modification on Medical Devices. Front. Bioeng. Biotechnol. 2020, 8, 910. [Google Scholar] [CrossRef]
- Borjihan, Q.; Dong, A. Design of nanoengineered antibacterial polymers for biomedical applications. Biomater. Sci. 2020, 8, 6867–6882. [Google Scholar] [CrossRef]
- Huang, T.; Qian, Y.; Wei, J.; Zhou, C. Polymeric Antimicrobial Food Packaging and Its Applications. Polymers 2019, 11, 560. [Google Scholar] [CrossRef] [Green Version]
- Mitra, D.; Kang, E.T.; Neoh, K.G. Polymer-Based Coatings with Integrated Antifouling and Bactericidal Properties for Targeted Biomedical Applications. ACS Appl. Polym. Mater. 2021, 3, 2233–2263. [Google Scholar] [CrossRef]
- Sun, J.; Li, M.; Lin, M.; Zhang, B.; Chen, X. High Antibacterial Activity and Selectivity of the Versatile Polysulfoniums that Combat Drug Resistance. Adv. Mater. 2021, 33, 2104402. [Google Scholar] [CrossRef]
- Mukherjee, S.; Barman, S.; Mukherjee, R.; Haldar, J. Amphiphilic Cationic Macromolecules Highly Effective against Multi-Drug Resistant Gram-Positive Bacteria and Fungi with No Detectable Resistance. Front. Bioeng. Biotechnol. 2020, 8, 55. [Google Scholar] [CrossRef]
- Sadrearhami, Z.; Nguyen, T.K.; Namivandi-Zangeneh, R.; Jung, K.; Wong, E.H.H.; Boyer, C. Recent advances in nitric oxide delivery for antimicrobial applications using polymer-based systems. J. Mater. Chem. B 2018, 6, 2945–2959. [Google Scholar] [CrossRef]
- Namivandi-Zangeneh, R.; Wong, E.H.H.; Boyer, C. Synthetic Antimicrobial Polymers in Combination Therapy: Tackling Antibiotic Resistance. ACS Infect. Dis. 2021, 7, 215–253. [Google Scholar] [CrossRef]
- Ding, X.; Duan, S.; Ding, X.; Liu, R.; Xu, F.J. Versatile Antibacterial Materials: An Emerging Arsenal for Combatting Bacterial Pathogens. Adv. Funct. Mater. 2018, 28, 1802140. [Google Scholar] [CrossRef]
- Ding, X.; Wang, A.; Tong, W.; Xu, F.J. Biodegradable Antibacterial Polymeric Nanosystems: A New Hope to Cope with Multidrug-Resistant Bacteria. Small 2019, 15, e1900999. [Google Scholar] [CrossRef]
- Gong, C.; Sun, J.; Xiao, Y.; Qu, X.; Lang, M. Synthetic Mimics of Antimicrobial Peptides for the Targeted Therapy of Multidrug-Resistant Bacterial Infection. Adv. Healthc. Mater. 2021, 451, 2101244. [Google Scholar] [CrossRef]
- Ganewatta, M.S.; Wang, Z.; Tang, C. Chemical syntheses of bioinspired and biomimetic polymers toward biobased materials. Nat. Rev. Chem. 2021, 5, 753–772. [Google Scholar] [CrossRef]
- Chin, W.; Yang, C.; Ng, V.W.L.; Huang, Y.; Cheng, J.; Tong, Y.W.; Coady, D.J.; Fan, W.; Hedrick, J.L.; Yang, Y.Y. Biodegradable Broad-Spectrum Antimicrobial Polycarbonates: Investigating the Role of Chemical Structure on Activity and Selectivity. Macromolecules 2013, 46, 8797–8807. [Google Scholar] [CrossRef]
- Chiloeches, A.; Funes, A.; Cuervo-Rodríguez, R.; López-Fabal, F.; Fernández-García, M.; Echeverría, C.; Muñoz-Bonilla, A. Biobased polymers derived from itaconic acid bearing clickable groups with potent antibacterial activity and negligible hemolytic activity. Polym. Chem. 2021, 12, 3190–3200. [Google Scholar] [CrossRef]
- Munoz-Bonilla, A.; Echeverria, C.; Sonseca, A.; Arrieta, M.P.; Fernandez-Garcia, M. Bio-Based Polymers with Antimicrobial Properties towards Sustainable Development. Materials 2019, 12, 641. [Google Scholar] [CrossRef] [Green Version]
- Kalelkar, P.P.; Geng, Z.; Finn, M.G.; Collard, D.M. Azide-Substituted Polylactide: A Biodegradable Substrate for Antimicrobial Materials via Click Chemistry Attachment of Quaternary Ammonium Groups. Biomacromolecules 2019, 20, 3366–3374. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, L.; Zhang, B.; Feng, W.; Xu, M.; Gao, L.; Liu, N.; Wang, Q.; Huang, X.; Li, P.; et al. Design and synthesis of biocompatible, hemocompatible, and highly selective antimicrobial cationic peptidopolysaccharides via click chemistry. Biomacromolecules 2019, 20, 2230–2240. [Google Scholar] [CrossRef]
- Yuan, Y.; Lim, D.S.W.; Wu, H.; Lu, H.; Zheng, Y.; Wan, A.C.A.; Ying, J.Y.; Zhang, Y. pH-Degradable imidazolium oligomers as antimicrobial materials with tuneable loss of activity. Biomater. Sci. 2019, 7, 2317–2325. [Google Scholar] [CrossRef]
- Lim, D.S.W.; Yuan, Y.; Zhang, Y. pH-Degradable Polymers as Impermanent Antimicrobial Agents for Environmental Sustainability. ACS Appl. Bio Mater. 2021, 4, 1544–1551. [Google Scholar] [CrossRef]
- Cuervo-Rodríguez, R.; Muñoz-Bonilla, A.; Araujo, J.; Echeverría, C.; Fernández-García, M. Influence of side chain structure on the thermal and antimicrobial properties of cationic methacrylic polymers. Eur. Polym. J. 2019, 117, 86–93. [Google Scholar] [CrossRef]
- Zare, M.; Bigham, A.; Zare, M.; Luo, H.; Rezvani Ghomi, E.; Ramakrishna, S. pHEMA: An Overview for Biomedical Applications. Int. J. Mol. Sci. 2021, 22, 6376. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, Approved Standard-Ninth Edition; CLSI Document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Cuervo-Rodriguez, R.; Munoz-Bonilla, A.; Lopez-Fabal, F.; Fernandez-Garcia, M. Hemolytic and Antimicrobial Activities of a Series of Cationic Amphiphilic Copolymers Comprised of Same Centered Comonomers with Thiazole Moieties and Polyethylene Glycol Derivatives. Polymers 2020, 12, 972. [Google Scholar] [CrossRef]
- Kasmi, S.; Louage, B.; Nuhn, L.; Verstraete, G.; Van Herck, S.; van Steenbergen, M.J.; Vervaet, C.; Hennink, W.E.; De Geest, B.G. Acrylamides with hydrolytically labile carbonate ester side chains as versatile building blocks for well-defined block copolymer micelles via RAFT polymerization. Polym. Chem. 2017, 8, 6544–6557. [Google Scholar] [CrossRef] [Green Version]
- Lienkamp, K.; Tew, G.N. Synthetic mimics of antimicrobial peptides—A versatile ring-opening metathesis polymerization based platform for the synthesis of selective antibacterial and cell-penetrating polymers. Chemistry 2009, 15, 11784–11800. [Google Scholar] [CrossRef]
- Krumm, C.; Harmuth, S.; Hijazi, M.; Neugebauer, B.; Kampmann, A.L.; Geltenpoth, H.; Sickmann, A.; Tiller, J.C. Antimicrobial poly(2-methyloxazoline)s with bioswitchable activity through satellite group modification. Angew. Chem. Int. Ed. Engl. 2014, 53, 3830–3834. [Google Scholar] [CrossRef]
- Monteiro, C.; Pinheiro, M.; Fernandes, M.; Maia, S.; Seabra, C.L.; Ferreira-da-Silva, F.; Reis, S.; Gomes, P.; Martins, M.C.L. Synthetic Mimic of Antimicrobial Peptide with Nonmembrane-Disrupting Antibacterial Properties A 17-mer Membrane-Active MSI-78 Derivative with Improved Selectivity toward Bacterial Cells. Mol. Pharm. 2015, 12, 2904–2911. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhang, K.; Reghu, S.; Lin, Y.; Chan-Park, M.B.; Liu, X.W. Synthesis of Antibacterial Glycosylated Polycaprolactones Bearing Imidazoliums with Reduced Hemolytic Activity. Biomacromolecules 2019, 20, 949–958. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuervo-Rodríguez, R.; López-Fabal, F.; Muñoz-Bonilla, A.; Fernández-García, M. Antibacterial Polymers Based on Poly(2-hydroxyethyl methacrylate) and Thiazolium Groups with Hydrolytically Labile Linkages Leading to Inactive and Low Cytotoxic Compounds. Materials 2021, 14, 7477. https://doi.org/10.3390/ma14237477
Cuervo-Rodríguez R, López-Fabal F, Muñoz-Bonilla A, Fernández-García M. Antibacterial Polymers Based on Poly(2-hydroxyethyl methacrylate) and Thiazolium Groups with Hydrolytically Labile Linkages Leading to Inactive and Low Cytotoxic Compounds. Materials. 2021; 14(23):7477. https://doi.org/10.3390/ma14237477
Chicago/Turabian StyleCuervo-Rodríguez, Rocío, Fátima López-Fabal, Alexandra Muñoz-Bonilla, and Marta Fernández-García. 2021. "Antibacterial Polymers Based on Poly(2-hydroxyethyl methacrylate) and Thiazolium Groups with Hydrolytically Labile Linkages Leading to Inactive and Low Cytotoxic Compounds" Materials 14, no. 23: 7477. https://doi.org/10.3390/ma14237477
APA StyleCuervo-Rodríguez, R., López-Fabal, F., Muñoz-Bonilla, A., & Fernández-García, M. (2021). Antibacterial Polymers Based on Poly(2-hydroxyethyl methacrylate) and Thiazolium Groups with Hydrolytically Labile Linkages Leading to Inactive and Low Cytotoxic Compounds. Materials, 14(23), 7477. https://doi.org/10.3390/ma14237477







