An Improved Squirrel Search Algorithm for Global Function Optimization
Abstract
1. Introduction
2. The Squirrel Search Algorithm
2.1. Initialize the Population
2.2. Classify the Population
2.3. Update the Position
2.4. Seasonal Transition Judgement and Random Updating
3. The improved Squirrel Search Algorithm
3.1. Motivation
| Algorithm 1. Pseudo Code of ISSA |
| Input:pop |
| Output:fbest (fbest is the best fitness value optimized by the algorithm) |
| for t = 1 to T (T is the total generation of the algorithm to be executed) |
| evaluate the fitness values of the population |
| update the population through the jumping search method introduced in Section 3.2 |
| if t == T/n (n is the total substages of the whole optimization, details in Section 3.4) |
| calculate the corresponding linear regression equations introduced in Section 3.4 |
| if two or more calculated slopes are positive |
| continue optimizing through the progressive search method introduced in Section 3.3 |
| else |
| continue optimizing through the jumping search method mentioned in Section 3.2 |
| end |
| end |
| end |
3.2. The Jumping Search Method
3.2.1. ‘Escape’ Operation in Winter
3.2.2. ‘Death’ Operation in Summer
3.2.3. Characters of the Jumping Search Method
| Algorithm 2. Pseudo Code of the Jumping Search Method |
| Input:pop |
| Output:popnew |
| if |
| season = winter |
| else |
| season = summer |
| end |
| if season == winter |
| forp1 = 1 to popsize (popsize is the total number of squirrels) |
| if |
| end |
| if |
| for p2 = 1 to nt1 (nt1 is the total number of threatened squirrels) |
| end |
| end |
| end |
| end |
| if season == summer |
| forp1 = 1 to popsize |
| if |
| end |
| if |
| for p2 = 1 to nt2 (nt2 is the total number of dead squirrels) |
| end |
| end |
| end |
3.3. The Progressive Search Method
3.3.1. The Principle of the Progressive Search Method
3.3.2. The Analysis of the Progressive Search Method
| Algorithm 3. Pseudo Code of the Progressive Search Method |
| Input:pop |
| Output:popnew |
| if |
| if |
| season = winter |
| else |
| season = summer |
| end |
| if season == winter |
| forp1 = 1 to popsize |
| end |
| if |
| select one dimension randomly and change it between FSL and FSH |
| end |
| end |
| end |
| if season == summer |
| forp1 = 1 to popsize |
| if |
| end |
| if |
| select one dimension randomly and change it between U and L |
| end |
| end |
3.4. Linear Regression Selection Strategy
4. Analysis of the Experimental Results
4.1. Benchmark Functions
4.2. The Influence of Parameter on ISSA
4.3. The Efficiency of the Proposed Methods
4.3.1. Comparison of Convergence Speed on Four Methods
4.3.2. Comparison of Population Diversity on Four Methods
4.3.3. Comparison of Comprehensive Performance on Four Methods
4.4. Performance Compared with Other Algorithms
- ISSA: n = 10; Nfs = 3;
- MDE: the crossover probability CR = 0.4, the mutation probability F is determined by the random number between 0 and 1.
- IGSA/PSO: the gravitational constant G0 = 100, α = 20;
- distABC: limit = (the population number × dimension)/2;
- ADN-RSN-PSO: the weight factor w = 0.7298, c1 = c2 = 2.05;
- PSO-GWO: the weight factor c1 = c2 = 2.05; aini = 2, afin = 0; r1, r2, r3, r4 are all the random numbers between 0 and 1.
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- He, J.; Lin, G. Average Convergence Rate of Evolutionary Algorithms. IEEE Trans. Evol. Comput. 2016, 20, 316–321. [Google Scholar] [CrossRef]
- Chugh, T.; Sindhya, K.; Hakanen, J. A survey on handling computationally expensive multiobjective optimization problems with evolutionary algorithms. Soft Comput. 2017, 23, 3137–3166. [Google Scholar] [CrossRef]
- Bhattacharyya, B.; Raj, S. Swarm intelligence based algorithms for reactive power planning with Flexible AC transmission system devices. Int. J. Electr. Syst. 2016, 78, 158–164. [Google Scholar] [CrossRef]
- Laina, R.; Lamzouri, F.E.-Z.; Boufounas, E.-M.; El Amrani, A.; Boumhidi, I. Intelligent control of a DFIG wind turbine using a PSO evolutionary algorithm. Procedia Comput. Sci. 2018, 127, 471–480. [Google Scholar] [CrossRef]
- Manjunath, P.G.C.; Krishna, P.; Parappagoudar, M.B.; Vundavilli, P.R. Multi-Objective Optimization of Squeeze Casting Process using Evolutionary Algorithms. IJSIR 2016, 7, 55–74. [Google Scholar]
- Ntouni, G.D.; Paschos, A.E.; Kapinas, V.M.; Karagiannidis, G.K.; Hadjileontiadis, L.J. Optimal detector design for molecular communication systems using an improved swarm intelligence algorithm. Micro Nano Lett. 2018, 13, 383–388. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, F.; Ma, Y.; Pota, H.R.; Shen, W. Evacuation path optimization based on quantum ant colony algorithm. Adv. Eng. Inform. 2016, 30, 259–267. [Google Scholar] [CrossRef]
- Yang, J.H.; Honavar, V. Feature Subset Selection Using a Genetic Algorithm. IEEE Intell. Syst. 1998, 13, 44–49. [Google Scholar] [CrossRef]
- Storn, R.; Price, K. Differential Evolution—A Simple and Efficient Heuristic for global Optimization over Continuous Spaces. J. Glob. Optim. 1997, 11, 341–359. [Google Scholar] [CrossRef]
- Hansen, N.; Ostermeier, A. Completely Derandomized Self-Adaptation in Evolution Strategies. Evol. Comput. 2001, 9, 159–195. [Google Scholar] [CrossRef]
- De Castro, L.N.; Von Zuben, F.J. Learning and Optimization Using the Clonal Selection Principle. IEEE Trans. Evol. Comput. 2002, 6, 239–251. [Google Scholar] [CrossRef]
- Xie, X.F.; Zhang, W.J.; Yang, Z.L. Social cognitive optimization for nonlinear programming problems. In Proceedings of the 2002 International Conference on Machine Learning and Cybernetics, Beijing, China, 4–5 November 2002; pp. 779–783. [Google Scholar]
- Ashrafi, S.M.; Dariane, A.B. A novel and effective algorithm for numerical optimization: Melody Search (MS). In Proceedings of the 2011 11th International Conference on Hybrid Intelligent Systems, Malacca, Malaysia, 5–8 December 2011; pp. 109–114. [Google Scholar]
- Rao, R.V.; Patel, V. An elitist teaching-learning-based optimization algorithm for solving complex constrained optimization problems. Int. J. Ind. Eng. Comput. 2012, 3, 535–560. [Google Scholar] [CrossRef]
- Cheng, M.Y.; Prayogo, D. Symbiotic Organisms Search: A new metaheuristic optimization algorithm. Comput. Struct. 2014, 139, 98–112. [Google Scholar] [CrossRef]
- Jahani, E.; Chizari, M. Tackling global optimization problems with a novel algorithm—Mouth Brooding Fish algorithm. Appl. Soft Comput. 2018, 62, 987–1002. [Google Scholar] [CrossRef]
- Kirkpatrick, S.; Gelatt, C.D., Jr.; Vecchi, M.P. Optimization by Simulated Annealing. Science 1983, 220, 671–680. [Google Scholar] [CrossRef]
- Rashedi, E.; Nezamabadi-Pour, H.; Saryazdi, S. GSA: A Gravitational Search Algorithm. Inf. Sci. 2009, 179, 2232–2248. [Google Scholar] [CrossRef]
- Mirjalili, S.; Sadiq, A.S. Magnetic Optimization Algorithm for training Multi Layer Perceptron. In Proceedings of the IEEE 3rd International Conference on Communication Software and Networks, Xi’an, China, 27–29 May 2011; pp. 42–46. [Google Scholar]
- Kaveh, A.; Khayatazad, M. A new meta-heuristic method: Ray Optimization. Comput. Struct. 2012, 112–113, 283–294. [Google Scholar] [CrossRef]
- Moein, S.; Logeswaran, R. KGMO: A swarm optimization algorithm based on the kinetic energy of gas molecule. Inf. Sci. 2014, 275, 127–144. [Google Scholar] [CrossRef]
- Kaveh, A.; Bakhshpoori, T. Water Evaporation Optimization: A novel physically inspired optimization algorithm. Comput. Struct. 2016, 167, 69–85. [Google Scholar] [CrossRef]
- Nematollahi, A.F.; Rahiminejad, A.; Vahidi, B. A novel physical based meta-heuristic optimization method known as Lightning Attachment Procedure Optimization. Appl. Soft Comput. 2017, 59, 596–621. [Google Scholar] [CrossRef]
- Colorni, A. Distributed Optimization by Ant Colonies. In Proceedings of the 1st European Conference on Artificial Life, Paris, France, 11–13 December 1991; pp. 134–142. [Google Scholar]
- Kennedy, J.; Eberhart, R.C. Particle swarm optimization. In Proceedings of the IEEE International Conference on Neural Networks, Perth, Australia, 27 November–1 December 1995; pp. 1942–1948. [Google Scholar]
- Basturk, B.; Karaboga, D. An artificial bee colony (ABC) algorithm for numeric function optimization. In Proceedings of the Swarm Intelligence Symposium, Indianapolis, IN, USA, 12–14 May 2006; pp. 687–697. [Google Scholar]
- Cuevas, E.; Cienfuegos, M. A swarm optimization algorithm inspired in the behavior of the social-spider. Expert. Syst. Appl. 2013, 40, 6374–6384. [Google Scholar] [CrossRef]
- Fausto, F.; Cuevas, E.; Valdivia, A. A global optimization algorithm inspired in the behavior of selfish herds. Biosystems 2017, 160, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, C.; Huang, H.; Tang, S.; Dong, K. An optimization method: Hummingbirds optimization algorithm. J. Syst. Eng. Electron. 2018, 29, 168–186. [Google Scholar] [CrossRef]
- Jain, M.; Singh, V.; Rani, A. A novel nature-inspired algorithm for optimization: Squirrel search algorithm. Swarm Evol. Comput. 2018, 44, 148–175. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, M. An improved genetic algorithm encoded by adaptive degressive ary number. Soft Comput. 2018, 22, 6861–6875. [Google Scholar] [CrossRef]
- Gomes, W.C.; dos Santos Filho, R.C.; de Sales Junior, C.D.S. An Improved Artificial Bee Colony Algorithm with Diversity Control. In Proceedings of the 2018 Brazilian Conference on Intelligent Systems, Sao Paulo, Brazil, 22–25 October 2018; pp. 19–24. [Google Scholar]
- Wang, S.; Li, Y.; Yang, H. Self-adaptive differential evolution algorithm with improved mutation strategy. Appl. Intell. 2017, 47, 644–658. [Google Scholar] [CrossRef]
- Kiran, M.S.; Hakli, H.; Gunduz, M. Artificial bee colony algorithm with variable search strategy for continuous optimization. Inf. Sci. 2015, 300, 140–157. [Google Scholar] [CrossRef]
- Feng, X.; Xu, H.; Wang, Y. The social team building optimization algorithm. Appl. Soft Comput. 2018, 2, 1–22. [Google Scholar] [CrossRef]
- Demišar, J.; Schuurmans, D. Statistical Comparisons of Classifiers over Multiple Data Sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Li, X.; Yin, M. Modified differential evolution with self-adaptive parameters method. J. Comb. Optim. 2016, 31, 546–576. [Google Scholar] [CrossRef]
- Xiao, J.; Niu, Y.; Chen, P. An improved gravitational search algorithm for green partner selection in virtual enterprises. Neurocomputing 2016, 217, 103–109. [Google Scholar] [CrossRef]
- Babaoglu, I. Artificial bee colony algorithm with distribution-based update rule. Appl. Soft Comput. 2015, 34, 851–861. [Google Scholar] [CrossRef]
- Sun, W.; Lin, A.; Yu, H. All-dimension neighborhood based particle swarm optimization with randomly selected neighbors. Inf. Sci. 2017, 405, 141–156. [Google Scholar] [CrossRef]
- Teng, Z.J.; Lv, J.L.; Guo, L.W. An improved hybrid grey wolf optimization algorithm. Soft Comput. 2018, 22, 1–15. [Google Scholar] [CrossRef]






| Name | Function | D | Range | Optimal |
|---|---|---|---|---|
| F1 Easom | 2 | [−100,100] | −1 | |
| F2 Matyas | 2 | [−10,10] | 0 | |
| F3 Bohachevsky1 | 2 | [−100,100] | 0 | |
| F4 Bohachevsky2 | 2 | [−100,100] | 0 | |
| F5 Bohachevsky3 | 2 | [−100,100] | 0 | |
| F6 Booth | 2 | [−10,10] | 0 | |
| F7 Michalewicz2 | 2 | −1.8013 | ||
| F8 Schaffer | 2 | [−100,100] | 0 | |
| F9 Michalewicz5 | 5 | −4.6877 | ||
| F10 Michalewicz10 | 10 | −9.6602 | ||
| F11 Zakharov | 30/50/100 | [−5,10] | 0 | |
| F12 Sphere | 30/50/100 | [−100,100] | 0 | |
| F13 SumSquares | 30/50/100 | [−10,10] | 0 | |
| F14 Schwefel 1.2 | 30/50/100 | [−100,100] | 0 | |
| F15 Schwefel 2.21 | 30/50/100 | [−100,100] | 0 | |
| F16 Schwefel 2.22 | 30/50/100 | [−10,10] | 0 | |
| F17 Elliptic | 30/50/100 | [−100,100] | 0 | |
| F18 Griewank | 30/50/100 | [−600,600] | 0 | |
| F19 Salomon | 30/50/100 | [−100,100] | 0 | |
| F20 Alpine | 30/50/100 | [−10,10] | 0 | |
| F21 Powell | 32/52/100 | [4,5] | 0 |
| F | n | ||||
|---|---|---|---|---|---|
| 0 | 5 | 10 | 15 | 20 | |
| F2 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| F6 | 6.2502e−04 ± 1.9021e−04 | 1.7129e−10 ± 9.2634e−11 | 3.5323e−08 ± 1.2626e−09 | 7.7188e−07 ± 2.8698e−08 | 7.5656e−07 ± 1.9151e−08 |
| F10 | −7.6632 ± 0.7985 | −9.4261 ± 0.0680 | −9.2260 ± 0.7780 | −8.1919 ± 1.3054 | −8.0299 ± 1.1781 |
| F16 | 6.2909e−163 ± 0 | 7.4432e−162 ± 0 | 1.0302e−164 ± 0 | 2.0455e−165 ± 0 | 7.1391e−164 ± 0 |
| F17 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| F18 | 1.8504e−15 ± 1.0135e−15 | 4.4095e−08 ± 1.9301e−07 | 2.1945e−15 ± 8.2481e−16 | 2.9976e−15 ± 6.5204e−15 | 3.1826e−15 ± 7.1639e−15 |
| Fun Accuracy | Method | ||||
|---|---|---|---|---|---|
| SSA | Jumping Search | Progressive Search | ISSA | ||
| F2 | 10−6 | 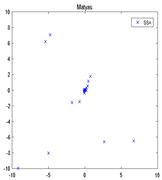 |  |  |  |
| 10−8 |  |  |  |  | |
| 10−10 |  |  |  |  | |
| F5 | 10−6 |  |  |  |  |
| 10−8 |  |  |  |  | |
| 10−10 |  |  |  |  | |
| Name | Method | |||
|---|---|---|---|---|
| SSA | Jumping Search Method | Progressive Search Method | ISSA | |
| F11 | 1.2837e+05 ± 7.3449e+04 | 2.5981e−04 ± 2.4867e−04 | 1.1115e+04 ± 3.3201e+03 | 0.0017 ± 0.0015 |
| F12 | 5.7955e+04 ± 2.1202e+04 | 7.1891e−05 ± 4.6142e−05 | 1.3107e+03 ± 543.9195 | 7.2366e−04 ± 6.0848e−04 |
| F20 | / | 6.0584e−05 ± 2.5350e−05 | 1.2767 ± 0.2946 | 2.2921e−04 ± 1.7366e−04 |
| F21 | / | 8.5775e−05 ± 8.4886e−05 | 7.2621e+04 ± 6.0527e+04 | 6.2753e−04 ± 4.7156e−04 |
| Name | Method | |||
|---|---|---|---|---|
| SSA | Jumping Search Method | Progressive Search Method | ISSA | |
| F1 | −1 ± 0 (=) | −1 ± 1.0909e−16 (=) | −1 ± 0 (=) | −1 ± 3.9171e−16 |
| F2 | 4.4212e−21 ± 1.5032e−20 (−) | 0 ± 0 (=) | 9.7986e−149 ± 5.3669e−148 (−) | 0 ± 0 |
| F3 | 7.4015e−18 ± 4.0540e−17 (−) | 0 ± 0 (=) | 0 ± 0 (=) | 0 ± 0 |
| F4 | 1.0191e−20 ± 2.9625e−20 (−) | 0 ± 0 (=) | 0 ± 0 (=) | 0 ± 0 |
| F5 | 1.2019e−20 ± 4.4285e−20 (−) | 0 ± 0 (=) | 0 ± 0 (=) | 0 ± 0 |
| F6 | 1.4402e−18 ± 3.0624e−18 (+) | 0.0202 ± 0.0238 (−) | 2.6295e−33 ± 1.4403e−32 (+) | 4.9331e−12 ± 1.6068e−11 |
| F7 | −1.8468 ± 0.0838 (−) | −1.7911 ± 0.0116 (−) | −1.8013 ± 6.8344e−16 (=) | −1.8013 ± 8.5739e−16 |
| F8 | 0 ± 0 (=) | 0 ± 0 (=) | 0 ± 0 (=) | 0 ± 0 |
| F9 | −4.6459 ± 2.7201e−15 (−) | −4.4543 ± 0.1818 (−) | −4.6808 ± 0.1336 (+) | −4.6617 ± 0.0304 |
| F10 | −9.5333 ± 0.2598 (−) | −8.2037 ± 0.3744 (−) | −9.6602 ± 0 (+) | −9.5882 ± 0.0891 |
| F11 | 2.7472e−13 ± 1.1640e−12 (−) | 0 ± 0 (=) | 2.1318e−16 ± 1.1676e−15 (−) | 0 ± 0 |
| F12 | 8.0478e−13 ± 4.3612e−12 (−) | 0 ± 0 (=) | 1.9301e−15 ± 1.0572e−14 (−) | 0 ± 0 |
| F13 | 2.0435e−10 ± 1.0129e−09 (−) | 0 ± 0 (=) | 3.5232e−11 ± 1.9297e−10 (−) | 0 ± 0 |
| F14 | 0.5472 ± 2.7161 (−) | 0 ± 0 (=) | 0.0865 ± 0.4149 (−) | 0 ± 0 |
| F15 | 7.2194e−08 ± 6.4247e−08 (−) | 0 ± 0 (=) | 0.0241 ± 0.1321 (−) | 0 ± 0 |
| F16 | 0.0978 ± 0.4017 (−) | 0 ± 0 (=) | 1.4322e−11 ± 7.8443e−11 (−) | 0 ± 0 |
| F17 | 9.7812e+03 ± 3.5168e+04 (−) | 0 ± 0 (=) | 2.3545e−15 ± 1.2896e−14 (−) | 0 ± 0 |
| F18 | 0.6323 ± 0.3594 (−) | 0 ± 0 (+) | 0.0129 ± 0.0650 (−) | 1.9614e−16 ± 3.6843e−16 |
| F19 | 0.1099 ± 0.0548 (−) | 0 ± 0 (=) | 0.0233 ± 0.0773 (−) | 0 ± 0 |
| F20 | 0.0025 ± 0.0109 (−) | 0 ± 0 (=) | 9.1222e−13 ± 4.9964e−12 (−) | 0 ± 0 |
| F21 | 8.6605 ± 38.6332 (−) | 0 ± 0 (=) | 7.3093e−05 ± 2.2255e−05 (−) | 0 ± 0 |
| Function | Rank | |||
|---|---|---|---|---|
| SSA | Jumping Search | Progressive Search | ISSA | |
| F1 | 1.5 | 3 | 1.5 | 4 |
| F2 | 4 | 1.5 | 3 | 1.5 |
| F3 | 4 | 2 | 2 | 2 |
| F4 | 4 | 2 | 2 | 2 |
| F5 | 4 | 2 | 2 | 2 |
| F6 | 2 | 4 | 1 | 3 |
| F7 | 4 | 3 | 1 | 2 |
| F8 | 2.5 | 2.5 | 2.5 | 2.5 |
| F9 | 3 | 4 | 1 | 2 |
| F10 | 3 | 4 | 1 | 2 |
| F11 | 4 | 1.5 | 3 | 1.5 |
| F12 | 4 | 1.5 | 3 | 1.5 |
| F13 | 4 | 1.5 | 3 | 1.5 |
| F14 | 4 | 1.5 | 3 | 1.5 |
| F15 | 3 | 1.5 | 4 | 1.5 |
| F16 | 4 | 1.5 | 3 | 1.5 |
| F17 | 4 | 1.5 | 3 | 1.5 |
| F18 | 4 | 1 | 3 | 2 |
| F19 | 4 | 1.5 | 3 | 1.5 |
| F20 | 4 | 1.5 | 3 | 1.5 |
| F21 | 4 | 1.5 | 3 | 1.5 |
| Total rank | 75 | 44 | 51 | 40 |
| Average rank | 3.5714 | 2.0952 | 2.4286 | 1.9048 |
| Sort | 1 | 3 | 2 | 4 |
| i | Algorithm | z = (Ri − R4)/ = (Ri − R4)/ = (Ri − R4)/0.3984 | Pi | |
|---|---|---|---|---|
| 1 | SSA | (3.5714 − 1.9048)/0.3984 = 4.1832 | 2e−05 | 0.0167 |
| 2 | progressive search | (2.4286 − 1.9048)/0.3984 = 1.3148 | 0.1885 | 0.0250 |
| 3 | jumping search | (2.0952 − 1.9048)/0.3984 = 0.4779 | 0.6348 | 0.0500 |
| Name | Method | Best | Worst | Mean | SD | R |
|---|---|---|---|---|---|---|
| F1 | Modified differential evolution (MDE) | −1 | −1 | −1 | 0 | 30 |
| Improved artificial bee colony (distABC) | −0.9957 | −0.4070 | −0.8023 | 0.1716 | 30 | |
| Improved gravitational search algorithm (IGSA/PSO) | −1 | 0 | −0.8333 | 0.3790 | 30 | |
| All-dimension neighborhood based particle swarm optimization with randomly selected neighbors (ADN-RSN-PSO) | −0.9126 | 0 | −0.0345 | 0.1668 | 30 | |
| Improved grey wolf optimization (PSO-GWO) | −1.0000 | −0.9983 | −0.9997 | 4.3750e−04 | 30 | |
| ISSA | −1 | −1.0000 | −1 | 4.1233e−17 | 30 | |
| F2 | MDE | 1.8981e−124 | 6.0392e−111 | 2.0134e−112 | 1.1026e−111 | 30 |
| distABC | 6.4478e−12 | 1.3143e−04 | 4.5038e−06 | 2.3974e−05 | 21 | |
| IGSA/PSO | 3.5151e−20 | 1.2328e−18 | 4.3235e−19 | 3.5081e−19 | 30 | |
| ADN-RSN-PSO | 1.5194e−14 | 9.3798e−06 | 3.8898e−07 | 1.7203e−06 | 20 | |
| PSO-GWO | 8.0797e−271 | 5.0693e−225 | 1.6898e−226 | 0 | 30 | |
| ISSA | 0 | 0 | 0 | 0 | 30 | |
| F3 | MDE | 0 | 0 | 0 | 0 | 30 |
| distABC | 0 | 6.6613e−16 | 2.2204e−17 | 1.2162e−16 | 30 | |
| IGSA/PSO | 0 | 6.6613e−16 | 5.1810e−17 | 5.1810e−17 | 30 | |
| ADN-RSN-PSO | 1.7529e−09 | 0.3443 | 0.0209 | 0.0670 | 1 | |
| PSO-GWO | 0 | 0 | 0 | 0 | 30 | |
| ISSA | 0 | 0 | 0 | 0 | 30 | |
| F4 | MDE | 0 | 0 | 0 | 0 | 30 |
| distABC | 0 | 0 | 0 | 0 | 30 | |
| IGSA/PSO | 0 | 5.5511e−17 | 1.8504e−18 | 1.0135e−17 | 30 | |
| ADN-RSN-PSO | 2.2204e−16 | 0.0025 | 1.5597e−04 | 4.6084e−04 | 6 | |
| PSO-GWO | 0 | 0 | 0 | 0 | 30 | |
| ISSA | 0 | 0 | 0 | 0 | 30 | |
| F5 | MDE | 0 | 0 | 0 | 0 | 30 |
| distABC | 0 | 2.2128e−10 | 7.3769e−12 | 4.0400e−11 | 30 | |
| IGSA/PSO | 0 | 1.6653e−16 | 2.9606e−17 | 4.3081e−17 | 30 | |
| ADN-RSN-PSO | 4.3178e−12 | 0.5315 | 0.0194 | 0.0968 | 3 | |
| PSO-GWO | 0 | 0 | 0 | 0 | 30 | |
| ISSA | 0 | 0 | 0 | 0 | 30 | |
| F6 | MDE | 0 | 0 | 0 | 0 | 30 |
| distABC | 9.3892e−04 | 0.9114 | 0.1484 | 0.1747 | 0 | |
| IGSA/PSO | 2.3141e−19 | 6.8399e−17 | 1.4187e−17 | 1.5244e−17 | 30 | |
| ADN-RSN-PSO | 4.6791e−13 | 3.3521e−04 | 2.3684e−05 | 7.2443e−05 | 7 | |
| PSO-GWO | 4.7856e−07 | 9.8150e−04 | 1.4743e−04 | 2.1193e−04 | 0 | |
| ISSA | 0 | 1.5479e−12 | 5.1596e−14 | 5.1596e−14 | 30 | |
| F7 | MDE | −1.8013 | −1.9996 | −1.8206 | 0.0587 | 30 |
| distABC | −1.7576 | −1.1419 | −1.4170 | 0.1852 | 12 | |
| IGSA/PSO | −1.8381 | −1.9969 | −1.9554 | 0.0443 | 30 | |
| ADN-RSN-PSO | −1.7829 | −1.4155 | −1.7056 | 0.1385 | 25 | |
| PSO-GWO | −1.8013 | −1.2138 | −1.7697 | 0.1218 | 27 | |
| ISSA | −1.8013 | −1.8013 | −1.8013 | 7.5040e−16 | 30 | |
| F8 | MDE | 0 | 0.0097 | 6.4773e−04 | 0.0025 | 28 |
| distABC | 0.0014 | 0.0103 | 0.0085 | 0.0027 | 0 | |
| IGSA/PSO | 5.7927e−04 | 0.0097 | 0.0071 | 0.0034 | 0 | |
| ADN-RSN-PSO | 0.2325 | 0.4767 | 0.3792 | 0.0769 | 0 | |
| PSO-GWO | 0 | 0 | 0 | 0 | 30 | |
| ISSA | 0 | 0 | 0 | 0 | 30 | |
| F9 | MDE | −4.6459 | −4.9833 | −4.7363 | 0.0866 | 30 |
| distABC | −2.6563 | −1.7364 | −2.2089 | 0.2644 | 0 | |
| IGSA/PSO | −4.3109 | −2.5452 | −3.6519 | 0.4646 | 19 | |
| ADN-RSN-PSO | −3.4226 | −2.1793 | −2.6879 | 0.3210 | 0 | |
| PSO-GWO | −4.4527 | −2.7246 | −3.4183 | 0.4289 | 14 | |
| ISSA | −4.6877 | −4.6459 | −4.6856 | 0.0093 | 30 | |
| F10 | MDE | −9.6552 | −9.1153 | −9.5328 | 0.1095 | 30 |
| distABC | −3.5674 | −2.7672 | −3.1627 | 0.1969 | 0 | |
| IGSA/PSO | −8.6839 | −3.8323 | −5.9454 | 1.2127 | 12 | |
| ADN-RSN-PSO | −4.9021 | −3.3798 | −3.7563 | 0.3143 | 0 | |
| PSO-GWO | −6.7936 | −4.1522 | −5.4923 | 0.6488 | 0 | |
| ISSA | −9.6602 | −9.5403 | −9.6241 | 0.0411 | 30 |
| Function | Rank | |||||
|---|---|---|---|---|---|---|
| MDE | distABC | IGSA/PSO | AND-RSN-PSO | PSO-GWO | ISSA | |
| F1 | 1 | 5 | 4 | 6 | 3 | 2 |
| F2 | 3 | 6 | 4 | 5 | 2 | 1 |
| F3 | 2 | 4 | 5 | 6 | 2 | 2 |
| F4 | 2.5 | 2.5 | 5 | 6 | 2.5 | 2.5 |
| F5 | 2 | 5 | 4 | 6 | 2 | 2 |
| F6 | 1 | 6 | 2 | 4 | 5 | 3 |
| F7 | 2 | 6 | 5 | 4 | 3 | 1 |
| F8 | 3 | 5 | 4 | 6 | 1.5 | 1.5 |
| F9 | 2 | 6 | 3 | 5 | 4 | 1 |
| F10 | 2 | 6 | 3 | 5 | 4 | 1 |
| Total rank | 20.5 | 51.5 | 39 | 53 | 29 | 17 |
| Average rank | 2.05 | 5.15 | 3.9 | 5.3 | 2.9 | 1.7 |
| Sort | 5 | 2 | 3 | 1 | 4 | 6 |
| i | Algorithm | z = (Ri − R6)/ = (Ri − R6)/ = (Ri − R6)/0.8367 | Pi | |
|---|---|---|---|---|
| 1 | AND-RSN-PSO | (5.3 − 1.7)/0.8367 = 4.3026 | 2e−05 | 0.01 |
| 2 | distABC | (5.15 − 1.7)/0.8367 = 4.1233 | 4e−05 | 0.0125 |
| 3 | IGSA/PSO | (3.9 − 1.7)/0.8367 = 2.6294 | 0.0087 | 0.0167 |
| 4 | PSO-GWO | (2.9 − 1.7)/0.8367 = 1.4342 | 0.1513 | 0.025 |
| 5 | MDE | (2.05 − 1.7)/0.8367 = 0.4183 | 0.6781 | 0.05 |
| Name | D | Method | Best | Worst | Mean | SD | R |
|---|---|---|---|---|---|---|---|
| F11 | 30 | MDE | 1.0401e−14 | 1.5722e−12 | 1.9227e−13 | 3.1621e−13 | 30 |
| distABC | 0.0366 | 2.2711 | 0.3127 | 0.4293 | 0 | ||
| IGSA/PSO | 7.9175e−07 | 0.0051 | 5.0970e−04 | 0.0011 | 0 | ||
| ADN-RSN-PSO | 5.7203e−17 | 27.6484 | 1.0388 | 5.0631 | 9 | ||
| PSO-GWO | 5.5036e−276 | 7.3227e−204 | 2.4409e−205 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 50 | MDE | 2.4111e−07 | 7.1216e−06 | 1.3533e−06 | 1.6316e−06 | 0 | |
| distABC | 5.3447e+03 | 1.5571e+05 | 5.4741e+04 | 3.8374e+04 | 0 | ||
| IGSA/PSO | 0.1664 | 3.5193 | 1.0541 | 0.7896 | 0 | ||
| ADN-RSN-PSO | 1.9193e−13 | 100.5956 | 6.6218 | 19.7265 | 2 | ||
| PSO-GWO | 1.6753e−281 | 1.4189e−221 | 4.7307e−223 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 100 | MDE | 0.7826 | 24.0248 | 5.3714 | 5.5151 | 0 | |
| distABC | 1.3689e+06 | 2.7723e+06 | 2.2532e+06 | 3.6327e+05 | 0 | ||
| IGSA/PSO | 637.5637 | 3.1652e+03 | 1.3765e+03 | 659.7935 | 0 | ||
| ADN-RSN-PSO | 1.8339e−12 | 0.4017 | 0.0410 | 0.0877 | 3 | ||
| PSO-GWO | 1.2087e−284 | 2.6558e−214 | 9.3853e−216 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| F12 | 30 | MDE | 9.2637e−14 | 7.6003e−11 | 5.8122e−12 | 1.3863e−11 | 30 |
| distABC | 0.0086 | 0.2122 | 0.0605 | 0.0567 | 0 | ||
| IGSA/PSO | 1.0212e−08 | 1.0867e−04 | 1.1358e−05 | 2.3307e−05 | 0 | ||
| ADN-RSN-PSO | 9.6397e−26 | 397.2135 | 14.0547 | 72.4037 | 3 | ||
| PSO-GWO | 6.2166e−292 | 6.3326e−231 | 2.1637e−232 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 50 | MDE | 1.6536e−07 | 9.7707e−06 | 2.9899e−06 | 2.6119e−06 | 0 | |
| distABC | 449.1668 | 2.6220e+03 | 1.2305e+03 | 564.3590 | 0 | ||
| IGSA/PSO | 0.0041 | 1.5093 | 0.2486 | 0.3447 | 0 | ||
| ADN-RSN-PSO | 8.2423e−17 | 403.1810 | 26.2589 | 82.8892 | 3 | ||
| PSO-GWO | 6.4181e−278 | 4.5968e−223 | 1.5336e−224 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 100 | MDE | 1.6536e−07 | 9.7707e−06 | 2.9899e−06 | 2.6119e−06 | 0 | |
| distABC | 2.2131e+05 | 2.8567e+05 | 2.5701e+05 | 1.4272e+04 | 0 | ||
| IGSA/PSO | 405.5180 | 2.5861e+03 | 1.0468e+03 | 448.9013 | 0 | ||
| ADN-RSN-PSO | 1.3639e−26 | 146.5426 | 5.9463 | 26.7914 | 6 | ||
| PSO-GWO | 7.3808e−273 | 3.3942e−226 | 1.1314e−227 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| F13 | 30 | MDE | 4.9141e−15 | 2.3773e−12 | 2.3077e−13 | 4.2943e−13 | 30 |
| distABC | 0.0019 | 0.0248 | 0.0103 | 0.0069 | 0 | ||
| IGSA/PSO | 1.6976e−05 | 0.0101 | 0.0018 | 0.0026 | 0 | ||
| ADN-RSN-PSO | 8.0743e−29 | 38.4053 | 2.4114 | 7.6095 | 9 | ||
| PSO-GWO | 2.0610e−290 | 2.3985e−230 | 8.0010e−232 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 50 | MDE | 4.3369e−08 | 2.3226e−06 | 5.8020e−07 | 5.5813e−07 | 0 | |
| distABC | 60.5866 | 437.6214 | 192.8170 | 90.6097 | 0 | ||
| IGSA/PSO | 0.2174 | 15.5868 | 2.6151 | 3.1442 | 0 | ||
| ADN-RSN-PSO | 2.3824e−19 | 35.9528 | 3.3225 | 9.7473 | 7 | ||
| PSO-GWO | 4.7500e−286 | 4.9925e−230 | 2.9195e−231 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 100 | MDE | 0.1475 | 4.8333 | 0.8994 | 0.9433 | 0 | |
| distABC | 7.3635e+04 | 1.2853e+05 | 1.0609e+05 | 1.4043e+04 | 0 | ||
| IGSA/PSO | 215.7341 | 673.2718 | 428.7446 | 119.2857 | 0 | ||
| ADN-RSN-PSO | 2.7413e−20 | 2.0323e+03 | 87.6861 | 369.4296 | 1 | ||
| PSO-GWO | 2.8630e−289 | 4.3016e−227 | 1.4339e−228 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| F14 | 30 | MDE | 5.2747e+03 | 3.2657e+04 | 1.3414e+04 | 6.1786e+03 | 0 |
| distABC | 1.3735e+04 | 9.6878e+04 | 5.2398e+04 | 1.8624e+04 | 0 | ||
| IGSA/PSO | 4.7593 | 149.2362 | 47.7658 | 37.3198 | 0 | ||
| ADN-RSN-PSO | 2.6758e−36 | 1.4738e+03 | 56.9252 | 268.1544 | 2 | ||
| PSO-GWO | 2.0189e−294 | 5.7304e−239 | 1.9102e−240 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 50 | MDE | 5.2107e+04 | 1.3519e+05 | 8.0055e+04 | 1.9715e+04 | 0 | |
| distABC | 1.7554e+05 | 4.6751e+05 | 2.8911e+05 | 6.8753e+04 | 0 | ||
| IGSA/PSO | 472.8938 | 2.5645e+03 | 1.1703e+03 | 534.2655 | 0 | ||
| ADN-RSN-PSO | 4.9794e−13 | 1.9352e+03 | 73.0431 | 352.4516 | 2 | ||
| PSO-GWO | 3.1521e−284 | 1.7900e−240 | 5.9668e−242 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 100 | MDE | 3.1074e+05 | 6.3577e+05 | 4.2536e+05 | 8.1732e+04 | 0 | |
| distABC | 9.0218e+05 | 2.2811e+06 | 1.4725e+06 | 3.2973e+05 | 0 | ||
| IGSA/PSO | 5.9127e+03 | 2.2013e+04 | 1.1214e+04 | 3.2236e+03 | 0 | ||
| ADN-RSN-PSO | 3.6628e−11 | 6.1001e+03 | 354.5833 | 1.1445e+03 | 2 | ||
| PSO-GWO | 5.5639e−279 | 3.6892e−238 | 6.3759e−240 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| F15 | 30 | MDE | 0.6789 | 15.6594 | 4.6249 | 3.3505 | 0 |
| distABC | 29.4236 | 88.1198 | 69.3981 | 14.3107 | 0 | ||
| IGSA/PSO | 0.1421 | 5.0155 | 1.3894 | 1.2303 | 0 | ||
| ADN-RSN-PSO | 3.5252e−06 | 3.2581 | 0.4436 | 0.8071 | 0 | ||
| PSO-GWO | 4.9641e−143 | 4.1240e−106 | 1.3747e−107 | 7.5293e−107 | 30 | ||
| ISSA | 6.8624e−294 | 1.9355e−221 | 6.4516e−223 | 0 | 30 | ||
| 50 | MDE | 7.3112 | 26.3407 | 15.3471 | 4.5763 | 0 | |
| distABC | 86.2687 | 95.1062 | 92.3951 | 2.4449 | 0 | ||
| IGSA/PSO | 5.0050 | 11.4719 | 8.6978 | 1.5023 | 0 | ||
| ADN-RSN-PSO | 5.6962e−09 | 2.5363 | 0.4027 | 0.6049 | 1 | ||
| PSO-GWO | 3.6326e−139 | 5.3333e−111 | 1.7782e−112 | 9.7371e−112 | 30 | ||
| ISSA | 2.2890e−275 | 5.3435e−222 | 1.7813e−223 | 0 | 30 | ||
| 100 | MDE | 24.5859 | 46.3713 | 33.8856 | 4.9749 | 0 | |
| distABC | 92.1930 | 97.9642 | 96.0475 | 1.1959 | 0 | ||
| IGSA/PSO | 14.1813 | 22.4854 | 17.8145 | 1.9772 | 0 | ||
| ADN-RSN-PSO | 3.5409e−13 | 2.3798 | 0.2541 | 0.5142 | 1 | ||
| PSO-GWO | 1.3222e−143 | 3.3096e−116 | 1.2837e−117 | 6.0495e−117 | 30 | ||
| ISSA | 1.3362e−271 | 1.3887e−230 | 4.8821e−232 | 0 | 30 | ||
| F16 | 30 | MDE | 6.3446e−14 | 2.0800e−11 | 2.1415e−12 | 4.0992e−12 | 30 |
| distABC | 0.0036 | 0.1511 | 0.0145 | 0.0262 | 0 | ||
| IGSA/PSO | 0.0016 | 0.2406 | 0.0451 | 0.0495 | 0 | ||
| ADN-RSN-PSO | 9.8349e−06 | 37.3092 | 2.4168 | 7.3907 | 0 | ||
| PSO-GWO | 5.6633e−141 | 4.9625e−113 | 1.6893e−114 | 9.0549e−114 | 30 | ||
| ISSA | 9.3727e−307 | 1.1611e−225 | 3.8704e−227 | 0 | 30 | ||
| 50 | MDE | 3.4911e−05 | 1.9115e−04 | 9.2416e−05 | 3.9599e−05 | 0 | |
| distABC | 3.5850 | 100.0040 | 16.9396 | 16.7889 | 0 | ||
| IGSA/PSO | 0.5307 | 13.3690 | 2.6048 | 2.4739 | 0 | ||
| ADN-RSN-PSO | 6.1043e−08 | 20.3200 | 2.5146 | 4.8767 | 0 | ||
| PSO-GWO | 4.9924e−140 | 1.5073e−107 | 5.0244e−109 | 2.7520e−108 | 30 | ||
| ISSA | 2.1080e−272 | 7.1760e−227 | 2.4359e−228 | 0 | 30 | ||
| 100 | MDE | 0.1146 | 2.2932 | 0.5097 | 0.4762 | 0 | |
| distABC | 3.5037e+13 | 6.5783e+27 | 2.9767e+26 | 1.2607e+27 | 0 | ||
| IGSA/PSO | 16.1544 | 40.1933 | 25.2979 | 5.3738 | 0 | ||
| ADN-RSN-PSO | 1.7105e−04 | 106.2857 | 8.2050 | 21.3386 | 0 | ||
| PSO-GWO | 3.4506e−142 | 4.3916e−113 | 1.5163e−114 | 8.0109e−114 | 30 | ||
| ISSA | 8.5914e−275 | 1.5030e−226 | 5.0101e−228 | 0 | 30 | ||
| F17 | 30 | MDE | 75.0038 | 75.0038 | 75.0038 | 2.4914e−06 | 0 |
| distABC | 14.1555 | 967.8401 | 263.9005 | 227.9731 | 0 | ||
| IGSA/PSO | 6.3987e+03 | 1.3092e+05 | 3.2932e+04 | 2.9068e+04 | 0 | ||
| ADN-RSN-PSO | 9.8556e−09 | 1.2370e+06 | 1.0690e+05 | 2.8807e+05 | 1 | ||
| PSO-GWO | 1.5518e−282 | 1.3001e−226 | 4.3877e−228 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 50 | MDE | 125.0040 | 125.0089 | 125.0061 | 0.0011 | 0 | |
| distABC | 8.7542e+05 | 8.0466e+06 | 3.7711e+06 | 1.7995e+06 | 0 | ||
| IGSA/PSO | 5.9214e+04 | 9.9143e+05 | 3.6022e+05 | 2.4168e+05 | 0 | ||
| ADN-RSN-PSO | 1.0970e−10 | 2.1368e+06 | 1.2419e+05 | 4.5201e+05 | 1 | ||
| PSO-GWO | 1.7465e−284 | 5.6730e−226 | 1.9183e−227 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 100 | MDE | 246.0984 | 254.7257 | 250.2010 | 1.7431 | 0 | |
| distABC | 7.9086e+08 | 1.7132e+09 | 1.1912e+09 | 2.6043e+08 | 0 | ||
| IGSA/PSO | 2.8580e+06 | 2.8682e+07 | 1.0857e+07 | 5.9353e+06 | 0 | ||
| ADN-RSN-PSO | 1.6856e−10 | 3.7886e+07 | 1.9511e+06 | 7.5422e+06 | 1 | ||
| PSO-GWO | 6.8705e−282 | 1.1504e−207 | 3.8348e−209 | 0 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| F18 | 30 | MDE | 5.6399e−14 | 0.0049 | 1.6441e−04 | 9.0052e−04 | 29 |
| distABC | 37.4773 | 52.3512 | 44.4924 | 3.8327 | 0 | ||
| IGSA/PSO | 2.2085 | 16.7038 | 7.2453 | 3.6023 | 0 | ||
| ADN-RSN-PSO | 473.9074 | 756.4108 | 618.1881 | 58.6785 | 0 | ||
| PSO-GWO | 1.4714e−04 | 0.5205 | 0.1391 | 0.1364 | 0 | ||
| ISSA | 0 | 1.8646e−11 | 6.2199e−13 | 3.4042e−12 | 30 | ||
| 50 | MDE | 2.1559e−07 | 0.0247 | 0.0020 | 0.0050 | 0 | |
| distABC | 88.2182 | 191.3685 | 142.3998 | 20.9568 | 0 | ||
| IGSA/PSO | 21.8794 | 81.5488 | 42.3904 | 14.2207 | 0 | ||
| ADN-RSN-PSO | 968.5157 | 1.3856e+03 | 1.1812e+03 | 107.6813 | 0 | ||
| PSO-GWO | 0.0012 | 0.5782 | 0.1944 | 0.1966 | 0 | ||
| ISSA | 0 | 2.0095e−14 | 3.8525e−15 | 5.4146e−15 | 30 | ||
| 100 | MDE | 0.1950 | 1.1188 | 0.6049 | 0.2438 | 0 | |
| distABC | 1.7147e+03 | 2.7003e+03 | 2.4415e+03 | 200.0542 | 0 | ||
| IGSA/PSO | 147.2077 | 249.3046 | 191.1295 | 25.7575 | 0 | ||
| ADN-RSN-PSO | 2.3436e+03 | 3.0052e+03 | 2.6587e+03 | 141.5865 | 0 | ||
| PSO-GWO | 0.0148 | 1.0070 | 0.4467 | 0.3082 | 0 | ||
| ISSA | 0 | 2.0095e−14 | 3.8525e−15 | 5.4146e−15 | 30 | ||
| F19 | 30 | MDE | 0.2008 | 0.3999 | 0.2899 | 0.0382 | 0 |
| distABC | 0.5839 | 1.3355 | 0.8775 | 0.1932 | 0 | ||
| IGSA/PSO | 1.0999 | 3.6999 | 1.8435 | 0.5102 | 0 | ||
| ADN-RSN-PSO | 1.8736e−05 | 2.4222 | 0.3108 | 0.5509 | 0 | ||
| PSO-GWO | 9.8614e−142 | 3.7041e−106 | 1.4517e−107 | 6.7762e−107 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 50 | MDE | 0.4999 | 1.1000 | 0.7342 | 0.1439 | 0 | |
| distABC | 3.2414 | 7.4063 | 5.6732 | 0.9657 | 0 | ||
| IGSA/PSO | 3.4999 | 7.0999 | 5.0539 | 0.7385 | 0 | ||
| ADN-RSN-PSO | 1.5276e−04 | 3.7168 | 0.2954 | 0.7185 | 0 | ||
| PSO-GWO | 1.1372e−147 | 7.0961e−103 | 2.3655e−104 | 1.2956e−103 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 100 | MDE | 2.2228 | 5.7138 | 3.5648 | 0.7504 | 0 | |
| distABC | 46.4171 | 55.0417 | 52.0511 | 1.8933 | 0 | ||
| IGSA/PSO | 9.4999 | 14.0488 | 11.1732 | 1.1383 | 0 | ||
| ADN-RSN-PSO | 1.0459e−07 | 3.5730 | 0.5094 | 0.9123 | 0 | ||
| PSO-GWO | 1.1118e−134 | 3.5210e−77 | 1.1737e−78 | 6.4285e−78 | 30 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| F20 | 30 | MDE | 2.0970e−07 | 0.0017 | 2.0093e−04 | 4.2588e−04 | 0 |
| distABC | 0.0349 | 0.9800 | 0.1546 | 0.2342 | 0 | ||
| IGSA/PSO | 0.0019 | 0.1676 | 0.0249 | 0.0340 | 0 | ||
| ADN-RSN-PSO | 1.9121e−07 | 18.8459 | 0.7225 | 3.4292 | 0 | ||
| PSO-GWO | 3.1745e−141 | 4.1132e−119 | 1.5246e−120 | 7.5032e−120 | 30 | ||
| ISSA | 1.1472e−268 | 1.7507e−232 | 6.9323e−234 | 0 | 30 | ||
| 50 | MDE | 4.9616e−05 | 0.0076 | 0.0017 | 0.0020 | 0 | |
| distABC | 23.2445 | 64.7154 | 36.0016 | 9.1027 | 0 | ||
| IGSA/PSO | 0.0772 | 3.2993 | 0.7989 | 0.6953 | 0 | ||
| ADN-RSN-PSO | 2.7451e−05 | 20.6995 | 1.0087 | 3.7981 | 0 | ||
| PSO-GWO | 1.1263e−143 | 1.1212e−110 | 3.7456e−112 | 2.0469e−111 | 30 | ||
| ISSA | 2.4273e−279 | 3.4792e−224 | 1.1597e−225 | 0 | 30 | ||
| 100 | MDE | 0.0418 | 0.2153 | 0.1025 | 0.0453 | 0 | |
| distABC | 178.0676 | 239.7760 | 209.6574 | 13.7426 | 0 | ||
| IGSA/PSO | 6.7693 | 16.2261 | 10.7582 | 2.3683 | 0 | ||
| ADN-RSN-PSO | 9.8116e−07 | 30.9275 | 2.5294 | 7.5049 | 0 | ||
| PSO-GWO | 1.7729e−144 | 5.6582e−121 | 2.2664e−122 | 1.0321e−121 | 30 | ||
| ISSA | 9.0375e−288 | 1.1851e−220 | 3.9503e−222 | 0 | 30 | ||
| F21 | 32 | MDE | 0.0204 | 38.6884 | 5.4340 | 10.5332 | 0 |
| distABC | 272.5527 | 8.4642e+04 | 1.9675e+04 | 2.1057e+04 | 0 | ||
| IGSA/PSO | 0.0468 | 20.9792 | 2.5257 | 3.8308 | 0 | ||
| ADN-RSN-PSO | 4.9194e−20 | 2.8246 | 0.1053 | 0.5147 | 9 | ||
| PSO-GWO | 1.2817e+07 | 1.2817e+07 | 1.2817e+07 | 0 | 0 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 52 | MDE | 0.3480 | 360.7218 | 22.4760 | 67.4348 | 0 | |
| distABC | 4.9677e+05 | 5.6793e+06 | 2.3595e+06 | 1.3184e+06 | 0 | ||
| IGSA/PSO | 1.2118 | 5.0887e+03 | 497.2259 | 1.0764e+03 | 0 | ||
| ADN-RSN-PSO | 3.3860e−31 | 0.0562 | 0.0020 | 0.0102 | 17 | ||
| PSO-GWO | 2.0828 e+07 | 2.0828 e+07 | 2.0828 e+07 | 0 | 0 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 | ||
| 100 | MDE | 61.9570 | 6.2051e+03 | 1.0242e+03 | 1.3454e+03 | 0 | |
| distABC | 3.7772e+07 | 6.2101e+07 | 5.8466e+07 | 4.7813e+06 | 0 | ||
| IGSA/PSO | 1.0160e+04 | 6.5678e+04 | 3.5827e+04 | 1.3363e+04 | 0 | ||
| ADN-RSN-PSO | 1.3081e−21 | 4.5014 | 0.2386 | 0.8825 | 13 | ||
| PSO-GWO | 4.0055 e+07 | 4.0055 e+07 | 4.0055 e+07 | 0 | 0 | ||
| ISSA | 0 | 0 | 0 | 0 | 30 |
| Function | Rank | |||||
|---|---|---|---|---|---|---|
| MDE | distABC | IGSA/PSO | AND-RSN-PSO | PSO-GWO | ISSA | |
| F11 | 3 | 5 | 4 | 6 | 2 | 1 |
| F12 | 3 | 5 | 4 | 6 | 2 | 1 |
| F13 | 3 | 5 | 4 | 6 | 2 | 1 |
| F14 | 5 | 6 | 3 | 4 | 2 | 1 |
| F15 | 5 | 6 | 4 | 3 | 2 | 1 |
| F16 | 3 | 4 | 5 | 6 | 2 | 1 |
| F17 | 3 | 4 | 5 | 6 | 2 | 1 |
| F18 | 2 | 5 | 4 | 6 | 3 | 1 |
| F19 | 3 | 5 | 6 | 4 | 2 | 1 |
| F20 | 3 | 5 | 4 | 6 | 2 | 1 |
| F21 | 4 | 5 | 3 | 2 | 6 | 1 |
| Total rank | 37 | 55 | 46 | 55 | 27 | 11 |
| Average rank | 3.3636 | 5 | 4.1818 | 5 | 2.4545 | 1 |
| Sort | 4 | 1 | 3 | 1 | 5 | 6 |
| i | Algorithm | z = (Ri − R6)/ = (Ri − R6)/ = (Ri − R6)/0.7977 | Pi | |
|---|---|---|---|---|
| 1 | AND-RSN-PSO | (5 − 1)/0.7977 = 5.0144 | 0 | 0.01 |
| 2 | distABC | (5 − 1)/0.7977 = 5.0144 | 0 | 0.01 |
| 3 | IGSA/PSO | (4.1818 − 1)/0.7977 = 3.9887 | 6e−05 | 0.0167 |
| 4 | MDE | (3.3636 − 1)/0.7977 = 2.9630 | 0.0030 | 0.025 |
| 5 | PSO-GWO | (2.4545 − 1)/0.7977 = 1.8234 | 0.068 | 0.05 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Du, T. An Improved Squirrel Search Algorithm for Global Function Optimization. Algorithms 2019, 12, 80. https://doi.org/10.3390/a12040080
Wang Y, Du T. An Improved Squirrel Search Algorithm for Global Function Optimization. Algorithms. 2019; 12(4):80. https://doi.org/10.3390/a12040080
Chicago/Turabian StyleWang, Yanjiao, and Tianlin Du. 2019. "An Improved Squirrel Search Algorithm for Global Function Optimization" Algorithms 12, no. 4: 80. https://doi.org/10.3390/a12040080
APA StyleWang, Y., & Du, T. (2019). An Improved Squirrel Search Algorithm for Global Function Optimization. Algorithms, 12(4), 80. https://doi.org/10.3390/a12040080





