Classical and Deep Learning Paradigms for Detection and Validation of Key Genes of Risky Outcomes of HCV
Abstract
:1. Introduction
2. Literature Review
3. Proposed Framework
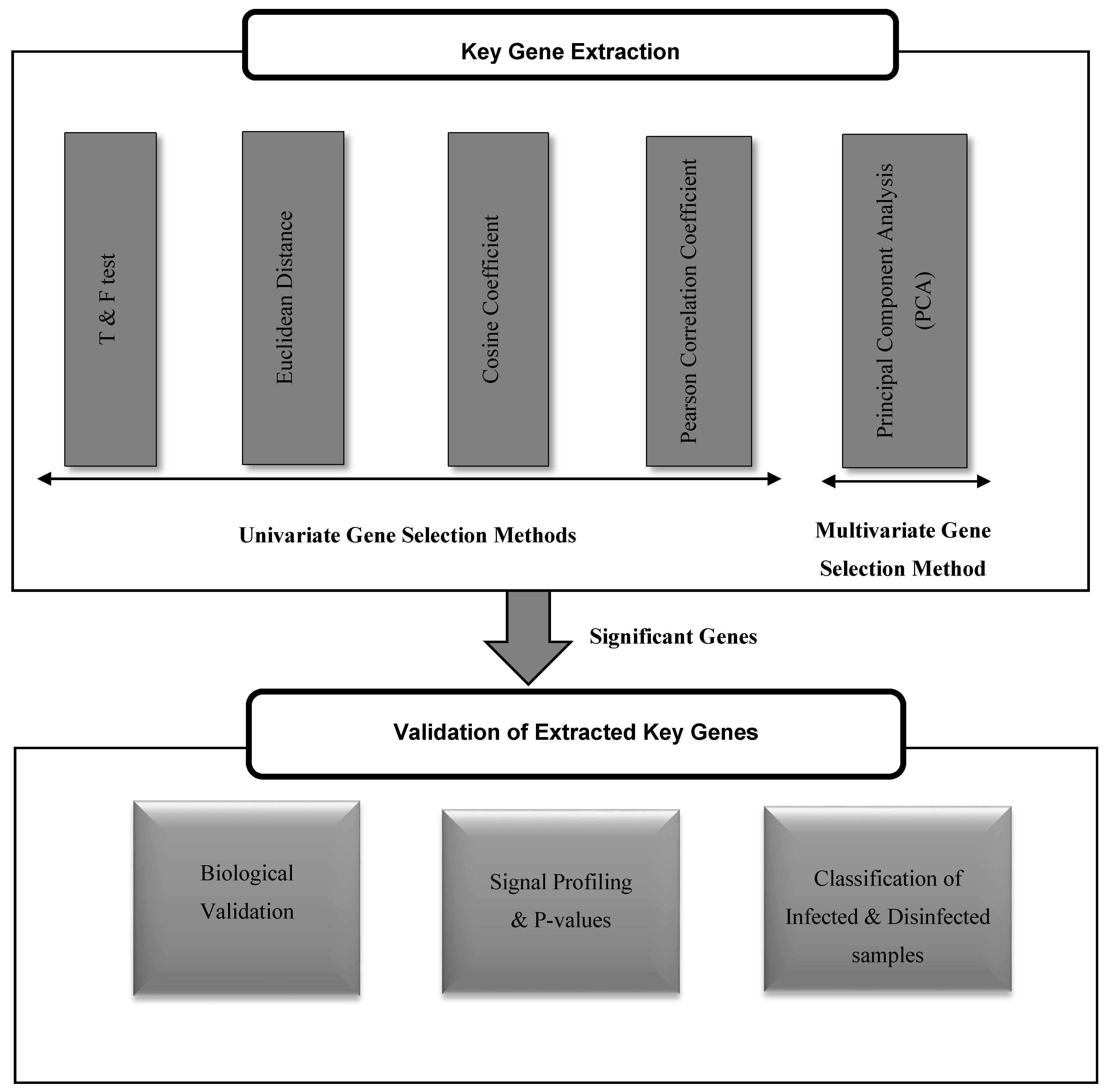
3.1. Key Genes Extraction
Ideal Up/Down Regulated Key Genes
3.2. Univariate Gene Selection Methods
3.3. Multivariate Gene Selection Method
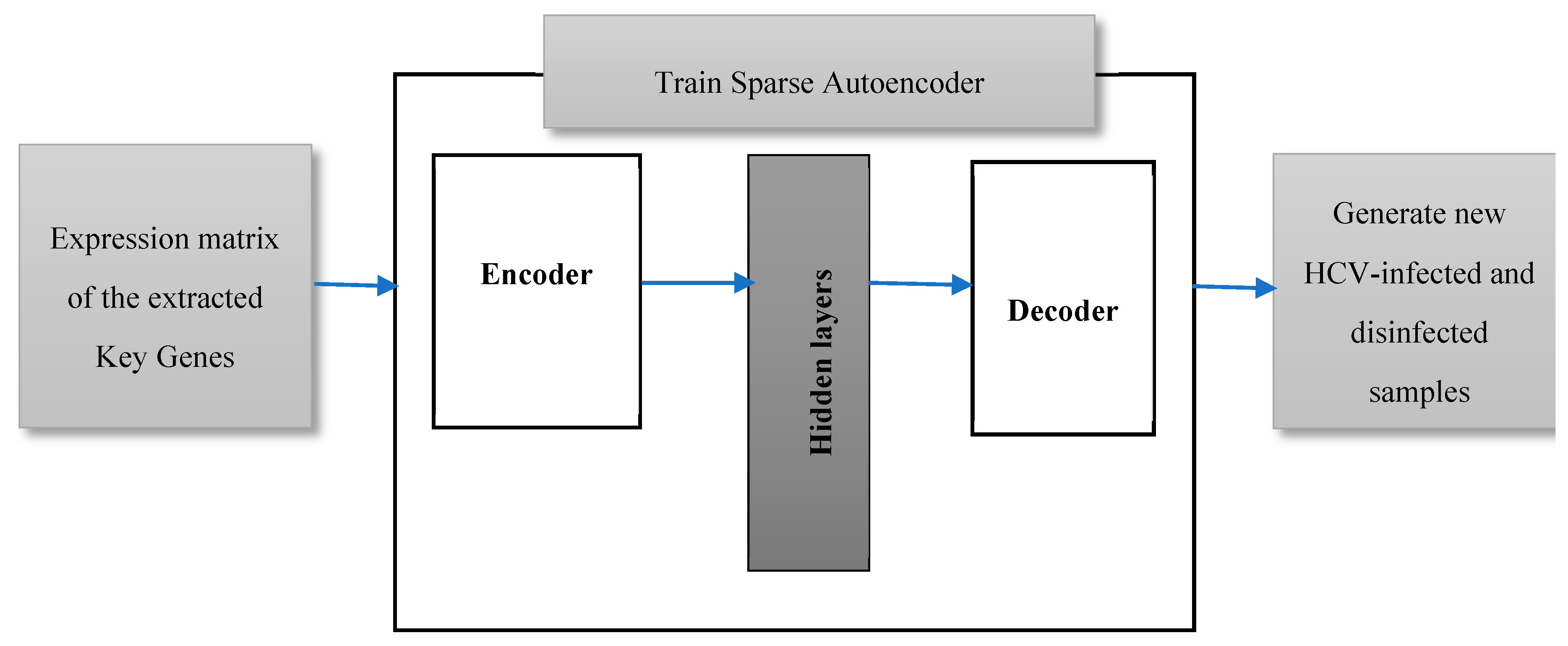
3.4. Validating the Extracted Key Genes
Learning Parameters
4. Results and Discussion
4.1. Biological validation of extracted Key Genes
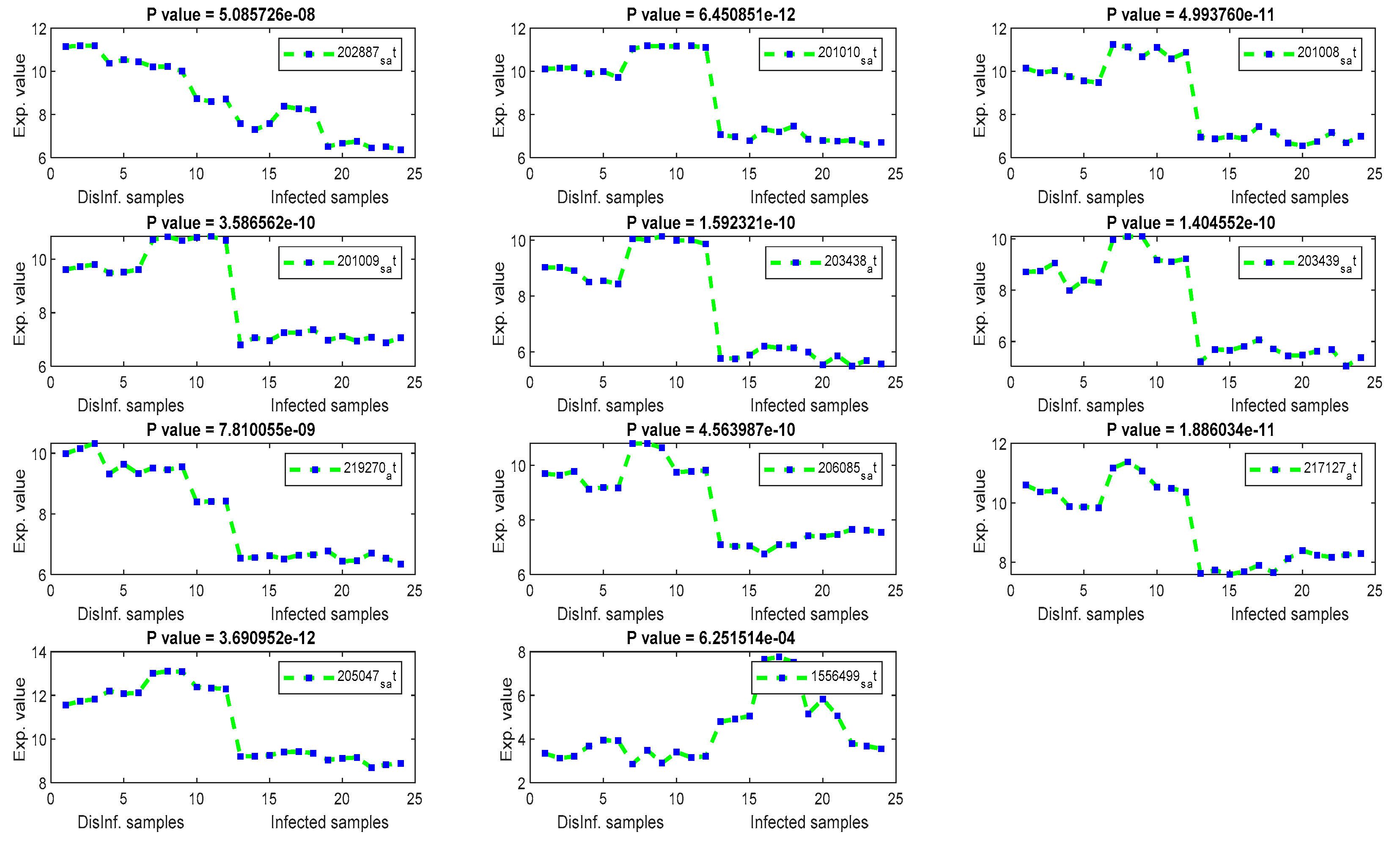
4.2. Signal Profiles and P-Values of Extracted Key Genes
4.3. Discussing the Relevance of Extracted Key Genes Based on Their Biological Examination and Signal Profiles
4.4. Examining the Relevance of Extracted Key Gens Using Conventional Classification & Data Augmentation
5. Conclusions
Funding
Conflicts of Interest
Appendix A
References
- Tang, J.; Jiang, R.; Deng, L.; Zhang, X.; Wang, K.; Sun, B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget 2015, 6, 4505–4515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartenschlager, R.; Cosset, F.L.; Lohmann, V. Hepatitis C virus replication cycle. J. Hepatol. 2010, 53, 583–585. [Google Scholar] [PubMed] [Green Version]
- Li, W.-Q.; Park, Y.; McGlynn, K.A.; Hollenbeck, A.R.; Taylor, P.R.; Goldstein, A.M.; Freedman, N.D. Index-based dietary patterns and risk of incident hepatocellular carcinoma and mortality from chronic liver disease in a prospective study. Hepatology 2014, 60, 588–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackham, S.; Baillie, A.; Al-Hababi, F.; Remlinger, K.; You, S.; Hamatake, R.; McGarvey, M.J. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J. Virol. 2010, 84, 5404–5414. [Google Scholar] [CrossRef] [Green Version]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar]
- Wang, X.; Simon, R. Microarray-based cancer prediction using single genes. BMC Bioinform. 2011, 12, 391. [Google Scholar] [CrossRef] [Green Version]
- Yoo, C.; Ramirez, L.; Liuzzi, J. Big data analysis using modern statistical and machine learning methods in medicine. Int. Neurourol. J. 2014, 18, 50–57. [Google Scholar]
- Sui, S.; Wang, X.; Zheng, H.; Guo, H.; Chen, T.; Ji, D.-M. Gene set enrichment and topological analyses based on interaction networks in pediatric acute lymphoblastic leukemia. Oncol. Lett. 2015, 10, 3354–3362. [Google Scholar] [CrossRef] [Green Version]
- Urda, D.; Luque-Baena, R.M.; Franco, L.; Jerez, J.M.; Sanchez-Marono, N. Machine learning models to search relevant genetic signatures in clinical context. In Proceedings of the International Joint Conference on Neural Networks, Anchorage, AK, USA, 14–19 May 2017; pp. 1649–1656. [Google Scholar]
- Yao, F.; Coquery, J.; Lê Cao, K.A. Independent Principal Component Analysis for biologically meaningful dimension reduction of large biological data sets. BMC Bioinform. 2012, 13, 24. [Google Scholar] [CrossRef] [Green Version]
- Bellinger, C.; Japkowicz, N.; Drummond, C. Synthetic oversampling for advanced radioactive threat detection. In Proceedings of the 2015 IEEE 14th International Conference on Machine Learning and Applications, ICMLA 2015, Miami, FL, USA, 9–11 December 2016; pp. 948–953. [Google Scholar]
- Bellinger, C.; Drummond, C.; Japkowicz, N. Manifold-based synthetic oversampling with manifold conformance estimation. Mach. Learn. 2018, 107, 605–637. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Xie, W.; Liu, T. Efficient feature selection and classification for microarray data. PLoS ONE 2018, 13, e0202167. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Li, Y.; Yi, K.; Wu, Z. Synthetic Data Approach for Classification and Regression. In Proceedings of the International Conference on Application-Specific Systems, Architectures and Processors, Milan, Italy, 10–12 July 2018. [Google Scholar]
- Lyu, B.; Haque, A. Deep Learning Based Tumor Type Classification Using Gene Expression Data. In Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics; Association for Computing Machinery: New York, NY, USA, 28 November 2018; pp. 89–96. [Google Scholar]
- Wang, S.; Minku, L.L.; Yao, X. A Systematic Study of Online Class Imbalance Learning with Concept Drift. IEEE Trans. Neural Networks Learn. Syst. 2018, 29, 4802–4821. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Wen, J.; Quitadamo, A.; Cheng, J.; Shi, X. A deep auto-encoder model for gene expression prediction. BMC Genom. 2017, 18, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ca, P.V.; Edu, L.T.; Lajoie, I.; Ca, Y.B.; Ca, P.-A.M. Stacked Denoising Autoencoders: Learning Useful Representations in a Deep Network with a Local Denoising Criterion Pascal Vincent Hugo Larochelle Yoshua Bengio Pierre-Antoine Manzagol. J. Mach. Learn. Res. 2010, 11, 3371–3408. [Google Scholar]
- Hsieh, C.B.; Chen, T.W.; Chu, C.M.; Chu, H.C.; Yu, C.P.; Chung, K.P. Is inconsistency of α-fetoprotein level a good prognosticator for hepatocellular carcinoma recurrence? World J. Gastroenterol. 2010, 16, 3049–3055. [Google Scholar] [CrossRef]
- Di Carlo, I.; Mannino, M.; Toro, A.; Ardiri, A.; Galia, A.; Cappello, G.; Bertino, G. Persistent increase in alpha-fetoprotein level in a patient without underlying liver disease who underwent curative resection of hepatocellular carcinoma. A case report and review of the literature. World J. Surg. Oncol. 2012, 10, 79. [Google Scholar] [CrossRef] [Green Version]
- You, Z.; Wang, S.; Gui, J.; Zhang, S. A novel hybrid method of gene selection and its application on tumor classification. In Advanced Intelligent Computing Theories and Applications. With Aspects of Artificial Intelligence; Huang, D.S., Wunsch, D.C., Levine, D.S., Jo, K.H., Eds.; Springer: Berlin, Germany, 2008; Volume 5227, pp. 1055–1068. [Google Scholar]
- Advances in Kernel Methods-Support Vector Learning. Available online: https://www.researchgate.net/publication/2346087_Advances_in_Kernel_Methods_-_Support_Vector_Learning (accessed on 10 October 2019).
- Park, T.; Casella, G. The Bayesian Lasso. J. Am. Stat. Assoc. 2008, 103, 681–686. [Google Scholar] [CrossRef]
- Meier, L.; Van De Geer, S.; Bühlmann, P. The Group Lasso for Logistic Regression. J. R. Stat. Soc. Ser. B 2008, 70, 53–71. [Google Scholar] [CrossRef] [Green Version]
- Reverter, F.; Vegas, E.; Sánchez, P. Mining Gene Expression Profiles: An Integrated Implementation of Kernel Principal Component Analysis and Singular Value Decomposition. Genom. Proteom. Bioinforma. 2010, 8, 200–210. [Google Scholar] [CrossRef] [Green Version]
- SchölkopfSch, B.; Smola, A.; Müller, K.R. Nonlinear Component Analysis as a Kernel Eigenvalue Problem. Neural Comput. 1998, 10, 1299–1319. [Google Scholar]
- Park, M.; Lee, J.W.; Bok Lee, J.; Heun Song, S. Several biplot methods applied to gene expression data. J. Stat. Plan. Inference 2008, 138, 500–515. [Google Scholar] [CrossRef]
- Nilashi, M.; Ahmadi, H.; Shahmoradi, L.; Ibrahim, O.; Akbari, E. A predictive method for hepatitis disease diagnosis using ensembles of neuro-fuzzy technique. J. Infect. Public Health 2019, 12, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xu, Q.; Xie, H.; Gu, G.; Jiang, J. Expression of serum miR-218 in hepatocellular carcinoma and its prognostic significance. Clin. Transl. Oncol. 2016, 18, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Abdel Samee, N.M.; Solouma, N.H.; Kadah, Y.M. Detection of biomarkers for Hepatocellular Carcinoma using a hybrid univariate gene selection methods. Theor. Biol. Med. Model. 2012, 9, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markopoulos, P.P.; Kundu, S.; Chamadia, S.; Pados, D.A. Efficient L1-Norm Principal-Component Analysis via Bit Flipping. IEEE Trans. Signal Process. 2017, 65, 4251–4264. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Factoextra: Extract and Visualize the Results of Multivariate Data Analyses Version 1.0.5 from CRAN. Available online: https://rdrr.io/cran/factoextra/ (accessed on 11 October 2019).
- Maglott, D.; Ostell, J.; Pruitt, K.D.; Tatusova, T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2005, 33, D54–D58. [Google Scholar] [CrossRef]
- Masaki, S.; Masutani, H.; Yoshihara, E.; Yodoi, J. Deficiency of thioredoxin binding protein-2 (TBP-2) enhances TGF-β signaling and promotes epithelial to mesenchymal transition. PLoS ONE 2012, 7, e39900. [Google Scholar] [CrossRef]
- Wu, F.; Li, T.-Y.; Su, S.-C.; Yu, J.-S.; Zhang, H.-L.; Tan, G.-Q.; Liu, J.-W.; Wang, B.-L. STC2 as a novel mediator for Mus81-dependent proliferation and survival in hepatocellular carcinoma. Cancer Lett. 2017, 388, 177–186. [Google Scholar] [CrossRef]
- Balasubramanian, M.N.; Butterworth, E.A.; Kilberg, M.S. Asparagine synthetase: Regulation by cell stress and involvement in tumor biology. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E789–E799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, J.A.; Rolfo, C.; Raez, L.E.; Prado, A.; Araujo, J.M.; Bravo, L.; Fajardo, W.; Morante, Z.D.; Aguilar, A.; Neciosup, S.P.; et al. In silico evaluation of DNA Damage Inducible Transcript 4 gene (DDIT4) as prognostic biomarker in several malignancies. Sci. Rep. 2017, 7, 1526. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallstrom, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Y.; Bowersock, A.; Badour, A.R.; Vij, N.; Juris, S.J.; Ash, D.E.; Mohanty, D.K. Dichotomous effects of isomeric secondary amines containing an aromatic nitrile and nitro group on human aortic smooth muscle cells via inhibition of cystathionine-γ-lyase. Biochimie 2017, 133, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.-P.; Chang, H.-L.; Bamodu, O.A.; Yadav, V.K.; Huang, T.-Y.; Wu, A.T.H.; Yeh, C.-T.; Tsai, S.-H.; Lee, W.-H. Collagen 1A1 (COL1A1) Is a Reliable Biomarker and Putative Therapeutic Target for Hepatocellular Carcinogenesis and Metastasis. Cancers 2019, 11, 786. [Google Scholar] [CrossRef] [Green Version]
- Zou, K.; Lu, X.; Ye, K.; Wang, C.; You, T.; Chen, J. Krüppel-like factor 2 promotes cell proliferation in hepatocellular carcinoma through up-regulation of c-myc. Cancer Biol. Ther. 2016, 17, 20–26. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.-X.; Jin, L.; Sun, S.-J.; Liu, P.; Feng, X.; Cheng, Z.-L.; Liu, W.-R.; Guan, K.-L.; Shi, Y.-H.; Yuan, H.-X.; et al. Metabolic reprogramming by PCK1 promotes TCA cataplerosis, oxidative stress and apoptosis in liver cancer cells and suppresses hepatocellular carcinoma. Oncogene 2018, 37, 1637–1653. [Google Scholar] [CrossRef]
- Forst, A.H.; Karlberg, T.; Herzog, N.; Thorsell, A.; Feijs, K.; Verheugd, P.; Kursula, P.; Nijmeijer, B.; Lippok, B.; Kleine, H.; et al. Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure 2013, 21, 462–475. [Google Scholar] [CrossRef] [Green Version]
- Sirivatanauksorn, Y.; Sirivatanauksorn, V.; Srisawat, C.; Khongmanee, A.; Tongkham, C. Differential expression of sprouty genes in hepatocellular carcinoma. J. Surg. Oncol. 2012, 105, 273–276. [Google Scholar] [CrossRef]
- Fu, P.; Yang, F.; Li, B.; Zhang, B.; Guan, L.; Sheng, J.; Ye, Y.; Wang, Z.; Li, P.; Xu, L.; et al. Meta-analysis of CYP2E1 polymorphisms in liver carcinogenesis. Dig. Liver Dis. 2017, 49, 77–83. [Google Scholar] [CrossRef]
- Katsuoka, F.; Motohashi, H.; Ishii, T.; Aburatani, H.; Engel, J.D.; Yamamoto, M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol. Cell. Biol. 2005, 25, 8044–8051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fénéant, L.; Levy, S.; Cocquerel, L. CD81 and hepatitis C virus (HCV) infection. Viruses 2014, 6, 535–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, P.; Sun, D.; Wang, L.; Fan, R.; Gao, Z. Deep sequencing and comprehensive expression analysis identifies several molecules potentially related to human poorly differentiated hepatocellular carcinoma. FEBS Open Bio 2017, 7, 1696–1706. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Liu, Q.; Jia, Y.; Tu, K.; Yao, Y.; Liu, Q.; Guo, C. BCAT1 promotes tumor cell migration and invasion in hepatocellular carcinoma. Oncol. Lett. 2016, 12, 2648–2656. [Google Scholar] [CrossRef] [Green Version]
- Dapito, D.H.; Mencin, A.; Gwak, G.Y.; Pradere, J.P.; Jang, M.K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of Hepatocellular Carcinoma by the Intestinal Microbiota and TLR4. Cancer Cell 2012, 21, 504–516. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.; Zheng, L.; Yoo, J.K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356.e16. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Zhao, J.; Lin, L.; Zhang, Y.; Zhong, F.; Liu, Y.; Yu, Y.; Shen, H.; Han, M.; He, F.; et al. Proteomic study explores AGR2 as pro-metastatic protein in HCC. Mol. Biosyst. 2012, 8, 2710–2718. [Google Scholar] [CrossRef]
- Chen, S.L.; Lu, S.X.; Liu, L.L.; Wang, C.H.; Yang, X.; Zhang, Z.Y.; Zhang, H.Z.; Yun, J. ping eEF1A1 Overexpression Enhances Tumor Progression and Indicates Poor Prognosis in Hepatocellular Carcinoma. Transl. Oncol. 2018, 11, 125–131. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Emelyanov, A.; Koh, C.H.V.; Spitsbergen, J.M.; Lam, S.H.; Mathavan, S.; Parinov, S.; Gong, Z. A high level of liver-specific expression of oncogenic Kras V12 drives robust liver tumorigenesis in transgenic zebrafish. DMM Dis. Model. Mech. 2011, 4, 801–813. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.-Y.; Jia, H.-L.; Dong, Q.-Z.; Wu, J.-C.; Zhao, Y.; Zhou, H.-J.; Ren, N.; Ye, Q.-H.; Qin, L.-X. Suitable reference genes for real-time PCR in human HBV-related hepatocellular carcinoma with different clinical prognoses. BMC Cancer 2009, 9, 49. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.; Wang, X.; Huang, L.; Tong, Y.; Chen, L.; Wu, H.; Xia, Q.; Kong, X. Deciphering the spectrum of mitochondrial DNA mutations in hepatocellular carcinoma using high-Throughput sequencing. Gene Expr. 2018, 18, 125–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strnad, P.; Lienau, T.C.; Tao, G.Z.; Lazzeroni, L.C.; Stickel, F.; Schuppan, D.; Omary, M.B. Keratin variants associate with progression of fibrosis during chronic hepatitis C infection. Hepatology 2006, 43, 1354–1363. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Z.; Du, F.; Yang, Q.; Hou, J.; Yan, X.; Geng, Y.; Zhao, Y.; Wang, H. Molecular mechanisms of pathogenesis in hepatocellular carcinoma revealed by RNA-sequencing. Mol. Med. Rep. 2017, 16, 6674–6682. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| AFFY ID | Gene Symbol | ENTREZ Gene ID | Oncology | Gene Pathway |
|---|---|---|---|---|
| 201010_s_at, 201008_s_at, 201009_s_at | TXNIP | 10628 | Breast cancer, prostate Carcinoma, colorectal carcinoma, Hepatocellular Carcinoma (HCC) | REACT_75808. The NLRP3 inflammasome. cellular response to tumor cell |
| 203438_at, 203439_s_at | STC2 | 8614 | Colorectecal cancer, Breast cancer, Mutation of HCC | KEGG: hsa: 8614. |
| 205047_s_at | ASNS | 440 | Cancer, Protein and/or amino acid deprivation | REACT_238. liver development. REACT_18355. ATF4 activates genes. |
| 202887_s_at | DDIT4 | 54541 | Pancreatic tumor, prostate cancer, lung carcinoma | REACT_355377. TP53 Regulates Metabolic Genes |
| 219270_at | CHAC1 | 79094 | downstream of the ATF4 | KEGG: hsa79094 CHAC1 is a component of the UPR, unfolded protein response pathway. |
| 206085_s_at, 217127_at | CTH | 1491 | Bladder Cancer | REACT_115589. Cysteine ormation from homocysteine |
| 1556499_s_at | COL1A1 | 1277 | Mutation in liver, infirative skin carcinoma, bendnar carcinoma | REACT_118779. Extracellular matrix organization. cascade. |
| 210587_at | INHBE | 83729 | hepatocellular carcinoma, HCC | REACT_15398. Glycoprotein hormones |
| 202672_s_at | ATF3 | 467 | Solid tumor | REACT_18355. ATF4 activates genes. |
| AFFY ID | Gene Symbol | ENTREZ Gene ID | Oncology | Gene Pathway |
|---|---|---|---|---|
| 213322_at | OARD1 | 221443 | infiltrating duct carcinoma | KEGG: hsa: 221443. |
| 36711_at | MAFF | 23764 | leukemia/lymphoma (BCR-ABL1) | REACT_24970. megakaryocyte, and platelet construction. |
| 205749_at | CYP1A1 | 1543. | hepatocellular carcinoma, NOS, unstated behavior | KEGG: hsa: 1543. REACT_116145. PPARA activates gene expression. |
| 219371_s_at | KLF2 | 10365. | chronic lymphocytic B-cell leukemia | KEGG: hsa: 10365. |
| 212558_at | SPRY1 | 10252 | HCC, gastrointestinal stromal sarcoma | REACT_12484. EGFR downregulation. |
| 205047_s_at | ASNS | 440 | Cancer, Protein and/or amino acid deprivation | REACT_238. liver development. REACT_18355. ATF4 activates genes. |
| 201010_s_at 201009_s_at | TXNIP | 10628 | HCC, Breast cancer, prostate Carcinoma, colorectal carcinoma | REACT_75808. The NLRP3 inflammasome. cellular response to tumor cell |
| 203119_at | CCDC86 | 79080 | HCV, squamous cell carcinoma | KEGG:hsa79080 |
| 232780_s_at | ZNF691 | 51058 | Infiltrating duct carcinoma | REACT_12627. Generic Transcription Pathway. |
| 202847_at | PCK2 | 5106 | HCC | KEGG:hsa00020Citrate cycle (TCA cycle) |
| AFFY ID | Gene Symbol | ENTREZ Gene ID | Oncology | -Gene Pathway |
|---|---|---|---|---|
| 36711_at | MAFF | 23764 | leukemia/lymphoma (BCR-ABL1) | REACT_24970. Megakaryocyte and platelet construction. |
| 205749_at | CYP1A1 | 1543. | hepatocellular carcinoma, NOS, unstated behavior | KEGG: hsa: 1543. REACT_116145. PPARA activates gene expression. |
| 209775_x_at | SLC19A1 | 6573. | anaplastic large cell lymphoma | KEGG: hsa 6573. REACT_11167. Metabolism of folate and pterines. |
| 205767_at | EREG | 2069. | chronic myelogenous leukemia (BCR/ABL-positive) | KEGG: hsa 2069. REACT_147727. Signaling by PI3K in Cancer. |
| 217996_at | PHLDA1 | 22822 | gastrointestinal stromal sarcoma | KEGG: hsa: 22822. |
| 226515_at | CCDC127 | 133957 | renal cell carcinoma | KEGG:hsa:133957 |
| 206085_s_at, 217127_at | CTH | 1491 | Bladder Cancer | REACT_115589. Cysteine ormation from homocysteine |
| 225285_at | BCAT1 | 586 | HCC | REACT_197. Branched-chain amino acid catabolism. |
| 202847_at | PCK2 | 5106 | HCC | KEGG:hsa00020Citrate cycle (TCA cycle) |
| 209173_at | AGR2 | 10551 | Breast Cancer | KEGG: hsa: 10551. |
| AFFY ID | Gene Symbol | ENTREZ Gene ID | Oncology | Gene Pathway |
|---|---|---|---|---|
| 204892_x_at | EEF1A1 | 1915 | HCC | REACT_1404. Peptide chain elongation |
| 1553567_s_at | ATP6 | 4508. | HCC, adenoma | REACT_6759. Development of ATP. |
| 200801_x_at | ACTP | 948575. | hematopoietic | KEGG: eco: b4067. |
| 212788_x_at | FTL | 2512. | HCC | REACT_163699. Scavenging by Class A Receptors. |
| 200801_x_at | ACTB | 60 | Langerhans-cell histiocytosis | REACT_20649. Cell-extracellular matrix interactions. |
| 201596_x_at | KRT18 | 3875. | HCV, adenocarcinoma | KEGG: hsa: 3875. |
| 1553570_x_at | COX2 | 5743 | Adenocarcinoma, Mutation in HCC | REACT_11213. Nicotinamide salvaging. |
| 224372_at | MTND4 | 4538. | Adenocarcinoma, Mutation in HCC | REACT_22393. Respiratory electron transport. |
| 212661_x_at | PPIA | 5478. | Burkitt lymphoma Mutation in HCC | REACT_9406. HIV-1 infection. |
| 221798_x_at | RPS2 | anaplastic large cell lymphoma | ||
| 1553538_s_at | COX1 | 5742. | Mutation gene in HCC | REACT_1396. COX reactions. |
| 211296_x_at | UBC | 7316. | leukemia/lymphoma | REACT_115852. Signaling by EGFR Cancer Variants. |
| Feature Selection Method | Mean Squared Reconstruction Error of the Generated Samples |
|---|---|
| T, and F test | 0.0891 |
| Pearson’s correlation, and cosine coefficient | 0.04966 |
| Euclidean distance | 0.05345 |
| Principal component analysis (PCA) | 0.005861 |
| Feature Selection Method | LDA-Linear | QDA-Quadratic | KNN | SVM | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| K = 1 | K = 3 | K = 5 | SVM-Linear | SVM-Quadratic | SVM-Cubic | SVM-RBF | SVM-RBF Optimized | |||
| T and F test | 66.7 | 95.83 | 79.1667 | 68.75 | 60.417 | 37.50 | 66.667 | 79.167 | 56.25 | 83.334 |
| Pearson’s correlation coefficient, cosine coefficient | 48 | 62.5 | 75 | 62.5 | 56.25 | 37.50 | 64.583 | 85.417 | 72.916 | 85.4167 |
| Euclidean distance) | 69.75 | 78.2 | 81.25 | 79.167 | 77.083 | 50 | 81.25 | 85.417 | 81.25 | 91.667 |
| Principal component analysis (PCA) | 70.8 | 93.75 | 85.41667 | 70.8334 | 72.9167 | 50 | 89.5833 | 87.50 | 77.083 | 91.667 |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdel Samee, N.M. Classical and Deep Learning Paradigms for Detection and Validation of Key Genes of Risky Outcomes of HCV. Algorithms 2020, 13, 73. https://doi.org/10.3390/a13030073
Abdel Samee NM. Classical and Deep Learning Paradigms for Detection and Validation of Key Genes of Risky Outcomes of HCV. Algorithms. 2020; 13(3):73. https://doi.org/10.3390/a13030073
Chicago/Turabian StyleAbdel Samee, Nagwan M. 2020. "Classical and Deep Learning Paradigms for Detection and Validation of Key Genes of Risky Outcomes of HCV" Algorithms 13, no. 3: 73. https://doi.org/10.3390/a13030073




