Deep Learning-Based Reconstruction of 3D Morphology of Geomaterial Particles from Single-View 2D Images
Abstract
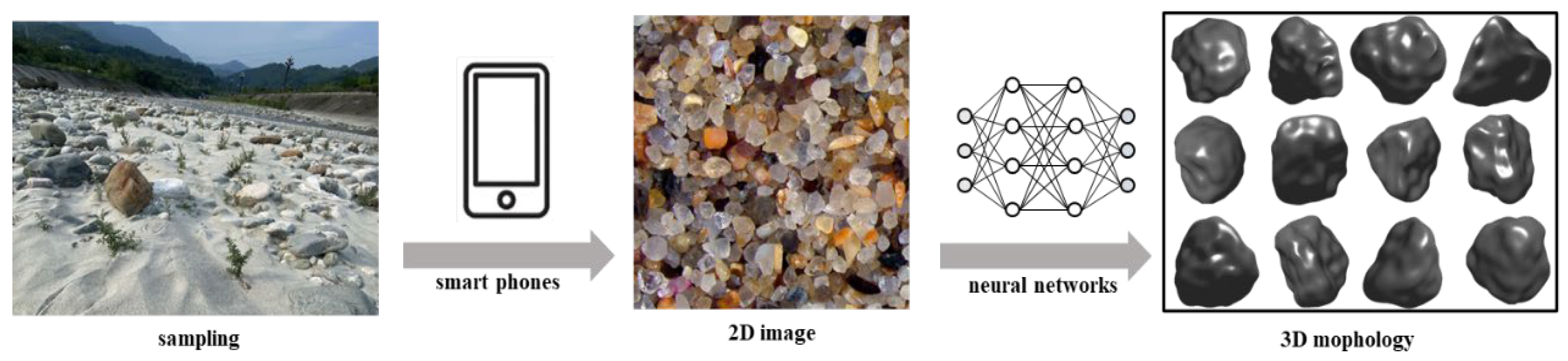
1. Introduction
2. Materials and Methods
2.1. Dataset
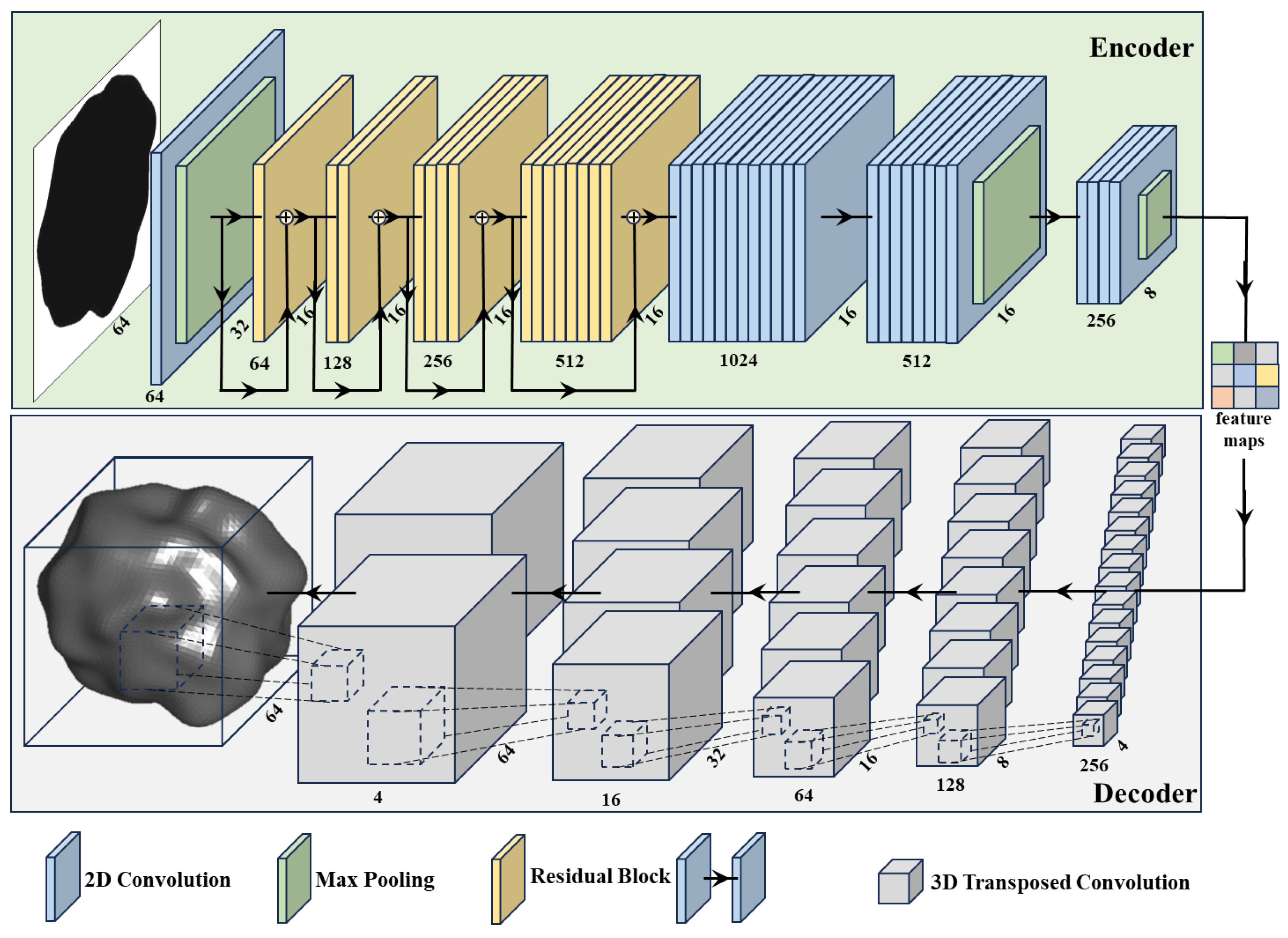
2.2. Network Architecture
2.2.1. Encoder
2.2.2. Decoder
2.3. Loss Function
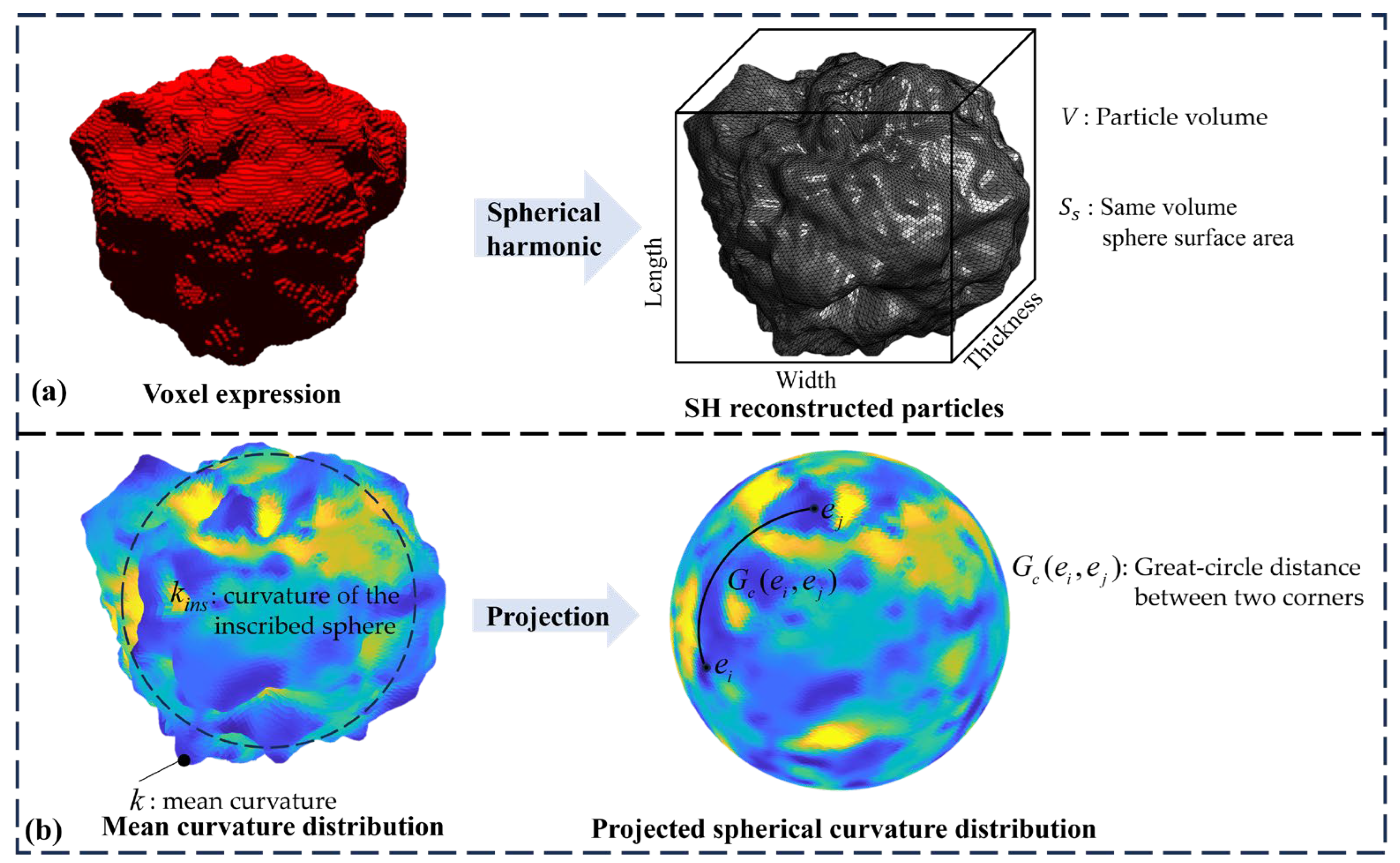
2.4. Evaluation Indicators
3. Results and Comparison with Other Models
3.1. Ablation Study
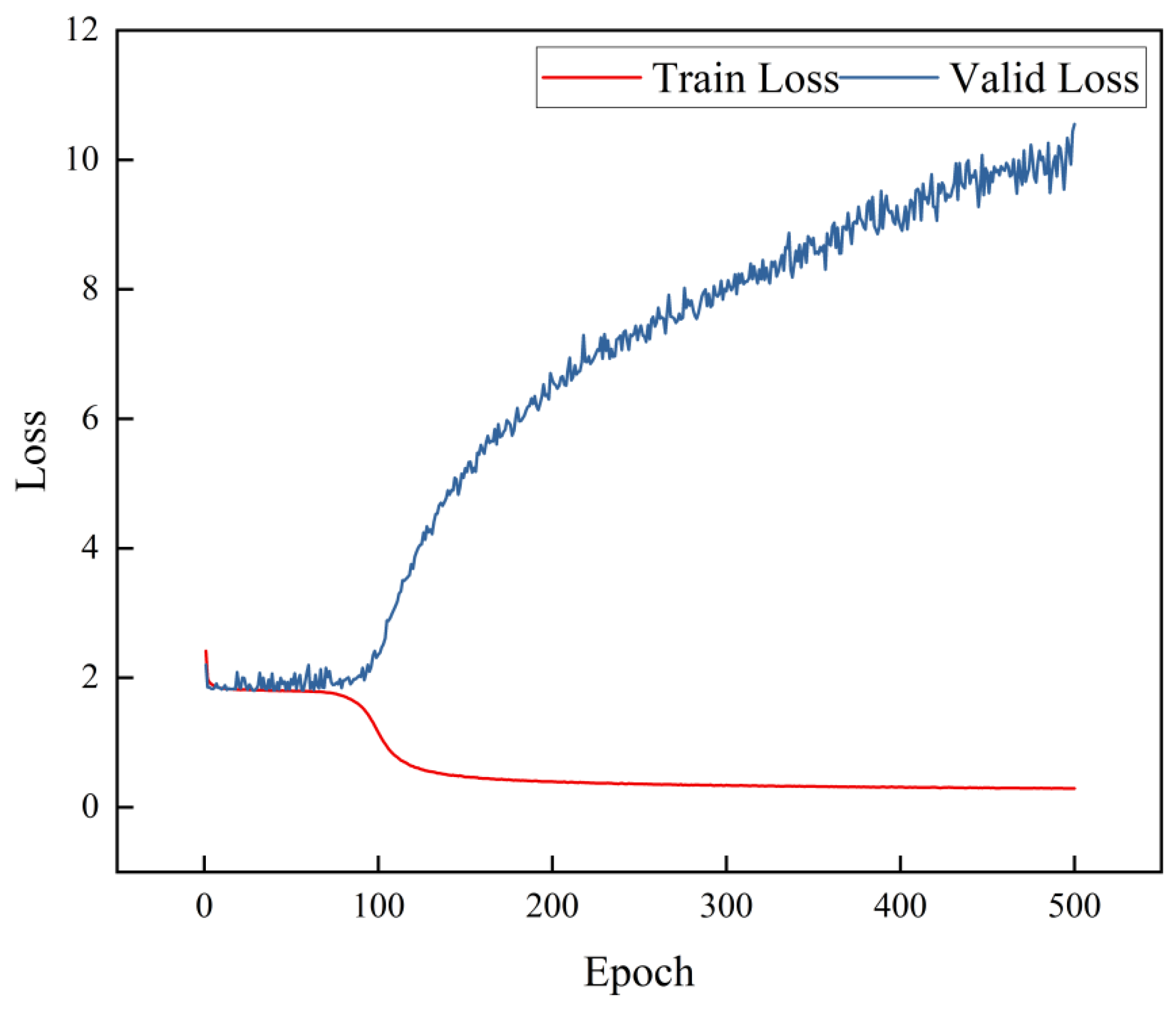
3.1.1. Loss Function
3.1.2. Learning Rate
3.2. Reconstruction Performance of Different Models
3.2.1. Lightweight Model
3.2.2. Comparison of Results from Different Models
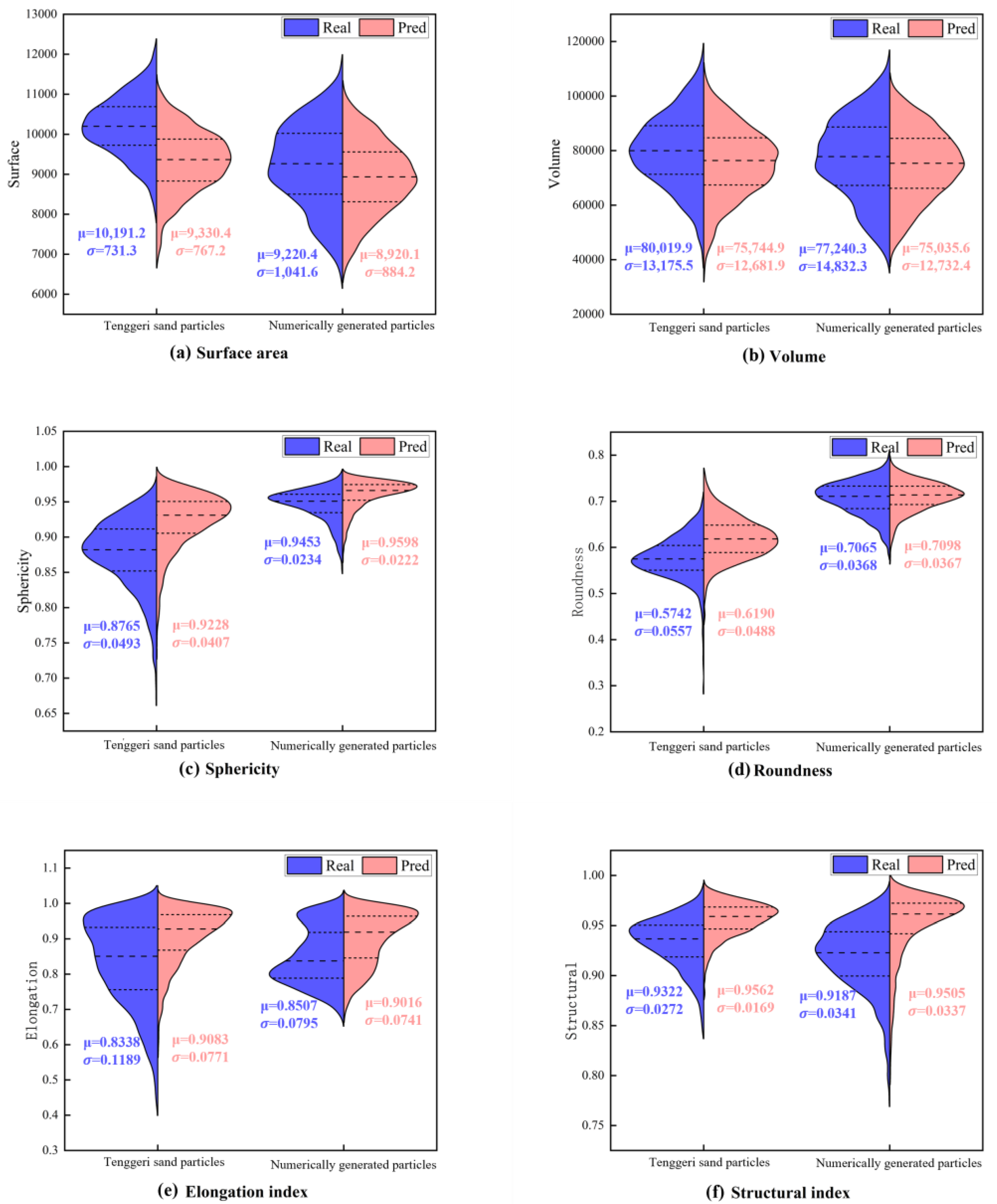
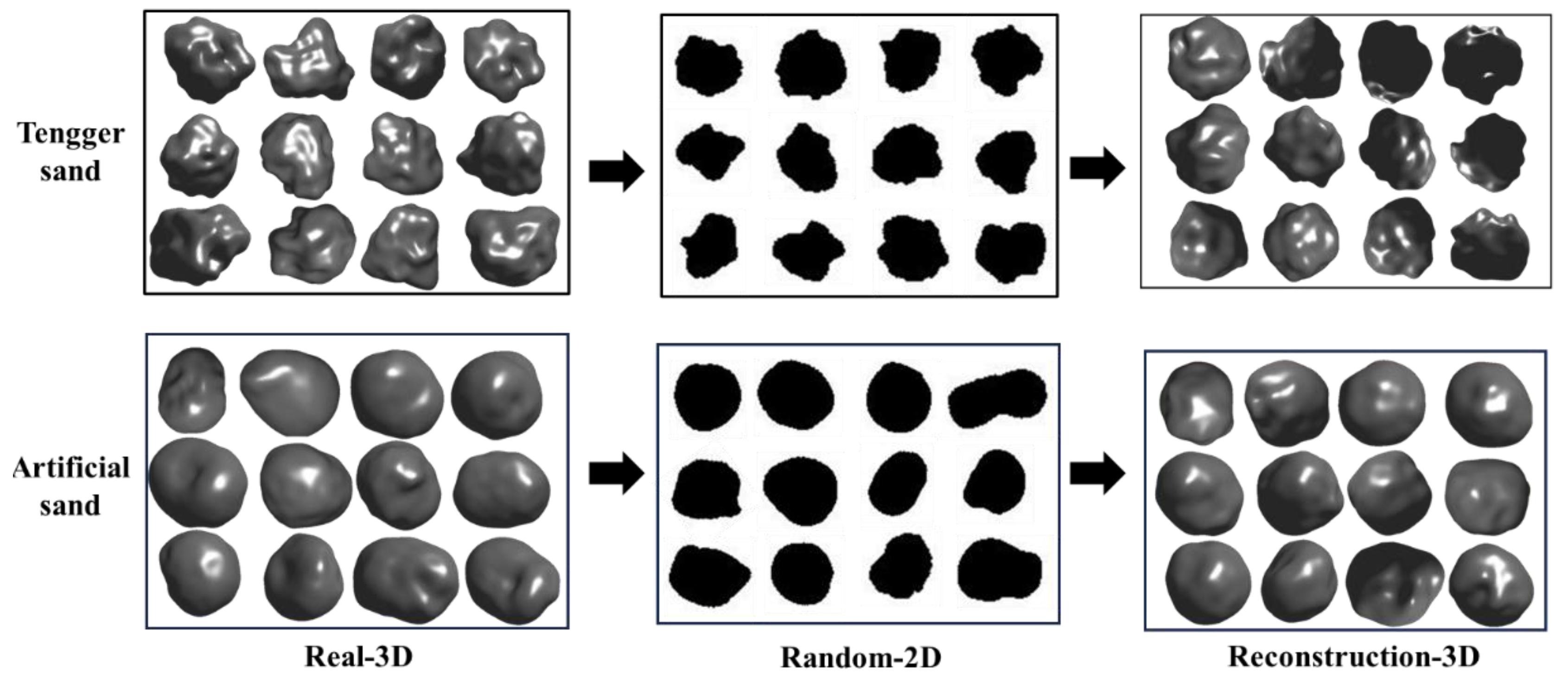
3.3. Reconstruction Results for Natural Sand Particles and Numerically Generated Digital Sand Particles
4. Discussion
5. Conclusions
- The distributions were similar for the reconstructed and real particles for the three sample types, indicating that upscaling from a single-view 2D image to 3D morphology was statistically feasible.
- The PVP model provided distributions of the reconstructed particles consistent with the real distributions. The surface area and volume were highly similar. The similarity between the distributions of the reconstructed and real particles for natural and numerically generated particles demonstrated the strong generalization ability of the model and its suitability for different particle types.
- Due to differences in formation, the reconstruction results were better for the HIT-LS1 lunar soil simulant than for the natural sand, but worse for the numerically generated sand particles, reflecting varying levels of difficulty for the AI model.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Altuhafi, F.N.; Coop, M.R.; Georgiannou, V.N. Effect of particle shape on the mechanical behavior of natural sands. J. Geotech. Geoenviron. Eng. 2016, 142, 04016071. [Google Scholar] [CrossRef]
- Deal, E.; Venditti, J.G.; Benavides, S.J.; Bradley, R.; Zhang, Q.; Kamrin, K.; Perron, J.T. Grain shape effects in bed load sediment transport. Nature 2023, 613, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Lawson, R.; Woods, S.; Jensen, E.; Erfani, E.; Gurganus, C.; Gallagher, M.; Connolly, P.; Whiteway, J.; Baran, A.; May, P.; et al. A review of ice particle shapes in cirrus formed in situ and in anvils. J. Geophys. Res. Atmos. 2019, 124, 10049–10090. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, S.; Luding, S. The role of particle shape in computational modelling of granular matter. Nat. Rev. Phys. 2023, 5, 505–525. [Google Scholar] [CrossRef]
- Bostanabad, R.; Zhang, Y.; Li, X.; Kearney, T.; Brinson, L.C.; Apley, D.W.; Liu, W.K.; Chen, W. Computational microstructure characterization and reconstruction: Review of the state-of-the-art techniques. Prog. Mater. Sci. 2018, 95, 1–41. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, J.; Zhao, B. Micromorphology characterization and reconstruction of sand particles using micro X-ray tomography and spherical harmonics. Eng. Geol. 2015, 184, 126–137. [Google Scholar] [CrossRef]
- Wyant, J.C. White light interferometry. In Holography: A Tribute to Yuri Denisyuk and Emmett Leith; SPIE: Warsaw, Poland, 2002; pp. 98–107. [Google Scholar]
- Ebrahim, M.A.B. 3D laser scanners’ techniques overview. Int. J. Sci. Res. 2015, 4, 323–331. [Google Scholar]
- Su, D.; Yan, W. Prediction of 3D size and shape descriptors of irregular granular particles from projected 2D images. Acta Geotech. 2020, 15, 1533–1555. [Google Scholar] [CrossRef]
- Kloss, C.; Goniva, C.; Hager, A.; Amberger, S.; Pirker, S. Models, algorithms and validation for opensource dem and cfd–dem. Prog. Comput. Fluid Dyn. Int. J. 2012, 12, 140–152. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Yin, Z.Y.; Su, D.; Liu, Z. Deep-learning-enhanced model reconstruction of realistic 3D rock particles by intelligent video tracking of 2D random particle projections. Acta Geotech. 2023, 18, 1407–1430. [Google Scholar] [CrossRef]
- Feng, J.; Teng, Q.; Li, B.; He, X.; Chen, H.; Li, Y. An end-to-end three-dimensional reconstruction framework of porous media from a single two-dimensional image based on deep learning. Comput. Methods Appl. Mech. Eng. 2020, 368, 113043. [Google Scholar] [CrossRef]
- Fu, J.; Xiao, D.; Li, D.; Thomas, H.R.; Li, C. Stochastic reconstruction of 3D microstructures from 2D cross-sectional images using machine learning-based characterization. Comput. Methods Appl. Mech. Eng. 2022, 390, 114532. [Google Scholar] [CrossRef]
- Holland, J.H. Genetic algorithms. Sci. Am. 1992, 267, 66–73. [Google Scholar] [CrossRef]
- Jaeger, H.M.; de Pablo, J.J. Perspective: Evolutionary design of granular media and block copolymer patterns. APL Mater. 2016, 4, 53209. [Google Scholar] [CrossRef]
- Miskin, M.Z.; Jaeger, H.M. Evolving design rules for the inverse granular packing problem. Soft Matter 2014, 10, 3708–3715. [Google Scholar] [CrossRef]
- Macedo, R.B.d.; Monfared, S.; Karapiperis, K.; Andrade, J. What is shape? characterizing particle morphology with genetic algorithms and deep generative models. Granul. Matter 2023, 25, 2. [Google Scholar] [CrossRef]
- Doersch, C. Tutorial on variational autoencoders. arXiv 2016, arXiv:1606.05908. [Google Scholar]
- Shi, J.J.; Zhang, W.; Wang, W.; Sun, Y.H.; Xu, C.Y.; Zhu, H.H.; Sun, Z.X. Randomly generating three-dimensional realistic schistous sand particles using deep learning: Variational autoencoder implementation. Eng. Geol. 2021, 291, 106235. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Jun, H.; Nichol, A. Shap-e: Generating conditional 3D implicit functions. arXiv 2023, arXiv:2305.02463. [Google Scholar]
- Liu, R.; Wu, R.; Van Hoorick, B.; Tokmakov, P.; Zakharov, S.; Vondrick, C. Zero-1-to-3: Zero-shot one image to 3D object. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–6 October 2023; pp. 9298–9309. [Google Scholar]
- Melas-Kyriazi, L.; Laina, I.; Rupprecht, C.; Vedaldi, A. Realfusion: 360deg reconstruction of any object from a single image. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Paris, France, 2–6 October 2023; pp. 8446–8455. [Google Scholar]
- Nichol, A.; Jun, H.; Dhariwal, P.; Mishkin, P.; Chen, M. Point-e: A system for generating 3D point clouds from complex prompts. arXiv 2022, arXiv:2212.08751. [Google Scholar]
- Long, X.; Guo, Y.C.; Lin, C.; Liu, Y.; Dou, Z.; Liu, L.; Ma, Y.; Zhang, S.H.; Habermann, M.; Theobalt, C.; et al. Wonder3D: Single image to 3D using cross-domain diffusion. arXiv 2023, arXiv:2310.15008. [Google Scholar]
- Fan, H.; Su, H.; Guibas, L.J. A point set generation network for 3D object reconstruction from a single image. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 605–613. [Google Scholar]
- Wang, N.; Zhang, Y.; Li, Z.; Fu, Y.; Liu, W.; Jiang, Y. Pixel2mesh: Generating 3D mesh models from single rgb images. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 52–67. [Google Scholar]
- Xu, Q.; Wang, W.; Ceylan, D.; Mech, R.; Neumann, U. Disn: Deep implicit surface network for high-quality single-view 3D reconstruction. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar] [CrossRef]
- Qian, G.; Mai, J.; Hamdi, A.; Ren, J.; Siarohin, A.; Li, B.; Lee, H.Y.; Skorokhodov, I.; Wonka, P.; Tulyakov, S.; et al. Magic123: One image to high-quality 3D object generation using both 2D and 3D diffusion priors. arXiv 2023, arXiv:2306.17843. [Google Scholar]
- Xie, H.; Yao, H.; Sun, X.; Zhou, S.; Zhang, S. Pix2vox: Context-aware 3D reconstruction from single and multi-view images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Replubic of Korea, 27 October–2 November 2019; pp. 2690–2698. [Google Scholar]
- Xie, H.; Yao, H.; Zhang, S.; Zhou, S.; Sun, W. Pix2vox++: Multi-scale context-aware 3D object reconstruction from single and multiple images. Int. J. Comput. Vis. 2020, 128, 2919–2935. [Google Scholar] [CrossRef]
- Deng, J.; Shi, S.; Li, P.; Zhou, W.; Zhang, Y.; Li, H. Voxel r-cnn: Towards high performance voxel-based 3D object detection. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual Conference, 2–9 February 2021; Volume 35, pp. 1201–1209. [Google Scholar]
- Shi, S.; Guo, C.; Jiang, L.; Wang, Z.; Shi, J.; Wang, X.; Li, H. Pv-rcnn: Point-voxel feature set abstraction for 3D object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10529–10538. [Google Scholar]
- Liu, Y.; Chen, Y.; Ding, B. Deep learning in frequency domain for inverse identification of nonhomogeneous material properties. J. Mech. Phys. Solids 2022, 168, 105043. [Google Scholar] [CrossRef]
- Wang, Y.; Chung, S.H.; Khan, W.A.; Wang, T. ALADA: A lite automatic data augmentation framework for industrial defect detection. Adv. Eng. Inform. 2023, 58, 102205. [Google Scholar] [CrossRef]
- Khan, W.A. Balanced weighted extreme learning machine for imbalance learning of credit default risk and manufacturing productivity. Ann. Oper. Res. 2023, 1–29. [Google Scholar] [CrossRef]
- Xie, H.; Wu, Q.; Liu, Y.; Xie, Y.; Gao, M.; Li, C. Direct measurement and theoretical prediction model of interparticle adhesion force between irregular planetary regolith particles. Int. J. Min. Sci. Technol. 2023, 33, 1425–1436. [Google Scholar] [CrossRef]
- Khan, W.A.; Masoud, M.; Eltoukhy, A.E.E.; Ullah, M. Stacked encoded cascade error feedback deep extreme learning machine network for manufacturing order completion time. J. Intell. Manuf. 2024, 1–27. [Google Scholar] [CrossRef]
- Simonyan, K. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Liu, Y.; Jeng, D.S.; Xie, H.; Li, C. On the particle morphology characterization of granular geomaterials. Acta Geotech. 2023, 18, 2321–2347. [Google Scholar] [CrossRef]
- Bagheri, G.; Bonadonna, C.; Manzella, I.; Vonlanthen, P. On the characterization of size and shape of irregular particles. Powder Technol. 2015, 270, 141–153. [Google Scholar] [CrossRef]
- Feng, Z.K.; Xu, W.J.; Lubbe, R. Three-dimensional morphological characteristics of particles in nature and its application for dem simulation. Powder Technol. 2020, 364, 635–646. [Google Scholar] [CrossRef]
- Xie, W.Q.; Zhang, X.P.; Yang, X.M.; Liu, Q.S.; Tang, S.H.; Tu, X.B. 3D size and shape characterization of natural sand particles using 2D image analysis. Eng. Geol. 2020, 279, 105915. [Google Scholar] [CrossRef]

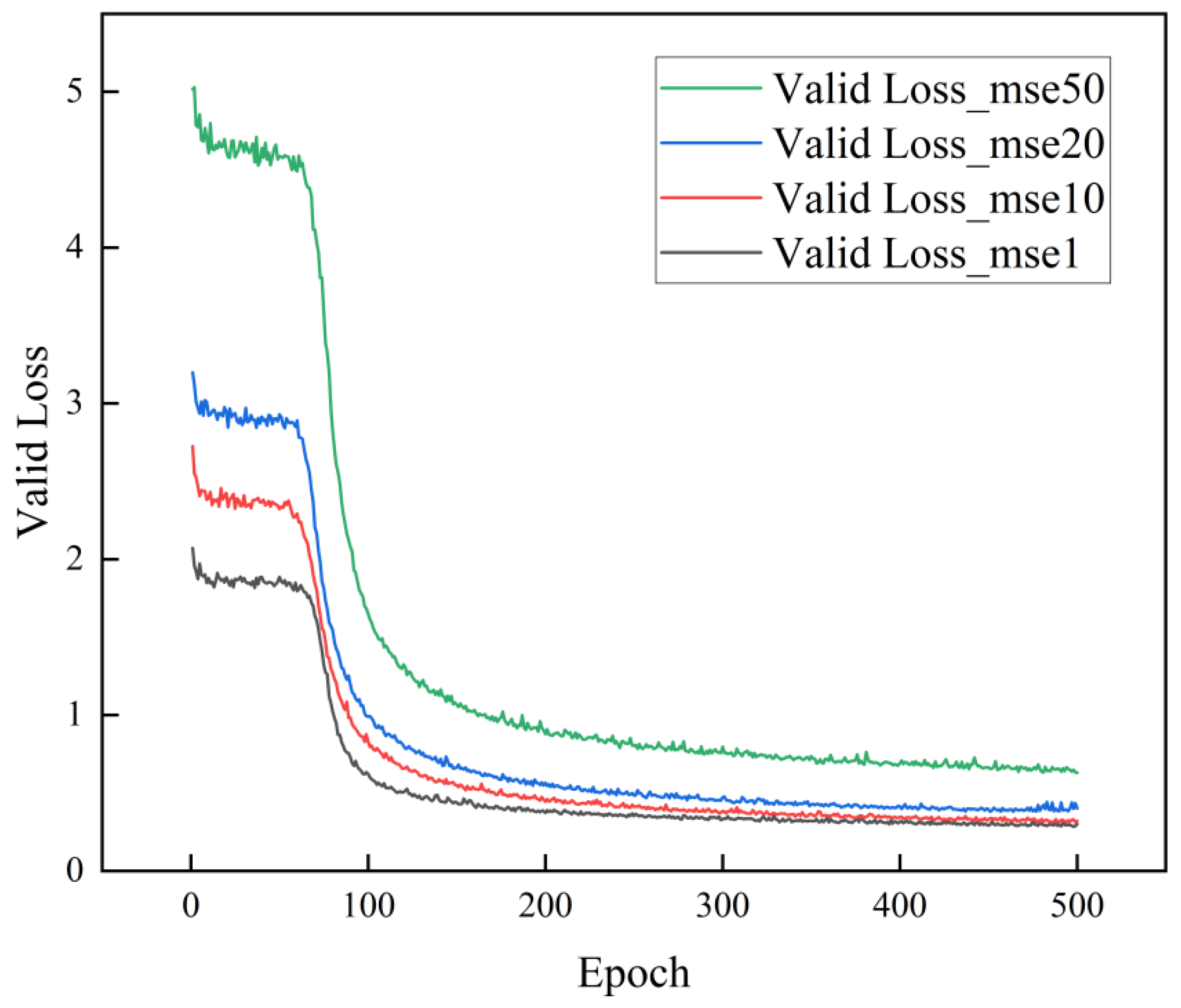
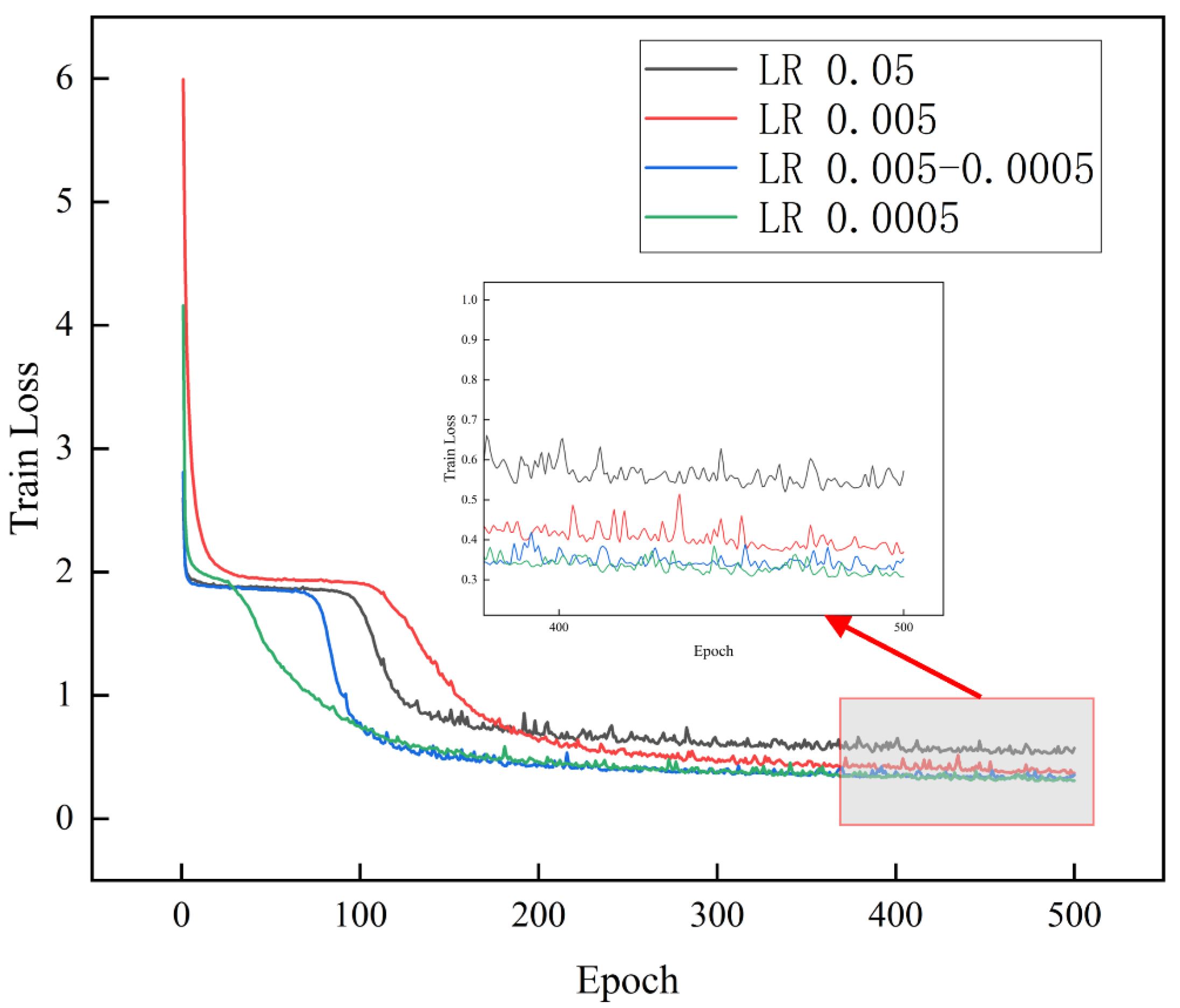

| Epoch | Learning Rate | Batch Size | Optimizer | Num_Workers |
|---|---|---|---|---|
| 500 | 0.005 | 256 | Adam | 12 |
| Surface Area | Volume | Sphericity | Roundness | Elongation Index | Structural Index | |
|---|---|---|---|---|---|---|
| PVP | 5.3% | 2.8% | 3.7% | 3.6% | 4.0% | 1.3% |
| PVP-S | 9.5% | 7.2% | 5.1% | 7.6% | 4.5% | 1.1% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Xie, H.; Li, C.; Liu, Y. Deep Learning-Based Reconstruction of 3D Morphology of Geomaterial Particles from Single-View 2D Images. Materials 2024, 17, 5100. https://doi.org/10.3390/ma17205100
Zhao J, Xie H, Li C, Liu Y. Deep Learning-Based Reconstruction of 3D Morphology of Geomaterial Particles from Single-View 2D Images. Materials. 2024; 17(20):5100. https://doi.org/10.3390/ma17205100
Chicago/Turabian StyleZhao, Jiangpeng, Heping Xie, Cunbao Li, and Yifei Liu. 2024. "Deep Learning-Based Reconstruction of 3D Morphology of Geomaterial Particles from Single-View 2D Images" Materials 17, no. 20: 5100. https://doi.org/10.3390/ma17205100
APA StyleZhao, J., Xie, H., Li, C., & Liu, Y. (2024). Deep Learning-Based Reconstruction of 3D Morphology of Geomaterial Particles from Single-View 2D Images. Materials, 17(20), 5100. https://doi.org/10.3390/ma17205100








