Controlled Formation of a Protein Corona Composed of Denatured BSA on Upconversion Nanoparticles Improves Their Colloidal Stability
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of UCNP-OA and UCNP-NOBF4
2.2. Characterization of UCNP-OA and UCNP-NOBF4
2.3. Optimization of dBSA Concentration for PC Formation on UCNP-NOBF4
2.4. Lyophilization of dBSA-UCNP-NOBF4 and Evaluation of Their Colloidal Stability
2.5. Hard PC Formation on the Surface of UCNP-NOBF4 and Lyophilized dBSA-UCNP-NOBF4
2.6. Protein Corona Quantification
3. Results
3.1. Synthesis and Characterization of UCNP-OA and UCNP-NOBF4
3.2. Optimization of dBSA Concentration for PC Formation on UCNP-NOBF4
3.3. Centrifugation of dBSA-UCNP-NOBF4 and Evaluation of Their Colloidal Stability
3.4. Lyophilization of dBSA-UCNP-NOBF4 and Evaluation of their Colloidal Stability
3.5. Colloidal Stability after Long-Term Storage
3.6. Hard Protein Corona Quantification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Safi, M.; Courtois, J.; Seigneuret, M.; Conjeaud, H.; Berret, J.-F. The effects of aggregation and protein corona on the cellular internalization of iron oxide nanoparticles. Biomaterials 2011, 32, 9353–9363. [Google Scholar] [CrossRef] [Green Version]
- Treuel, L.; Nienhaus, G.U. Toward a molecular understanding of nanoparticle–protein interactions. Biophys. Rev. 2012, 4, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Colvin, V.L. The potential environmental impact of engineered nanomaterials. Nat. Biotechnol. 2003, 21, 1166–1170. [Google Scholar] [CrossRef]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef]
- Patel, H.; Moghimi, S.M. Serum-mediated recognition of liposomes by phagocytic cells of the reticuloendothelial system—The concept of tissue specificity. Adv. Drug Deliv. Rev. 1998, 32, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, F.; Zhao, D. Lab on upconversion nanoparticles: Optical properties and applications engineering via designed nanostructure. Chem. Soc. Rev. 2015, 44, 1346–1378. [Google Scholar] [CrossRef] [PubMed]
- Rafique, R.; Kailasa, S.K.; Park, T.J. Recent advances of upconversion nanoparticles in theranostics and bioimaging applications. Trends Anal. Chem. 2019, 120, 115646. [Google Scholar] [CrossRef]
- Jin, J.; Gu, Y.-J.; Man, C.W.-Y.; Cheng, J.; Xu, Z.; Zhang, Y.; Wang, H.; Lee, V.H.-Y.; Cheng, S.H.; Wong, W.-T. Polymer-Coated NaYF4:Yb3+, Er3+ Upconversion Nanoparticles for Charge-Dependent Cellular Imaging. ACS Nano 2011, 5, 7838–7847. [Google Scholar] [CrossRef]
- Guller, A.E.; Generalova, A.N.; Petersen, E.V.; Nechaev, A.V.; Trusova, I.A.; Landyshev, N.N.; Nadort, A.; Grebenik, E.A.; Deyev, S.M.; Shekhter, A.B.; et al. Cytotoxicity and non-specific cellular uptake of bare and surface-modified upconversion nanoparticles in human skin cells. Nano Res. 2015, 8, 1546–1562. [Google Scholar] [CrossRef]
- Wang, F.; Wang, J.; Liu, X. Direct Evidence of a Surface Quenching Effect on Size-Dependent Luminescence of Upconversion Nanoparticles. Angew. Chem. Int. Ed. 2010, 49, 7456–7460. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, Z.; Li, F. Upconversion nanophosphors for small-animal imaging. Chem. Soc. Rev. 2012, 41, 1323–1349. [Google Scholar] [CrossRef]
- Chatterjee, D.K.; Rufaihah, A.J.; Zhang, Y. Upconversion fluorescence imaging of cells and small animals using lanthanide doped nanocrystals. Biomaterials 2008, 29, 937–943. [Google Scholar] [CrossRef]
- Song, Z.; Anissimov, Y.G.; Zhao, J.; Nechaev, A.V.; Nadort, A.; Jin, D.; Prow, T.W.; Roberts, M.S.; Zvyagin, A.V. Background free imaging of upconversion nanoparticle distribution in human skin. J. Biomed. Opt. 2012, 18, e061215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Zhu, X.; Lu, Y.; Zhao, J.; Feng, W.; Jia, G.; Wang, F.; Li, F.; Jin, D. High-Contrast Visualization of Upconversion Luminescence in Mice Using Time-Gating Approach. Anal. Chem. 2016, 88, 3449–3454. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Zhang, F.; Lv, L.; Han, Z.; Wang, Y.; Gu, Y.; Chen, H. Bovine Serum Albumin Coated Upconversion Nanoparticles for Near Infrared Fluorescence Imaging in Mouse Model. J. Nanosci. Nanotechnol. 2017, 17, 932–938. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Abbineni, G.; Clevenger, A.; Mao, C.; Xu, S. Upconversion nanoparticles: Synthesis, surface modification and biological applications. Nanomedicine 2011, 7, 710–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, A.; Ye, X.; Chen, J.; Kang, Y.; Gordon, T.; Kikkawa, J.M.; Murray, C.B. A Generalized Ligand-Exchange Strategy Enabling Sequential Surface Functionalization of Colloidal Nanocrystals. J. Am. Chem. Soc. 2011, 133, 998–1006. [Google Scholar] [CrossRef]
- Matsumura, Y.; Oda, T.; Maeda, H. General mechanism of intratumor accumulation of macromolecules: Advantage of mac-romolecular therapeutics. Gan To Kagaku Ryoho 1987, 14, 821–829. [Google Scholar]
- Elzoghby, A.O.; Samy, W.M.; Elgindy, N.A. Albumin-based nanoparticles as potential controlled release drug delivery systems. J. Control. Release 2012, 157, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Barua, S.; Mitragotri, S. Challenges associated with penetration of nanoparticles across cell and tissue barriers: A review of current status and future prospects. Nano Today 2014, 9, 223–243. [Google Scholar] [CrossRef] [PubMed]
- Sindhwani, S.; Syed, A.M.; Ngai, J.; Kingston, B.R.; Maiorino, L.; Rothschild, J.; Macmillan, P.; Zhang, Y.; Rajesh, N.U.; Hoang, T.; et al. The entry of nanoparticles into solid tumours. Nat. Mater. 2020, 19, 566–575. [Google Scholar] [CrossRef]
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggård, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef] [Green Version]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.M.V.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef]
- Hadjidemetriou, M.; Kostarelos, K. Evolution of the nanoparticle corona. Nat. Nanotechnol. 2017, 12, 288–290. [Google Scholar] [CrossRef]
- Bern, M.; Sand, K.M.K.; Nilsen, J.; Sandlie, I.; Andersen, J.T. The role of albumin receptors in regulation of albumin homeostasis: Implications for drug delivery. J. Control. Release 2015, 211, 144–162. [Google Scholar] [CrossRef]
- Loureiro, A.; Azoia, N.G.; Gomes, A.C.; Cavaco-Paulo, A. Albumin-Based Nanodevices as Drug Carriers. Curr. Pharm. Des. 2016, 22, 1371–1390. [Google Scholar] [CrossRef]
- An, F.-F.; Zhang, X.-H. Strategies for Preparing Albumin-based Nanoparticles for Multifunctional Bioimaging and Drug Delivery. Theranostics 2017, 7, 3667–3689. [Google Scholar] [CrossRef]
- Bakshi, M.S.; Thakur, P.; Kaur, G.; Kaur, H.; Banipal, T.S.; Possmayer, F.; Petersen, N.O. Stabilization of PbS Nanocrystals by Bovine Serum Albumin in its Native and Denatured States. Adv. Funct. Mater. 2009, 19, 1451–1458. [Google Scholar] [CrossRef]
- Jung, S.H.; Kim, S.K.; Jung, S.H.; Kim, E.H.; Cho, S.H.; Jeong, K.-S.; Seong, H.; Shin, B.C. Increased stability in plasma and enhanced cellular uptake of thermally denatured albumin-coated liposomes. Colloids Surf. B Biointerfaces 2010, 76, 434–440. [Google Scholar] [CrossRef]
- Park, J.H.; Jackman, J.A.; Ferhan, A.R.; Ma, G.J.; Yoon, B.K.; Cho, N.-J. Temperature-Induced Denaturation of BSA Protein Molecules for Improved Surface Passivation Coatings. ACS Appl. Mater. Interfaces 2018, 10, 32047–32057. [Google Scholar] [CrossRef]
- Baler, K.; Martin, O.A.; Carignano, M.A.; Ameer, G.A.; Vila, J.A.; Szleifer, I. Electrostatic Unfolding and Interactions of Albumin Driven by pH Changes: A Molecular Dynamics Study. J. Phys. Chem. B 2014, 118, 921–930. [Google Scholar] [CrossRef]
- Peters, T. All About Albumin: 2—The Albumin Molecule: Its Structure and Chemical Properties; Elsevier: New York, NY, USA, 1995; pp. 9–75. [Google Scholar] [CrossRef]
- Mai, H.-X.; Zhang, Y.-W.; Sun, L.-D.; Yan, C.-H. Highly Efficient Multicolor Up-Conversion Emissions and Their Mechanisms of Monodisperse NaYF4:Yb,Er Core and Core/Shell-Structured Nanocrystals. J. Phys. Chem. C 2007, 111, 13721–13729. [Google Scholar] [CrossRef]
- Guryev, E.L.; Smyshlyaeva, A.S.; Shilyagina, N.Y.; Sokolova, E.A.; Shanwar, S.; Kostyuk, A.B.; Lyubeshkin, A.V.; Schulga, A.A.; Konovalova, E.V.; Lin, Q.; et al. UCNP-based Photoluminescent Nanomedicines for Targeted Imaging and Theranostics of Cancer. Molecules 2020, 25, 4302. [Google Scholar] [CrossRef]
- Shanwar, S.; Liang, L.; Nechaev, A.V.; Balalaeva, I.V.; Vodeneev, V.A.; Zvyagin, A.V.; Guryev, E.L. The assembly of a pho-toluminescent nanocomplex based on upconversion nanoparticles. Opera Med. Physiol. 2020, 7, 42–48. [Google Scholar] [CrossRef]
- Kostyuk, A.; Liang, L.; Zhang, R.; Tretyakov, A.A.; Zvyagin, A.V. Protein corona formation on polymer-coated nanoparticles. In Proceedings of the “Science of the Future” Conference, Kazan, Russia, 20 September 2016; pp. 250–252. [Google Scholar]
- Generalova, A.; Chichkov, B.; Khaydukov, E. Multicomponent nanocrystals with anti-Stokes luminescence as contrast agents for modern imaging techniques. Adv. Colloid Interface Sci. 2017, 245, 1–19. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential—What they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Maurya, A.; Dwivedi, A.; Bahadur, A.; Rai, S. Enhanced upconversion and downshifting emissions from Tm3+, Yb3+ co-doped CaZrO3 phosphor in the presence of alkali ions (Li+, Na+ and K+). J. Alloy Compd. 2019, 786, 457–467. [Google Scholar] [CrossRef]
- Arppe, R.; Hyppänen, I.; Perälä, N.; Peltomaa, R.; Kaiser, M.; Würth, C.; Christ, S.; Resch-Genger, U.; Schäferling, M.; Soukka, T. Quenching of the upconversion luminescence of NaYF4:Yb3+, Er3+and NaYF4:Yb3+, Tm3+nanophosphors by water: The role of the sensitizer Yb3+in non-radiative relaxation. Nanoscale 2015, 7, 11746–11757. [Google Scholar] [CrossRef] [Green Version]
- Sedlmeier, A.; Gorris, H.H. Surface modification and characterization of photon-upconverting nanoparticles for bioanalytical applications. Chem. Soc. Rev. 2015, 44, 1526–1560. [Google Scholar] [CrossRef] [Green Version]
- Tong, R.; Yala, L.; Fan, T.M.; Cheng, J. The formulation of aptamer-coated paclitaxel–polylactide nanoconjugates and their targeting to cancer cells. Biomaterials 2010, 31, 3043–3053. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.-R.; Qiao, P. Drug Delivery in Cancer Therapy, Quo Vadis? Mol. Pharm. 2018, 15, 3603–3616. [Google Scholar] [CrossRef] [PubMed]
- Mirshafiee, V.; Kim, R.; Park, S.; Mahmoudi, M.; Kraft, M.L. Impact of protein pre-coating on the protein corona composition and nanoparticle cellular uptake. Biomaterials 2016, 75, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liu, Z. Albumin Carriers for Cancer Theranostics: A Conventional Platform with New Promise. Adv. Mater. 2016, 28, 10557–10566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.X.; Zhu, G.Y.; Lu, B.Y.; Zhang, C.L.; Peng, Q. Concentration-dependent protein adsorption at the nano-bio interfaces of polymeric nanoparticles and serum proteins. Nanomedicine 2017, 12, 2757–2769. [Google Scholar] [CrossRef]

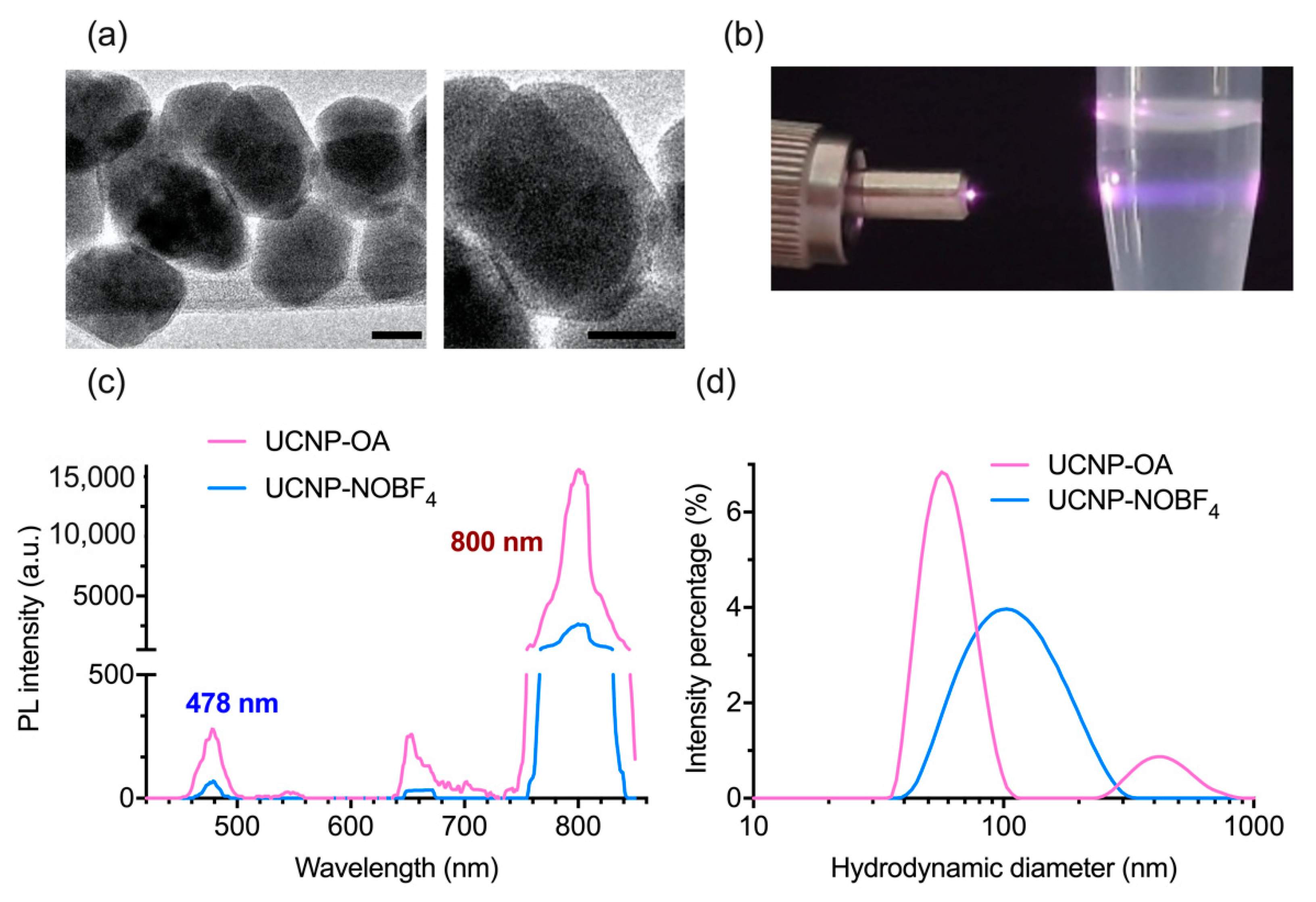
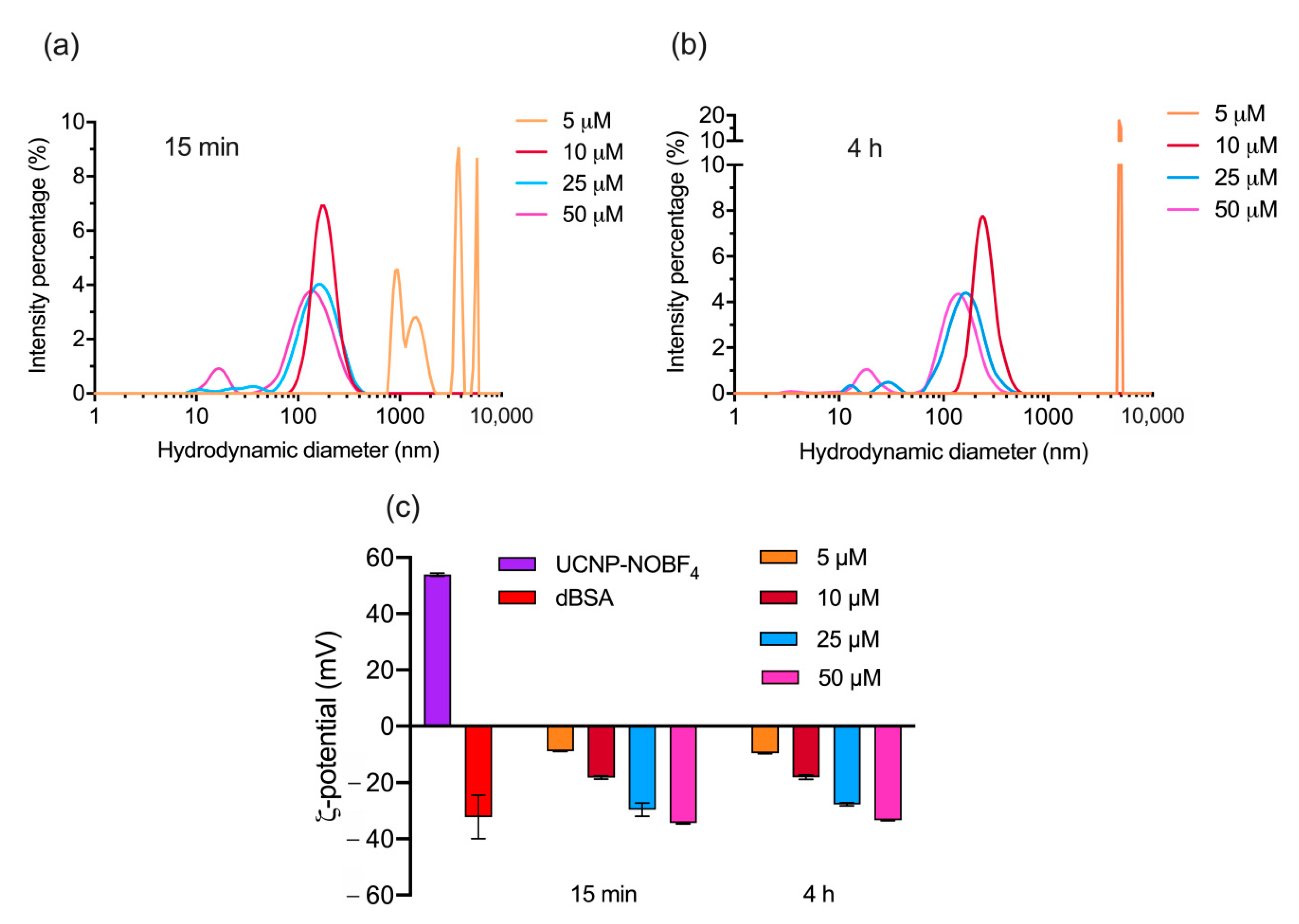
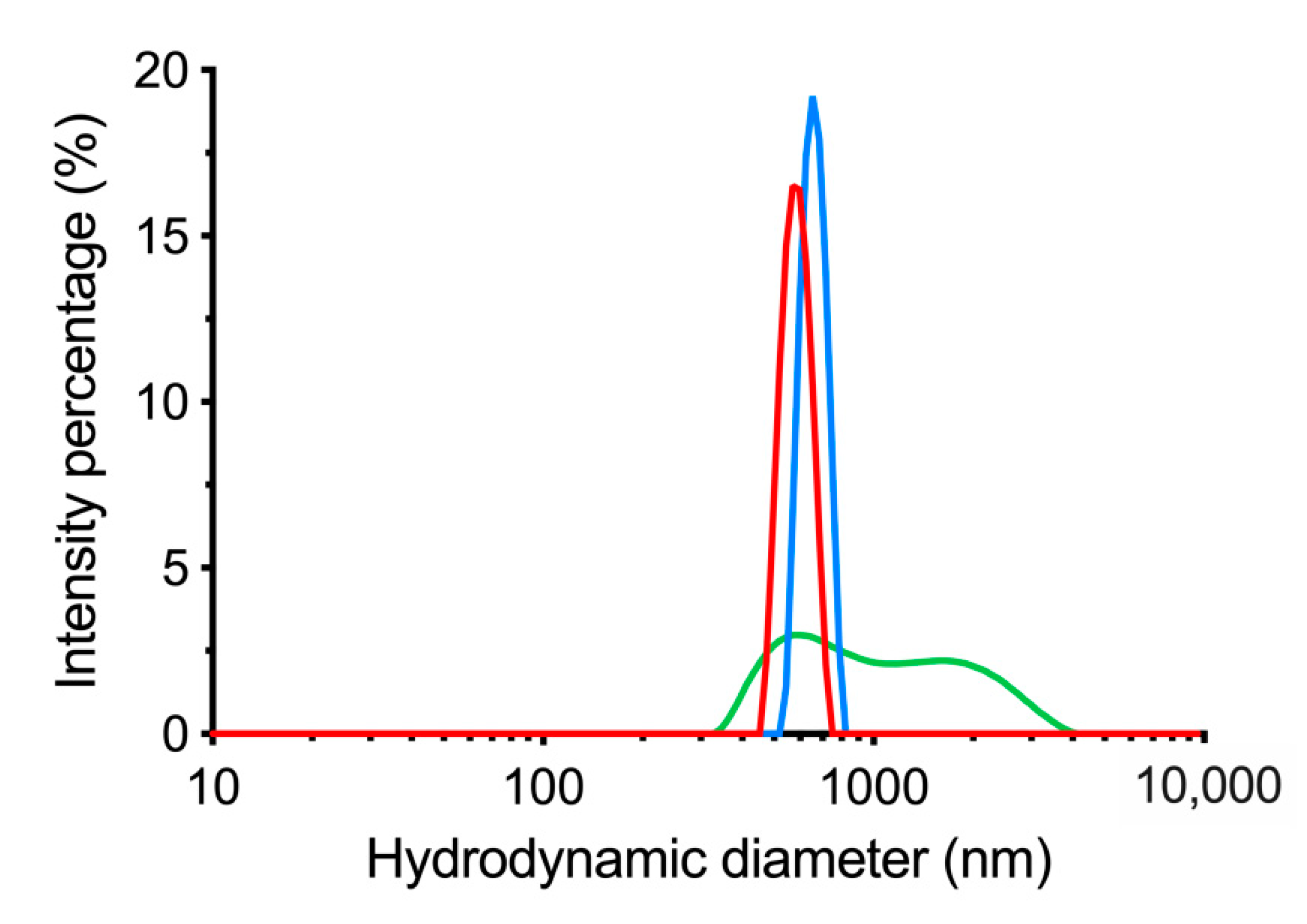

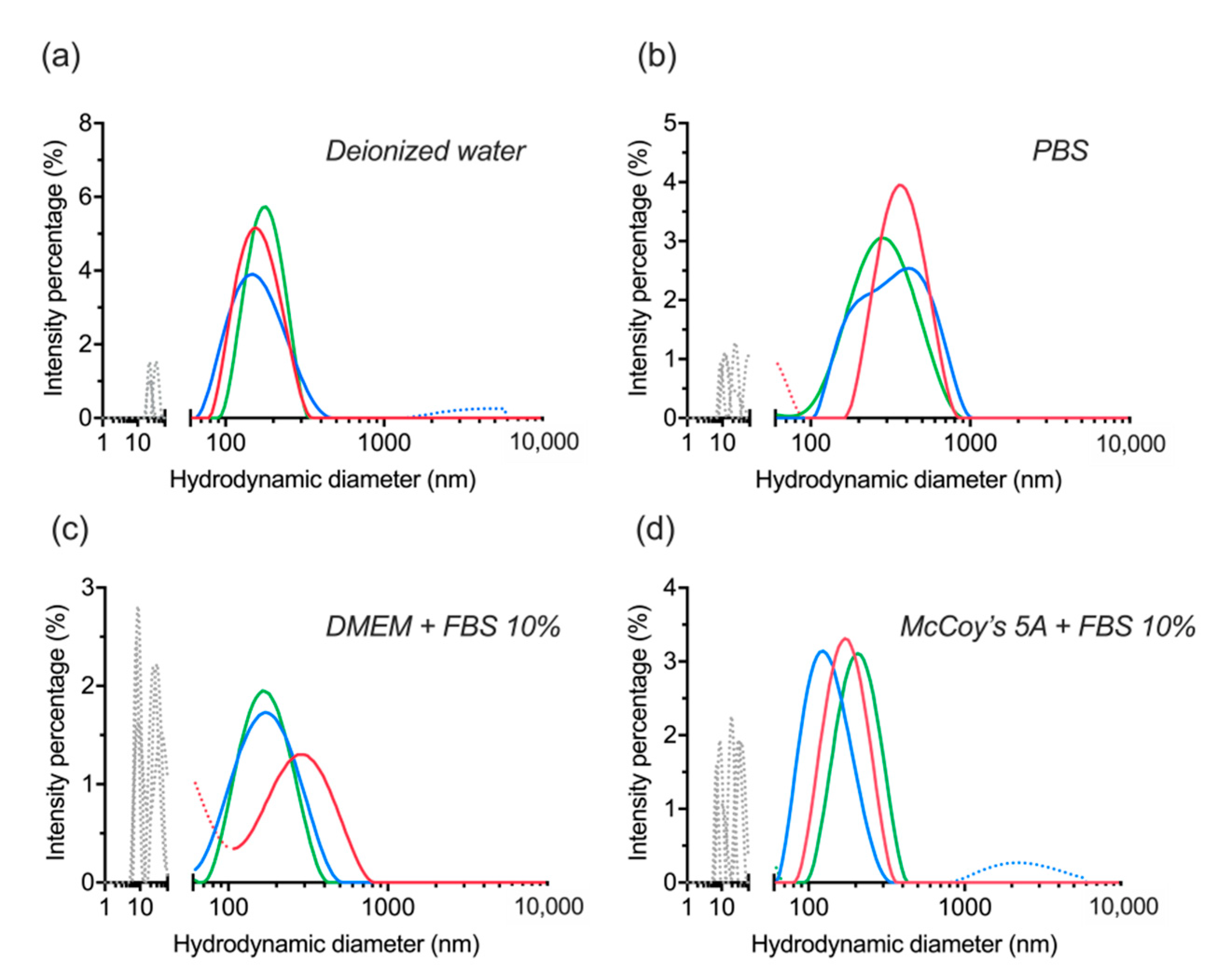
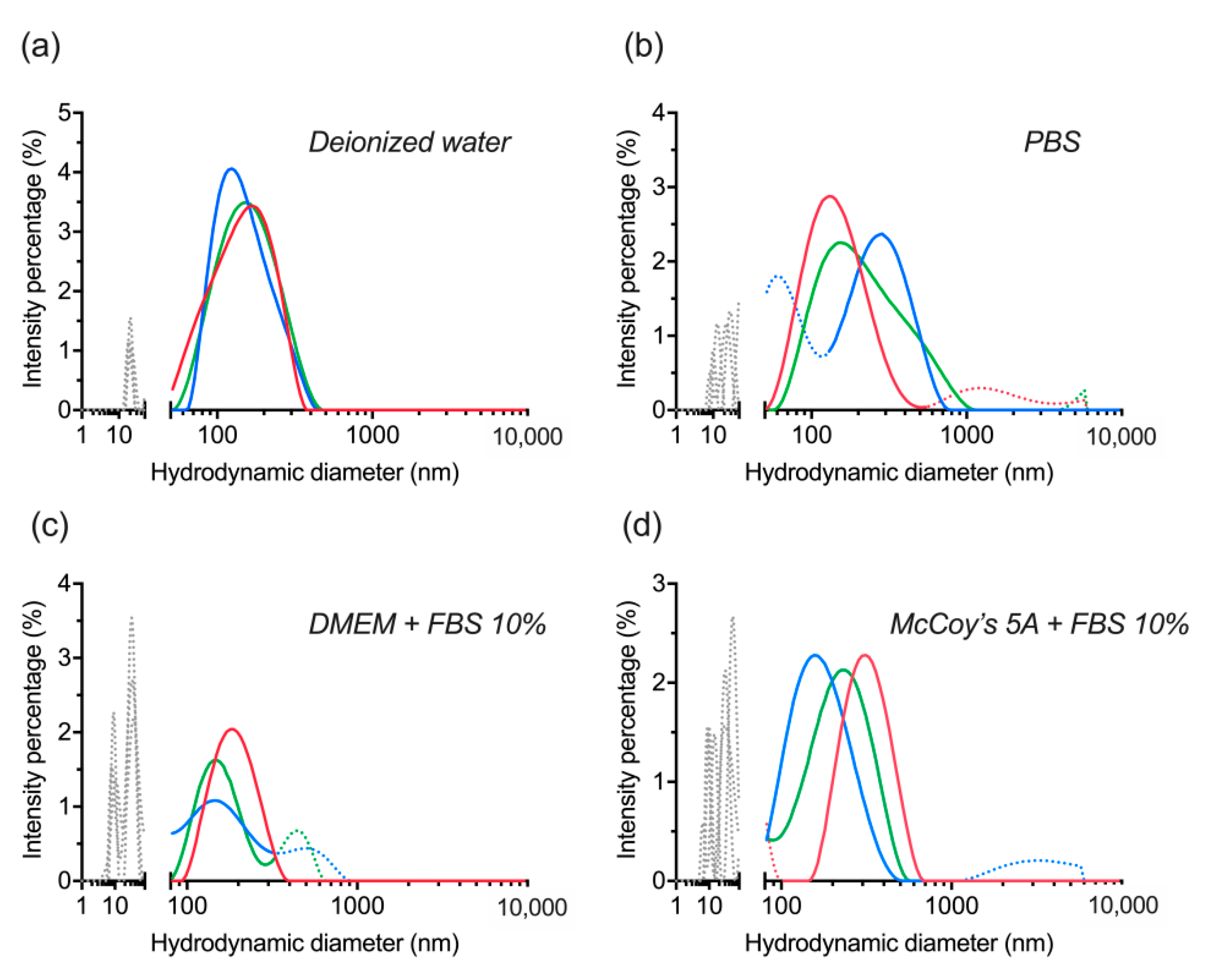
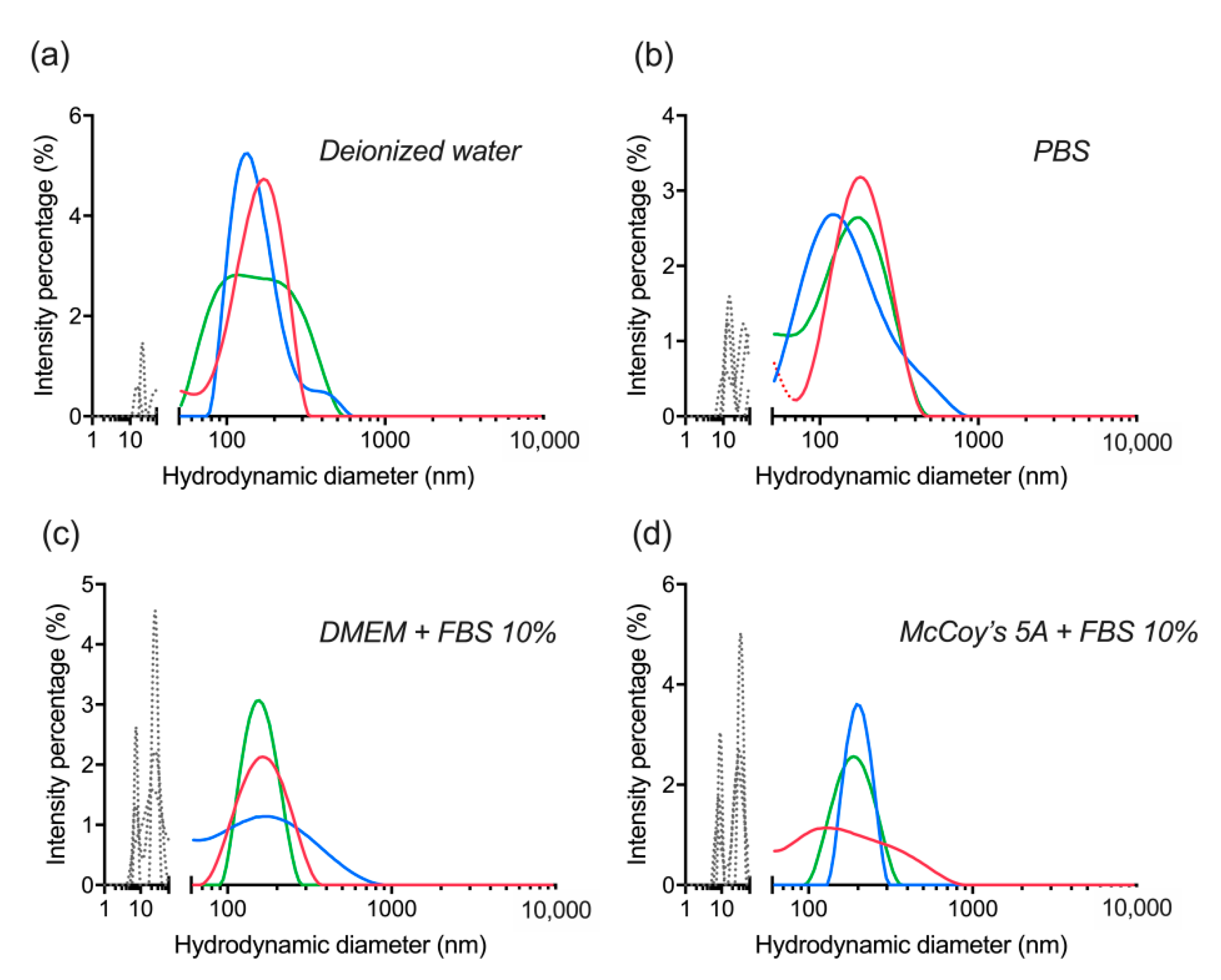
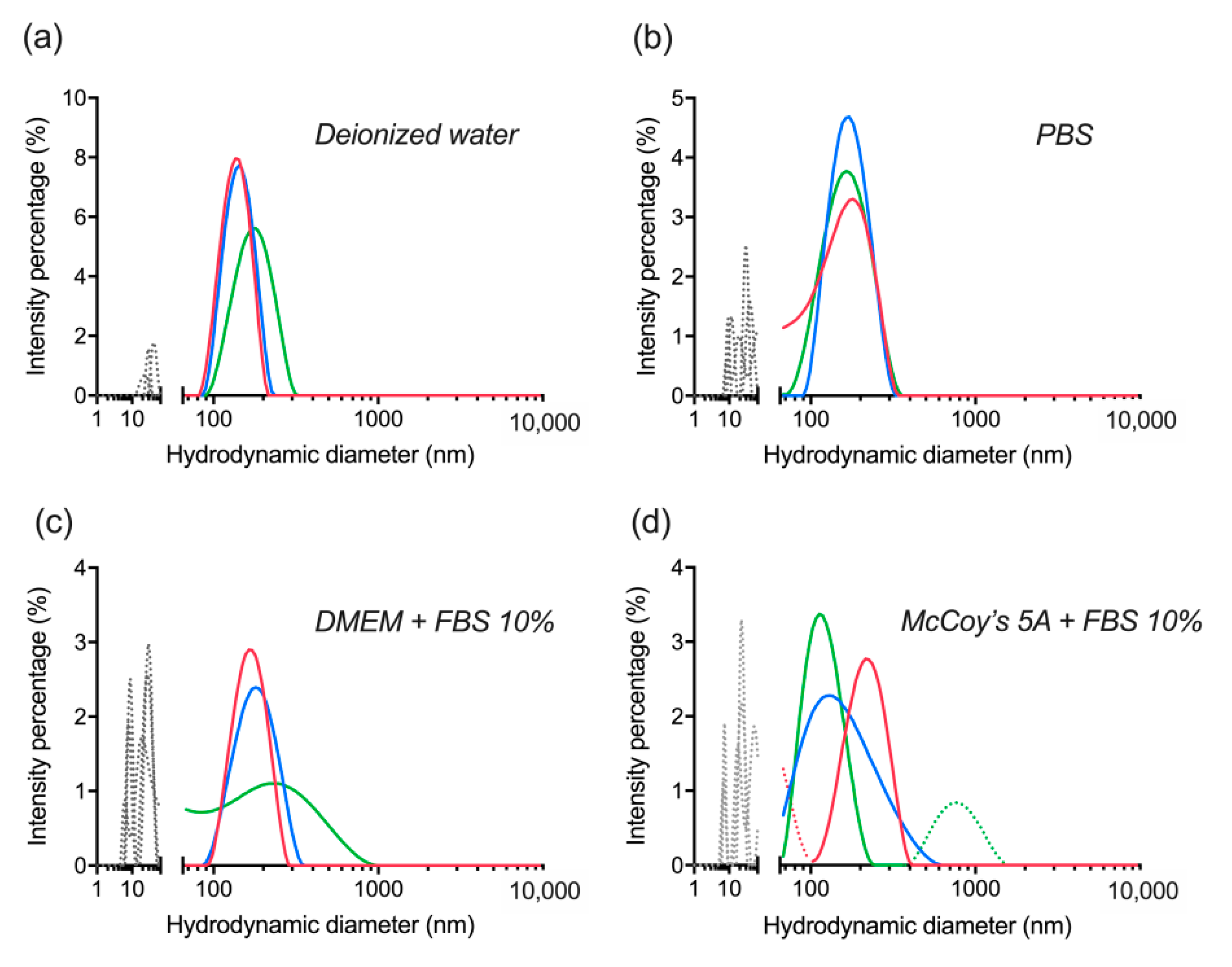
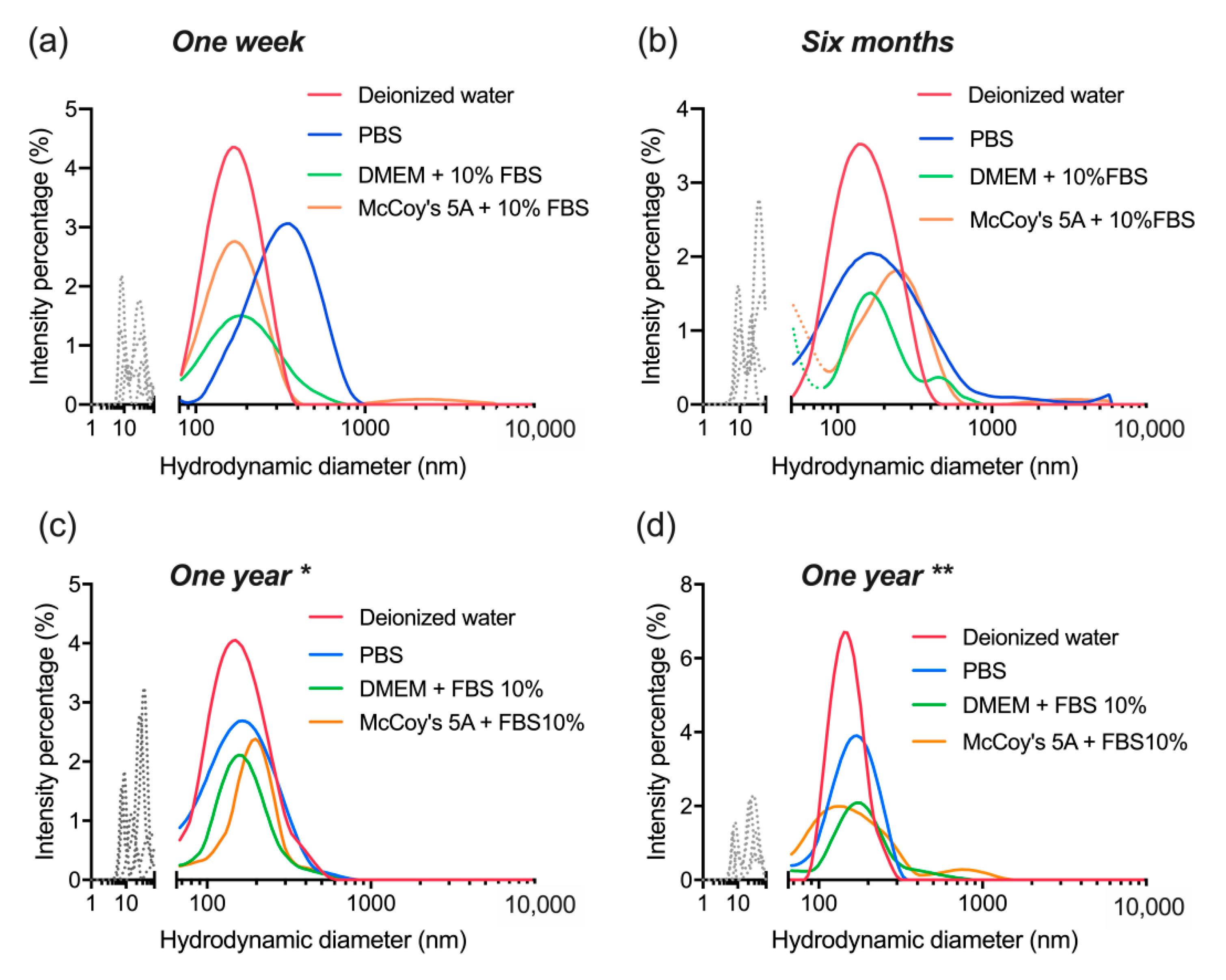

| UCNP | Average Size, nm | PDI |
|---|---|---|
| UCNP-OA | 67.94 ± 0.38 | 0.254 ± 0.011 |
| UCNP-NOBF4 | 101 ± 0.473 | 0.169 ± 0.011 |
| UCNP-NOBF4 (0.25 mg/mL) Incubated with dBSA | Average Size, nm | PDI |
|---|---|---|
| Incubation time: 15 min | ||
| 5 μM | 4755 ± 786.6 | 0.281 ± 0.159 |
| 10 μM | 176.2 ± 2.937 | 0.127 ± 0.006 |
| 25 μM | 112.3 ± 4.994 | 0.375 ± 0.071 |
| 50 μM | 80.27 ± 0.6862 | 0.56 ± 0.008 |
| Incubation time: 4 h | ||
| 5 μM | 7721 ± 383.9 | 0.13 ± 0.024 |
| 10 μM | 247 ± 5.918 | 0.139 ± 0.016 |
| 25 μM | 116.3 ± 2.468 | 0.333 ± 0.003 |
| 50 μM | 78.82 ± 0.8603 | 0.569 ± 0.005 |
| Centrifuged dBSA-UCNP-NOBF4 | ||
|---|---|---|
| Average Size, nm | PDI | ζ-Potential |
| 747.5 ± 76.71 | 0.292 ± 0.015 | +6.74 ± 1.21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shanwar, S.; Liang, L.; Nechaev, A.V.; Bausheva, D.K.; Balalaeva, I.V.; Vodeneev, V.A.; Roy, I.; Zvyagin, A.V.; Guryev, E.L. Controlled Formation of a Protein Corona Composed of Denatured BSA on Upconversion Nanoparticles Improves Their Colloidal Stability. Materials 2021, 14, 1657. https://doi.org/10.3390/ma14071657
Shanwar S, Liang L, Nechaev AV, Bausheva DK, Balalaeva IV, Vodeneev VA, Roy I, Zvyagin AV, Guryev EL. Controlled Formation of a Protein Corona Composed of Denatured BSA on Upconversion Nanoparticles Improves Their Colloidal Stability. Materials. 2021; 14(7):1657. https://doi.org/10.3390/ma14071657
Chicago/Turabian StyleShanwar, Samah, Liuen Liang, Andrey V. Nechaev, Daria K. Bausheva, Irina V. Balalaeva, Vladimir A. Vodeneev, Indrajit Roy, Andrei V. Zvyagin, and Evgenii L. Guryev. 2021. "Controlled Formation of a Protein Corona Composed of Denatured BSA on Upconversion Nanoparticles Improves Their Colloidal Stability" Materials 14, no. 7: 1657. https://doi.org/10.3390/ma14071657
APA StyleShanwar, S., Liang, L., Nechaev, A. V., Bausheva, D. K., Balalaeva, I. V., Vodeneev, V. A., Roy, I., Zvyagin, A. V., & Guryev, E. L. (2021). Controlled Formation of a Protein Corona Composed of Denatured BSA on Upconversion Nanoparticles Improves Their Colloidal Stability. Materials, 14(7), 1657. https://doi.org/10.3390/ma14071657








