A Soft-Voting Ensemble Based Co-Training Scheme Using Static Selection for Binary Classification Problems
Abstract
:1. Introduction
2. Related Works
2.1. Co-Training Studies
2.2. Ensemble Selection Strategies
3. The Proposed Co-Training Scheme
- We propose a multi-view SSL algorithm that handles efficiently both labeled (L) and unlabeled (U) data in the case of binary output variables.
- Instead of demanding two sufficient and redundant views, a random feature split is applied, thereby increasing the applicability and improving the performance of the finally formatted algorithm [38].
- We introduce a simple mechanism concerning the cardinality of unlabeled examples per different class that is mined for avoiding overfitting phenomena in cases where imbalanced datasets must be assessed.
- We insert a preprocess stage, where a pool of single learners is mined by a Static Ensemble Selection algorithm to extract a powerful soft-Voting ensemble per different classification problem, seeking to produce a more accurate and robust semi-supervised algorithm operating under small labeled ratio values.
| Algorithm 1. SSoftEC strategy |
| Input: L—labeled set f—number of folds to split the L C—pool of classification algorithms exporting class probabilities α—value of balancing parameter Main Procedure: For each i, j ∊ {0, 1, …, |C|} and i ≠ j do Set iter = 0, Split L to f separate folds: While iter ≤ f do Train Ci, Cj on Apply Ci, Cj on Update according to Equation (4) iter = iter + 1 Output: Return pair of indices i, j such that: . |
| Algorithm 2.Ensemble based co-training variant |
| Mode: Pool-based scenario over a provided dataset D = Xn × k ⋃ Yn × 1 xi—vector with k features <f1, f2, … fk> ∀ 1 ≤ i ≤ n yi—scalar class variable with yi ∊ {0, 1} ∀ 1 ≤ i ≤ n {xi, yi}—i-th labeled instance (li) with 1 ≤ i ≤ nl {xi}—i-th unlabeled instance (ui) with 1 ≤ i ≤ nu Fview—separate feature sets with view ∊ [1,2] learnerview—build of selected learner on corresponding View, ∀view = 1, 2 Input: Liter—labeled instances during iter-th iteration, Liter ⊂ D Uiter—unlabeled instances during iter-th iteration, Uiter ⊂ D iter—number of combined executed iterations MaxIter—maximum number of iterations C—pool of classifiers ≡ {SVM, kNN, DT, NB, LR} (f, α)—number of folds to split the validation set during SEC and value of Equation (4) Preprocess: k′—number of features after having converted each categorical feature into binary n′—number of instances after having removed instances with at least one missing value Cj—instance cardinalities of both existing classes with j ∊ {min, max} Minedc—define number of mined instances per class, where c ∊ {class0, class1} Main Procedure: Apply SSoftEC(L0, f, C, α) and obtain Construct Set iter = 0 While iter < MaxIter do For each view Train learnerview on Assign class probabilities for each ui ∊ Uiter For each class Detect the top Minedclass ≡ Indview Update: iter = iter + 1 Output: Use trained on LMaxIter to predict class labels of test data. |
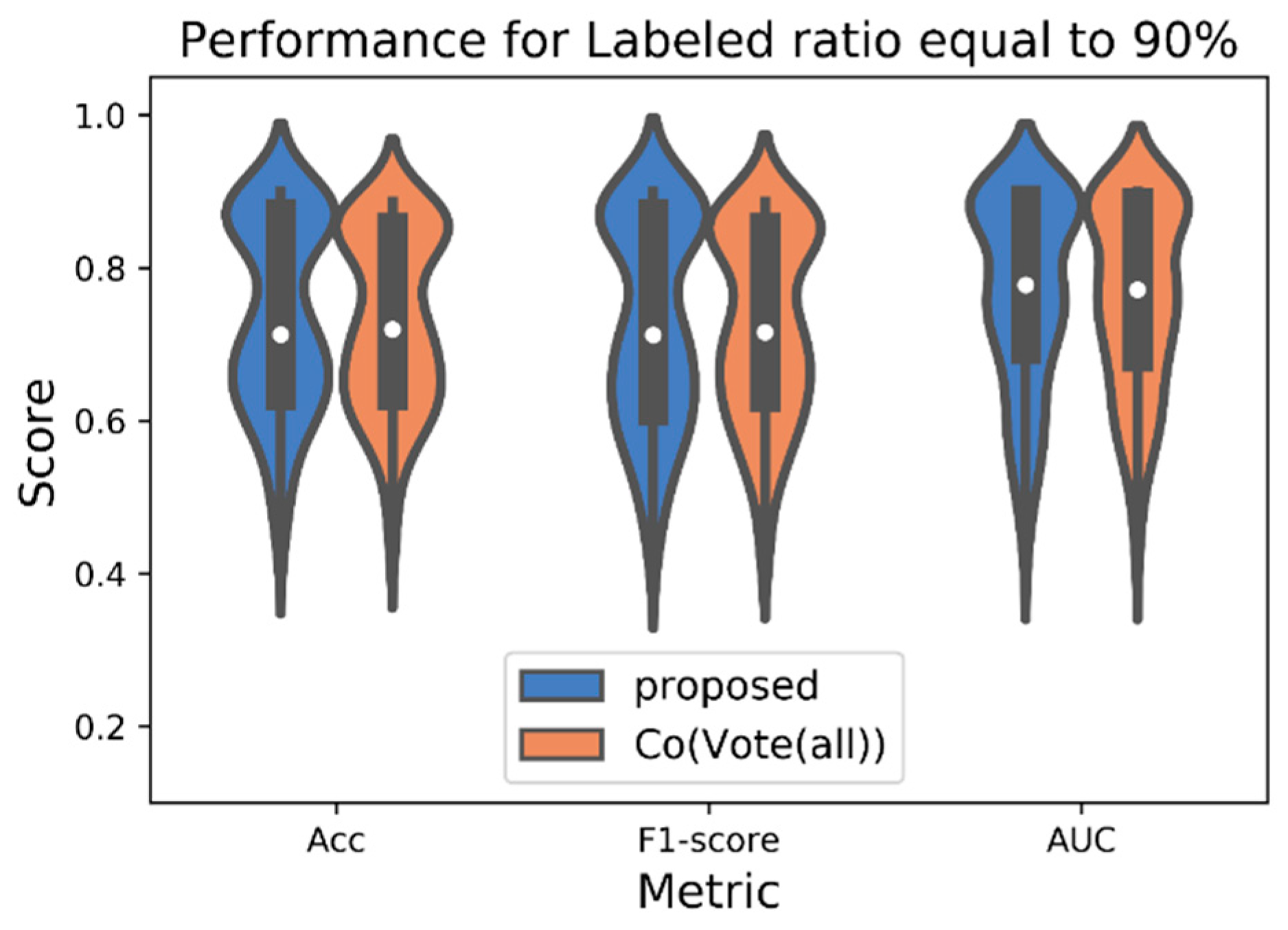
4. Experimental Procedure and Results
- The k-Nearest Neighbor (kNN) instance-based learner [53] with k equal to 5, a very effective method for classification problems, using the Euclidean metric as a similarity measure to determine the distance between two instances,
- A simple Decision Tree (DT) algorithm, a variant of tree induction algorithms with large depth that split the feature space using ‘gini’ criterion [44],
- The NB probabilistic classifier, a simple and quite efficient classification algorithm based on the assumption that features are independent of each other given the class label [54],
- The Logistic Regression (LR), a well-known discriminative algorithm that assumes the log likelihood ratio of class distributions is linear in the provided examples. Its main function supports the binomial case of the target variable, exporting posterior probabilities in a direct way. In our implementation, L2-norm during penalization stage was chosen [55].
- C ≡ {SVM, kNN, DT, NB, LR}, the list of participant classification algorithms,
- , where learner ∊ C,
- , where , ∊ C with ,
- , where all participants of C are exploited under the Voting scheme,
- , where this kind of approach corresponds to the case that learner1 ≡ learner2 ≡ learner, with learner ∊ C,
- , where , ∊ C with , and the ensemble Voting learner is the same for both views, similar with the previous scenario,
- , where all participants of C are exploited under the Voting scheme for each view, and finally,
- , which coincides with the proposed semi-supervised algorithm.
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Schwenker, F.; Trentin, E. Pattern classification and clustering: A review of partially supervised learning approaches. Pattern Recognit. Lett. 2014, 37, 4–14. [Google Scholar] [CrossRef]
- Kim, A.; Cho, S.-B. An ensemble semi-supervised learning method for predicting defaults in social lending. Eng. Appl. Artif. Intell. 2019, 81, 193–199. [Google Scholar] [CrossRef]
- Li, J.; Wu, S.; Liu, C.; Yu, Z.; Wong, H.-S. Semi-Supervised Deep Coupled Ensemble Learning With Classification Landmark Exploration. IEEE Trans. Image Process. 2020, 29, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Kostopoulos, G.; Karlos, S.; Kotsiantis, S.; Ragos, O. Semi-supervised regression: A recent review. J. Intell. Fuzzy Syst. 2018, 35, 1483–1500. [Google Scholar] [CrossRef]
- Alpaydin, E. Introduction to Machine Learning; MIT Press: Cambridge, MA, USA, 2010. [Google Scholar]
- Ng, V.; Cardie, C. Weakly supervised natural language learning without redundant views. In Proceedings of the 2003 Human Language Technology Conference of the North American Chapter of the Association for Computational Linguistics, Edmonton, AB, Canada, 27 May–1 June 2003. [Google Scholar]
- Blum, A.; Mitchell, T. Combining labeled and unlabeled data with co-training. In Proceedings of the Eleventh Annual Conference on Computational Learning Theory—COLT’ 98, New York, NY, USA, 24–26 July 1998; pp. 92–100. [Google Scholar]
- Zhou, Z.-H.; Li, M. Tri-training: Exploiting unlabeled data using three classifiers. IEEE Trans. Knowl. Data Eng. 2005, 17, 1529–1541. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Goldberg, A.B. Introduction to Semi-Supervised Learning; Morgan & Claypool Publishers: Williston, VN, USA, 2009. [Google Scholar]
- Zhou, Z.-H. Ensemble Methods: Foundations and Algorithms; Taylor & Francis: Abingdon, UK, 2012. [Google Scholar]
- Zhou, Z.-H. When semi-supervised learning meets ensemble learning. Front. Electr. Electron. Eng. China 2011, 6, 6–16. [Google Scholar] [CrossRef]
- Sinha, A.; Chen, H.; Danu, D.G.; Kirubarajan, T.; Farooq, M. Estimation and decision fusion: A survey. Neurocomputing 2008, 71, 2650–2656. [Google Scholar] [CrossRef]
- Wu, Y.; He, J.; Man, Y.; Arribas, J.I. Neural network fusion strategies for identifying breast masses. In Proceedings of the 2004 IEEE International Joint Conference on Neural Networks (IEEE Cat. No. 04CH37541), Budapest, Hungary, 25–29 July 2004; pp. 2437–2442. [Google Scholar]
- Wu, Y.; Arribas, J.I. Fusing output information in neural networks: Ensemble performs better. In Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE Cat. No. 03CH37439), Cancun, Mexico, 17–21 September 2003; pp. 2265–2268. [Google Scholar]
- Livieris, I.; Kanavos, A.; Tampakas, V.; Pintelas, P. An auto-adjustable semi-supervised self-training algorithm. Algorithms 2018, 11, 139. [Google Scholar] [CrossRef] [Green Version]
- Britto, A.S.; Sabourin, R.; Oliveira, L.E.S. Dynamic selection of classifiers—A comprehensive review. Pattern Recognit. 2014, 47, 3665–3680. [Google Scholar] [CrossRef]
- Hou, C.; Xia, Y.; Xu, Z.; Sun, J. Semi-supervised learning competence of classifiers based on graph for dynamic classifier selection. In Proceedings of the IEEE 2016 23rd International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016; pp. 3650–3654. [Google Scholar]
- Jiang, Z.; Zhang, S.; Zeng, J. A hybrid generative/discriminative method for semi-supervised classification. Knowl. Based Syst. 2013, 37, 137–145. [Google Scholar] [CrossRef]
- Ceci, M.; Pio, G.; Kuzmanovski, V.; Džeroski, S. Semi-supervised multi-view learning for gene network reconstruction. PLoS ONE 2015, 10, e0144031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Goldberg, A.B. Introduction to Semi-Supervised Learning. Synth. Lect. Artif. Intell. Mach. Learn. 2009, 3, 1–130. [Google Scholar] [CrossRef] [Green Version]
- Nigam, K.; Ghani, R. Analyzing the effectiveness and applicability of co-training. In Proceedings of the Ninth International Conference on Information and Knowledge Management, New York, NY, USA, 6–11 November 2000; pp. 86–93. [Google Scholar] [CrossRef]
- Yu, N. Exploring C o-training strategies for opinion detection. J. Assoc. Inf. Sci. Technol. 2014, 65, 2098–2110. [Google Scholar] [CrossRef]
- Lin, W.-Y.; Lo, C.-F. Co-training and ensemble based duplicate detection in adverse drug event reporting systems. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine, Shanghai, China, 18–21 December 2013; pp. 7–8. [Google Scholar]
- Culp, M.; Michailidis, G. A co-training algorithm for multi-view data with applications in data fusion. J. Chemom. 2009, 23, 294–303. [Google Scholar] [CrossRef] [Green Version]
- Wehrens, R.; Mevik, B.-H. The pls package: Principal component and partial least squares regression in R. J. Stat. Softw. 2007, 18, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Levatić, J.; Ceci, M.; Kocev, D.; Džeroski, S. Self-training for multi-target regression with tree ensembles. Knowl. Based Syst. 2017, 123, 41–60. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Z.-H. Improve Computer-Aided Diagnosis With Machine Learning Techniques Using Undiagnosed Samples. IEEE Trans. Syst. Man Cybern. Part A Syst. Hum. 2007, 37, 1088–1098. [Google Scholar] [CrossRef] [Green Version]
- Deng, C.; Guo, M.Z. A new co-training-style random forest for computer aided diagnosis. J. Intell. Inf. Syst. 2011, 36, 253–281. [Google Scholar] [CrossRef]
- Liu, C.; Yuen, P.C. A Boosted Co-Training Algorithm for Human Action Recognition. IEEE Trans. Circuits Syst. Video Technol. 2011, 21, 1203–1213. [Google Scholar] [CrossRef]
- Kostopoulos, G.; Karlos, S.; Kotsiantis, S.B. Multi-view Learning for Early Prognosis of Academic Performance: A Case Study. IEEE Trans. Learn. Technol. 2019, 12, 212–224. [Google Scholar] [CrossRef]
- Pio, G.; Malerba, D.; D’Elia, D.; Ceci, M. Integrating microRNA target predictions for the discovery of gene regulatory networks: A semi-supervised ensemble learning approach. BMC Bioinform. 2014, 15, S4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietterich, T.G. Ensemble Methods in Machine Learning. Mult. Classif. Syst. 2000, 1857, 1–15. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Alonso-Betanzos, A. Recent Advances in Ensembles for Feature Selection; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar]
- Azizi, N.; Farah, N. From static to dynamic ensemble of classifiers selection: Application to Arabic handwritten recognition. Int. J. Knowl. Based Intell. Eng. Syst. 2012, 16, 279–288. [Google Scholar] [CrossRef]
- Mousavi, R.; Eftekhari, M.; Rahdari, F. Omni-Ensemble Learning (OEL): Utilizing Over-Bagging, Static and Dynamic Ensemble Selection Approaches for Software Defect Prediction. Int. J. Artif. Intell. Tools 2018, 27, 1850024. [Google Scholar] [CrossRef]
- Tsoumakas, G.; Partalas, I.; Vlahavas, I. An Ensemble Pruning Primer. Appl. Supervised Unsupervised Ensemble Methods 2009, 245, 1–13. [Google Scholar] [CrossRef]
- Cruz, R.M.O.; Sabourin, R.; Cavalcanti, G.D.C. Analyzing different prototype selection techniques for dynamic classifier and ensemble selection. In Proceedings of the IEEE 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 3959–3966. [Google Scholar]
- Zhao, J.; Xie, X.; Xu, X.; Sun, S. Multi-view learning overview: Recent progress and new challenges. Inf. Fusion 2017, 38, 43–54. [Google Scholar] [CrossRef]
- Au, T.C. Random Forests, Decision Trees, and Categorical Predictors: The “Absent Levels” Problem. J. Mach. Learn. Res. 2018, 19, 1–30. [Google Scholar]
- Ling, C.X.; Du, J.; Zhou, Z.-H. When does Co-training Work in Real Data? Adv. Knowl. Discov. Data Min. Proc. 2009, 5476, 596–603. [Google Scholar]
- Ni, Q.; Zhang, L.; Li, L. A Heterogeneous Ensemble Approach for Activity Recognition with Integration of Change Point-Based Data Segmentation. Appl. Sci. 2018, 8, 1695. [Google Scholar] [CrossRef] [Green Version]
- Platt, J.C. Probabilistic Outputs for Support Vector Machines and Comparisons to Regularized Likelihood Methods. Adv. Large Margin Classif. 1999, 10, 61–74. [Google Scholar]
- Garcia, E.K.; Feldman, S.; Gupta, M.R.; Srivastava, S. Completely lazy learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1274–1285. [Google Scholar] [CrossRef]
- Loh, W.-Y. Classification and regression trees. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2011, 1, 14–23. [Google Scholar] [CrossRef]
- Zheng, F.; Webb, G. A comparative study of semi-naive Bayes methods in classification learning. In Proceedings of the 4th Australas Data Mining Conference AusDM05 2005, Sydney, Australia, 5–6 December 2005; pp. 141–156. [Google Scholar]
- Samworth, R.J. Optimal weighted nearest neighbour classifiers. arXiv 2011, arXiv:1101.5783v3. [Google Scholar] [CrossRef]
- Giacinto, G.; Roli, F. Design of effective neural network ensembles for image classification purposes. Image Vis. Comput. 2001, 19, 699–707. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, Z.-H. Theoretical Foundation of Co-Training and Disagreement-Based Algorithms. arXiv 2017, arXiv:1708.04403. [Google Scholar]
- Dua, D.; Graff, C. UCI Machine Learning Repository. Available online: http://archive.ics.uci.edu/ml (accessed on 1 November 2019).
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning: with Applications in R; Springer: New York, NY, USA, 2013. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Chang, C.; Lin, C. LIBSVM: A Library for Support Vector Machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–39. [Google Scholar] [CrossRef]
- Aha, D.W.; Kibler, D.; Albert, M.K. Instance-Based Learning Algorithms. Mach. Learn. 1991, 6, 37–66. [Google Scholar] [CrossRef] [Green Version]
- Rish, I. An empirical study of the naive Bayes classifier. In Proceedings of the IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence, Seattle, WA, USA, 4–6 August 2001; pp. 41–46. [Google Scholar]
- Sperandei, S. Understanding logistic regression analysis. Biochem. Medica 2014, 24, 12–18. [Google Scholar] [CrossRef]
- Hodges, J.L.; Lehmann, E.L. Rank methods for combination of independent experiments in analysis of variance. Ann. Math. Stat. 1962, 33, 482–497. [Google Scholar] [CrossRef]
- Hollander, M.; Wolfe, D.A.; Chicken, E. Nonparametric Statistical Methods; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014. [Google Scholar]
- Kumar, G.; Kumar, K. The Use of Artificial-Intelligence-Based Ensembles for Intrusion Detection: A Review. Appl. Comput. Intell. Soft Comput. 2012, 2012, 850160. [Google Scholar] [CrossRef] [Green Version]
- Karlos, S.; Kaleris, K.; Fazakis, N. Optimized Active Learning Strategy for Audiovisual Speaker Recognition. In Proceedings of the 20th International Conference on Speech and Computer SPECOM 2018, Leipzig, Germany, 18–22 September 2018; pp. 281–290. [Google Scholar]
- Tencer, L.; Reznakova, M.; Cheriet, M. Summit-Training: A hybrid Semi-Supervised technique and its application to classification tasks. Appl. Soft Comput. J. 2017, 50, 1–20. [Google Scholar] [CrossRef]
- Tanha, J.; van Someren, M.; Afsarmanesh, H. Semi-supervised self-training for decision tree classifiers. Int. J. Mach. Learn. Cybern. 2017, 8, 355–370. [Google Scholar] [CrossRef] [Green Version]
- Chapelle, O.; Schölkopf, B.; Zien, A. Metric-Based Approaches for Semi-Supervised Regression and Classification. In Semi-Supervised Learning; MIT Press: Cambridge, MA, USA, 2006; pp. 420–451. [Google Scholar]
- Wainer, J. Comparison of 14 different families of classification algorithms on 115 binary datasets. arXiv 2016, arXiv:1606.00930. [Google Scholar]
- Yaslan, Y.; Cataltepe, Z. Co-training with relevant random subspaces. Neurocomputing 2010, 73, 1652–1661. [Google Scholar] [CrossRef]
- Zhang, M.-L.; Zhou, Z.-H. Exploiting unlabeled data to enhance ensemble diversity. Data Min. Knowl. Discov. 2013, 26, 98–129. [Google Scholar] [CrossRef] [Green Version]
- Karlos, S.; Fazakis, N.; Kotsiantis, S.; Sgarbas, K. Self-Trained Stacking Model for Semi-Supervised Learning. Int. J. Artif. Intell. Tools 2017, 26. [Google Scholar] [CrossRef]
- Barua, S.; Islam, M.M.; Yao, X.; Murase, K. MWMOTE--Majority Weighted Minority Oversampling Technique for Imbalanced Data Set Learning. IEEE Trans. Knowl. Data Eng. 2014, 26, 405–425. [Google Scholar] [CrossRef]
- Guo, H.; Diao, X.; Liu, H. Embedding Undersampling Rotation Forest for Imbalanced Problem. Comput. Intell. Neurosci. 2018, 2018, 6798042. [Google Scholar] [CrossRef]
- Vluymans, S. Learning from Imbalanced Data. In Dealing with Imbalanced and Weakly Labelled Data in Machine Learning Using Fuzzy and Rough Set Methods. Studies in Computational Intelligence; Springer: Cham, Switzerland, 2019; pp. 81–110. [Google Scholar]
- Tanha, J. MSSBoost: A new multiclass boosting to semi-supervised learning. Neurocomputing 2018, 314, 251–266. [Google Scholar] [CrossRef]
- Chuang, C.-L. Application of hybrid case-based reasoning for enhanced performance in bankruptcy prediction. Inf. Sci. 2013, 236, 174–185. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining—KDD’16, San Francisco, CA, USA, 13–17 August 2016; ACM Press: New York, NY, USA, 2016; pp. 1135–1144. [Google Scholar]
- Kale, D.; Liu, Y. Accelerating Active Learning with Transfer Learning. In Proceedings of the 2013 IEEE 13th International Conference on Data Mining, Dallas, TX, USA, 7–10 December 2013; pp. 1085–1090. [Google Scholar] [CrossRef]
- Nielsen, M.A. Neural Networks and Deep Learning. 2015. Available online: http://neuralnetworksanddeeplearning.com (accessed on 1 November 2019).
- Chen, D.; Che, N.; Le, J.; Pan, Q. A co-training based entity recognition approach for cross-disease clinical documents. Concurr. Comput. Pract. Exp. 2018, e4505. [Google Scholar] [CrossRef]

| Dataset | # Instances | # Features | Dataset | # Instances | # Features |
|---|---|---|---|---|---|
| bands | 365 | 19 | monk-2 | 432 | 6 |
| breast | 277 | 48 | pima | 768 | 8 |
| bupa | 345 | 6 | saheart | 468 | 9 |
| chess | 3196 | 38 | sick | 3772 | 33 |
| colic.orig | 368 | 471 | tic-tac-toe | 958 | 27 |
| diabetes | 768 | 8 | vote | 435 | 16 |
| heart-statlog | 270 | 13 | wdbc | 569 | 30 |
| kr-vs-kp | 3196 | 40 | wisconsin | 683 | 9 |
| mammographic | 830 | 5 |
| Algorithm | Parameters |
|---|---|
| k-NN | Number of neighbors: 5 |
| Distance function: Euclidean distance | |
| SVM | Kernel function: RBF |
| DT | Splitting criterion: gini |
| Min instances per leaf = 2 | |
| LR | Norm: L2 |
| NB | Gaussian distribution |
| Self-training | MaxIter = 20 |
| Co-training | MaxIter = 20 |
| Dataset | Algorithms | ||||
|---|---|---|---|---|---|
| bands | 0.668 ± 0.082 | 0.668 ± 0.051 | 0.649 ± 0.036 | 0.627 ± 0.036 | 0.657 ± 0.084 |
| breast | 0.679 ± 0.065 | 0.696 ± 0.056 | 0.7 ± 0.038 | 0.693 ± 0.056 | 0.686 ± 0.114 |
| bupa | 0.571 ± 0.052 | 0.603 ± 0.085 | 0.529 ± 0.041 | 0.537 ± 0.042 | 0.591 ± 0.057 |
| chess | 0.966 ± 0.013 | 0.948 ± 0.007 | 0.955 ± 0.008 | 0.947 ± 0.011 | 0.962 ± 0.01 |
| colic.ORIG | 0.759 ± 0.055 | 0.757 ± 0.044 | 0.724 ± 0.077 | 0.73 ± 0.053 | 0.768 ± 0.036 |
| diabetes | 0.773 ± 0.038 | 0.777 ± 0.032 | 0.738 ± 0.048 | 0.765 ± 0.03 | 0.681 ± 0.059 |
| h-statlog | 0.719 ± 0.036 | 0.7 ± 0.027 | 0.707 ± 0.051 | 0.77 ± 0.034 | 0.656 ± 0.08 |
| kr-vs-kp | 0.966 ± 0.019 | 0.947 ± 0.008 | 0.953 ± 0.008 | 0.946 ± 0.012 | 0.963 ± 0.012 |
| mammographic | 0.788 ± 0.019 | 0.796 ± 0.023 | 0.793 ± 0.024 | 0.787 ± 0.025 | 0.78 ± 0.039 |
| monk-2 | 1 ± 0 | 0.918 ± 0.019 | 0.88 ± 0.047 | 0.784 ± 0.029 | 1 ± 0 |
| pima | 0.691 ± 0.02 | 0.683 ± 0.025 | 0.686 ± 0.038 | 0.697 ± 0.032 | 0.652 ± 0.033 |
| saheart | 0.715 ± 0.021 | 0.713 ± 0.067 | 0.702 ± 0.036 | 0.728 ± 0.02 | 0.672 ± 0.038 |
| sick | 0.976 ± 0.003 | 0.967 ± 0.005 | 0.963 ± 0.006 | 0.95 ± 0.004 | 0.978 ± 0.004 |
| tic-tac-toe | 0.804 ± 0.07 | 0.805 ± 0.038 | 0.735 ± 0.037 | 0.824 ± 0.036 | 0.796 ± 0.057 |
| vote | 0.914 ± 0.026 | 0.891 ± 0.026 | 0.916 ± 0.024 | 0.889 ± 0.02 | 0.889 ± 0.033 |
| wdbc | 0.958 ± 0.021 | 0.963 ± 0.01 | 0.972 ± 0.012 | 0.956 ± 0.021 | 0.942 ± 0.031 |
| wisconsin | 0.974 ± 0.006 | 0.98 ± 0.01 | 0.972 ± 0.011 | 0.97 ± 0.016 | 0.936 ± 0.039 |
| Dataset | Algorithms | ||||
|---|---|---|---|---|---|
| bands | 0.646 ± 0.096 | 0.632 ± 0.068 | 0.643 ± 0.082 | 0.603 ± 0.049 | 0.594 ± 0.028 |
| breast | 0.652 ± 0.07 | 0.664 ± 0.076 | 0.67 ± 0.118 | 0.669 ± 0.032 | 0.649 ± 0.072 |
| bupa | 0.554 ± 0.049 | 0.588 ± 0.085 | 0.586 ± 0.059 | 0.502 ± 0.042 | 0.512 ± 0.032 |
| chess | 0.966 ± 0.013 | 0.947 ± 0.007 | 0.962 ± 0.01 | 0.955 ± 0.008 | 0.947 ± 0.011 |
| colic.ORIG | 0.745 ± 0.056 | 0.761 ± 0.043 | 0.762 ± 0.039 | 0.708 ± 0.083 | 0.735 ± 0.049 |
| diabetes | 0.765 ± 0.05 | 0.769 ± 0.034 | 0.68 ± 0.054 | 0.723 ± 0.055 | 0.75 ± 0.042 |
| h-statlog | 0.717 ± 0.036 | 0.7 ± 0.027 | 0.653 ± 0.082 | 0.706 ± 0.052 | 0.77 ± 0.034 |
| kr-vs-kp | 0.966 ± 0.019 | 0.947 ± 0.008 | 0.963 ± 0.012 | 0.953 ± 0.008 | 0.946 ± 0.012 |
| mammographic | 0.786 ± 0.019 | 0.795 ± 0.022 | 0.779 ± 0.039 | 0.791 ± 0.025 | 0.785 ± 0.026 |
| monk-2 | 1 ± 0 | 0.918 ± 0.019 | 1 ± 0 | 0.879 ± 0.047 | 0.78 ± 0.03 |
| pima | 0.673 ± 0.018 | 0.664 ± 0.027 | 0.652 ± 0.034 | 0.664 ± 0.033 | 0.685 ± 0.03 |
| saheart | 0.65 ± 0.034 | 0.7 ± 0.073 | 0.675 ± 0.038 | 0.688 ± 0.039 | 0.716 ± 0.019 |
| sick | 0.974 ± 0.003 | 0.962 ± 0.007 | 0.976 ± 0.004 | 0.959 ± 0.008 | 0.939 ± 0.006 |
| tic-tac-toe | 0.807 ± 0.069 | 0.805 ± 0.037 | 0.799 ± 0.056 | 0.733 ± 0.038 | 0.822 ± 0.036 |
| vote | 0.914 ± 0.026 | 0.891 ± 0.026 | 0.888 ± 0.033 | 0.916 ± 0.024 | 0.888 ± 0.02 |
| wdbc | 0.958 ± 0.021 | 0.963 ± 0.01 | 0.943 ± 0.031 | 0.972 ± 0.012 | 0.956 ± 0.02 |
| wisconsin | 0.974 ± 0.006 | 0.98 ± 0.01 | 0.935 ± 0.04 | 0.972 ± 0.011 | 0.969 ± 0.016 |
| Dataset | Algorithms | ||||
|---|---|---|---|---|---|
| bands | 0.716 ± 0.083 | 0.652 ± 0.079 | 0.764 ± 0.063 | 0.716 ± 0.052 | 0.69 ± 0.06 |
| breast | 0.701 ± 0.077 | 0.671 ± 0.105 | 0.652 ± 0.062 | 0.661 ± 0.071 | 0.621 ± 0.081 |
| bupa | 0.584 ± 0.09 | 0.567 ± 0.065 | 0.602 ± 0.08 | 0.602 ± 0.08 | 0.54 ± 0.082 |
| chess | 0.99 ± 0.004 | 0.992 ± 0.004 | 0.985 ± 0.007 | 0.981 ± 0.006 | 0.992 ± 0.005 |
| colic.ORIG | 0.835 ± 0.044 | 0.817 ± 0.029 | 0.722 ± 0.079 | 0.813 ± 0.026 | 0.762 ± 0.097 |
| diabetes | 0.835 ± 0.037 | 0.772 ± 0.028 | 0.854 ± 0.011 | 0.824 ± 0.035 | 0.801 ± 0.039 |
| h-statlog | 0.765 ± 0.05 | 0.779 ± 0.053 | 0.802 ± 0.024 | 0.73 ± 0.051 | 0.769 ± 0.033 |
| kr-vs-kp | 0.99 ± 0.004 | 0.991 ± 0.004 | 0.985 ± 0.007 | 0.979 ± 0.006 | 0.992 ± 0.005 |
| mammographic | 0.857 ± 0.016 | 0.876 ± 0.021 | 0.856 ± 0.015 | 0.87 ± 0.021 | 0.874 ± 0.016 |
| monk-2 | 0.996 ± 0.005 | 1 ± 0 | 1 ± 0 | 0.956 ± 0.018 | 0.958 ± 0.025 |
| pima | 0.655 ± 0.041 | 0.681 ± 0.031 | 0.68 ± 0.02 | 0.647 ± 0.041 | 0.651 ± 0.041 |
| saheart | 0.784 ± 0.057 | 0.764 ± 0.033 | 0.772 ± 0.019 | 0.791 ± 0.028 | 0.767 ± 0.046 |
| sick | 0.956 ± 0.011 | 0.961 ± 0.013 | 0.904 ± 0.024 | 0.917 ± 0.011 | 0.963 ± 0.025 |
| tic-tac-toe | 0.883 ± 0.031 | 0.903 ± 0.036 | 0.852 ± 0.054 | 0.914 ± 0.035 | 0.804 ± 0.041 |
| vote | 0.963 ± 0.005 | 0.964 ± 0.006 | 0.953 ± 0.018 | 0.958 ± 0.007 | 0.963 ± 0.007 |
| wdbc | 0.995 ± 0.003 | 0.992 ± 0.006 | 0.993 ± 0.004 | 0.995 ± 0.003 | 0.993 ± 0.006 |
| wisconsin | 0.998 ± 0.002 | 0.995 ± 0.007 | 0.996 ± 0.003 | 0.997 ± 0.003 | 0.997 ± 0.002 |
| Friedman Ranking | |||||
|---|---|---|---|---|---|
| Acc | F1-Score | AUC | |||
| Algorithm | Rank | Algorithm | Rank | Algorithm | Rank |
| 9.96 | 10.43 | 10.43 | |||
| Co(Vote(all)) | 11.59 | Co(Vote(all)) | 11.29 | Co(Vote(DT,LR) | 11.33 |
| Self(Vote(all)) | 13.19 | Co(Vote(DT,LR)) | 12.81 | Co(Vote(all)) | 12.88 |
| Co(LR) | 13.36 | Self(Vote(all)) | 13.28 | Co(Vote(kNN,LR) | 13.17 |
| Co(Vote(DT,LR)) | 13.89 | Co(LR) | 13.34 | Self(Vote(all)) | 13.66 |
| Co(Vote(SVM,LR)) | 14.13 | Co(DT) | 13.44 | Co(Vote(DT,GNB)) | 14.17 |
| Co(DT) | 14.41 | Co(Vote(SVM,DT)) | 13.54 | Self(Vote(DT,LR) | 14.28 |
| Co(Vote(SVM,DT)) | 14.58 | Co(Vote(DT,GNB)) | 13.64 | Co(Vote(SVM,LR) | 14.98 |
| Co(Vote(DT,GNB)) | 14.58 | Self(Vote(DT,GNB)) | 14.31 | Co(SVM) | 14.98 |
| Co(Vote(kNN,LR)) | 14.92 | Co(Vote(kNN,DT)) | 14.54 | Co(LR) | 14.98 |
| Self(Vote(DT,GNB)) | 15.33 | Co(Vote(kNN,LR)) | 15.06 | Self(Vote(DT,GNB)) | 15.24 |
| Self(LR) | 15.44 | Self(DT) | 15.12 | Co(Vote(GNB,LR)) | 15.29 |
| Co(Vote(kNN,DT)) | 15.64 | Self(LR) | 15.31 | Self(LR) | 16.01 |
| Self(Vote(SVM,LR)) | 16.02 | Self(Vote(kNN,DT)) | 15.46 | Self(Vote(kNN,LR) | 16.59 |
| Self(DT) | 16.04 | Self(Vote(DT,LR)) | 15.78 | Co(Vote(kNN,DT) | 16.74 |
| Self(Vote(kNN,DT)) | 16.61 | Self(Vote(SVM,DT)) | 16.33 | Co(Vote(kNN,GNB)) | 16.81 |
| Self(Vote(DT,LR)) | 16.91 | Co(Vote(SVM,LR)) | 16.33 | Co(Vote(SVM,DT) | 16.84 |
| Self(Vote(kNN,LR)) | 16.96 | Co(Vote(kNN,GNB)) | 17.05 | Co(DT) | 16.84 |
| Co(Vote(SVM,kNN)) | 17.49 | Self(Vote(kNN,LR)) | 17.21 | Co(Vote(SVM,GNB)) | 17.06 |
| Self(Vote(SVM,DT)) | 17.56 | Self(Vote(SVM,LR)) | 17.38 | Co(GNB) | 17.06 |
| Co(Vote(kNN,GNB)) | 18.06 | Self(Vote(kNN,GNB)) | 17.69 | Self(Vote(GNB,LR)) | 17.59 |
| Co(kNN) | 18.18 | Co(kNN) | 17.95 | Self(Vote(SVM,LR) | 18.09 |
| Self(Vote(kNN,GNB)) | 18.34 | Co(Vote(SVM,kNN)) | 19.20 | Self(SVM) | 18.09 |
| Co(Vote(SVM,GNB)) | 19.45 | Co(Vote(SVM,GNB)) | 19.59 | Self(Vote(kNN,GNB)) | 18.42 |
| Self(Vote(SVM,kNN)) | 19.46 | Self(Vote(SVM,GNB)) | 19.83 | Self(Vote(kNN,DT) | 18.54 |
| Self(Vote(SVM,GNB)) | 20.01 | Self(Vote(GNB,LR)) | 19.88 | Self(Vote(SVM,GNB)) | 18.62 |
| Self(Vote(GNB,LR)) | 20.29 | Co(Vote(GNB,LR)) | 20.02 | Co(Vote(SVM,kNN) | 19.41 |
| Co(Vote(GNB,LR)) | 20.43 | Self(kNN) | 21.06 | Co(kNN) | 19.41 |
| Co(SVM) | 20.74 | Self(GNB) | 21.15 | Self(GNB) | 19.52 |
| Self(kNN) | 20.85 | Self(Vote(SVM,kNN)) | 21.61 | Self(Vote(SVM,DT) | 21.03 |
| Self(SVM) | 21.89 | Co(GNB) | 21.84 | Self(Vote(SVM,kNN)) | 22.62 |
| Self(GNB) | 22.14 | Co(SVM) | 24.21 | Self(kNN) | 24.67 |
| Co(GNB) | 22.56 | Self(SVM) | 25.33 | Self(DT) | 25.67 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karlos, S.; Kostopoulos, G.; Kotsiantis, S. A Soft-Voting Ensemble Based Co-Training Scheme Using Static Selection for Binary Classification Problems. Algorithms 2020, 13, 26. https://doi.org/10.3390/a13010026
Karlos S, Kostopoulos G, Kotsiantis S. A Soft-Voting Ensemble Based Co-Training Scheme Using Static Selection for Binary Classification Problems. Algorithms. 2020; 13(1):26. https://doi.org/10.3390/a13010026
Chicago/Turabian StyleKarlos, Stamatis, Georgios Kostopoulos, and Sotiris Kotsiantis. 2020. "A Soft-Voting Ensemble Based Co-Training Scheme Using Static Selection for Binary Classification Problems" Algorithms 13, no. 1: 26. https://doi.org/10.3390/a13010026
APA StyleKarlos, S., Kostopoulos, G., & Kotsiantis, S. (2020). A Soft-Voting Ensemble Based Co-Training Scheme Using Static Selection for Binary Classification Problems. Algorithms, 13(1), 26. https://doi.org/10.3390/a13010026






