1. Introduction
Skin cancer is one of the most common types of cancer in the global Caucasian population [
1]. It is one of the three most dangerous and fastest-growing types of cancer and is therefore a serious public health problem [
2]. Skin tumors can be benign [
3] or malignant; the main malignant cancers are basalioma, squamous cell carcinoma, and malignant melanoma [
4]. Of these, melanoma is the least common but at the same time is the most aggressive, and it can lead to death if diagnosed late. Therefore, it is critical to detect it early, which increases the chances of treatment and cure [
5]. Its diagnosis follows some simple rules [
6]: the ABCDE rule, which is based on morphological features of the lesions such as asymmetry (A), border irregularity (B), color variety (C), diameter (D), and evolution (E); the Seven Point Checklist, which is based on seven dermoscopic features representative of melanoma; and the Menzies method, which scores based on the presence/absence of certain positive or negative characteristics. The ABCDE rule is the most widely used because of the winning combination of ease of application and diagnostic efficacy. In fact, while nevi generally are symmetrical (round or oval), have regular edges (smooth and uniform), show homogeneous color, small size (diameter less than 6 mm), and do not evolve over time (control in follow-up), most melanomas have an asymmetrical shape with irregular edges, the color is not uniform but can vary in shades of brown (as the melanoma grows, it can also take on red, blue, or white colors), they are larger than nevi, and their features change over time. In some cases, however, melanomas and nevi show similar features to each other, and it is therefore difficult to distinguish them by simple visual inspection. The identification of some characteristics, such as symmetry, size, broths, and the presence and distribution of color features (but also blue–white areas, atypical pigmented networks, and globules) is essential for the diagnosis of skin lesions [
7]; however, it is a complex and time-consuming procedure that shows a strong dependence on the subjectivity and experience of the physician. These problems necessitate the development of computer-aided diagnostic systems (CAD systems), which generally involve several steps for the analysis and classification of dermoscopic images of skin lesions [
8]: preprocessing, segmentation, feature extraction, and classification using ML and DL approaches. Preprocessing aims to attenuate artifacts in the images, which are mainly due to the presence of hairs and marker marks on the lesions, while the segmentation phase aims to isolate the lesion from the surrounding skin and thus extract clinically relevant features. Several solutions have been proposed for these two phases by researchers [
9,
10,
11,
12,
13,
14], many of which are laborious and require the training of additional machine and deep learning models. The feature extraction step can be manual [
15] or can be automated using machine learning algorithms. Manual feature extraction in the case of skin lesion classification is based on the methodologies devised by dermatologists for skin cancer diagnosis and, in particular, the ABCDE rule of dermatology. Using machine learning methods, learned features are derived automatically from the datasets and require no prior knowledge of the problem. A variety of approaches are also possible for the final stage of classification: from classical machine learning approaches to state-of-the-art methodologies based on deep convolutional neural networks.
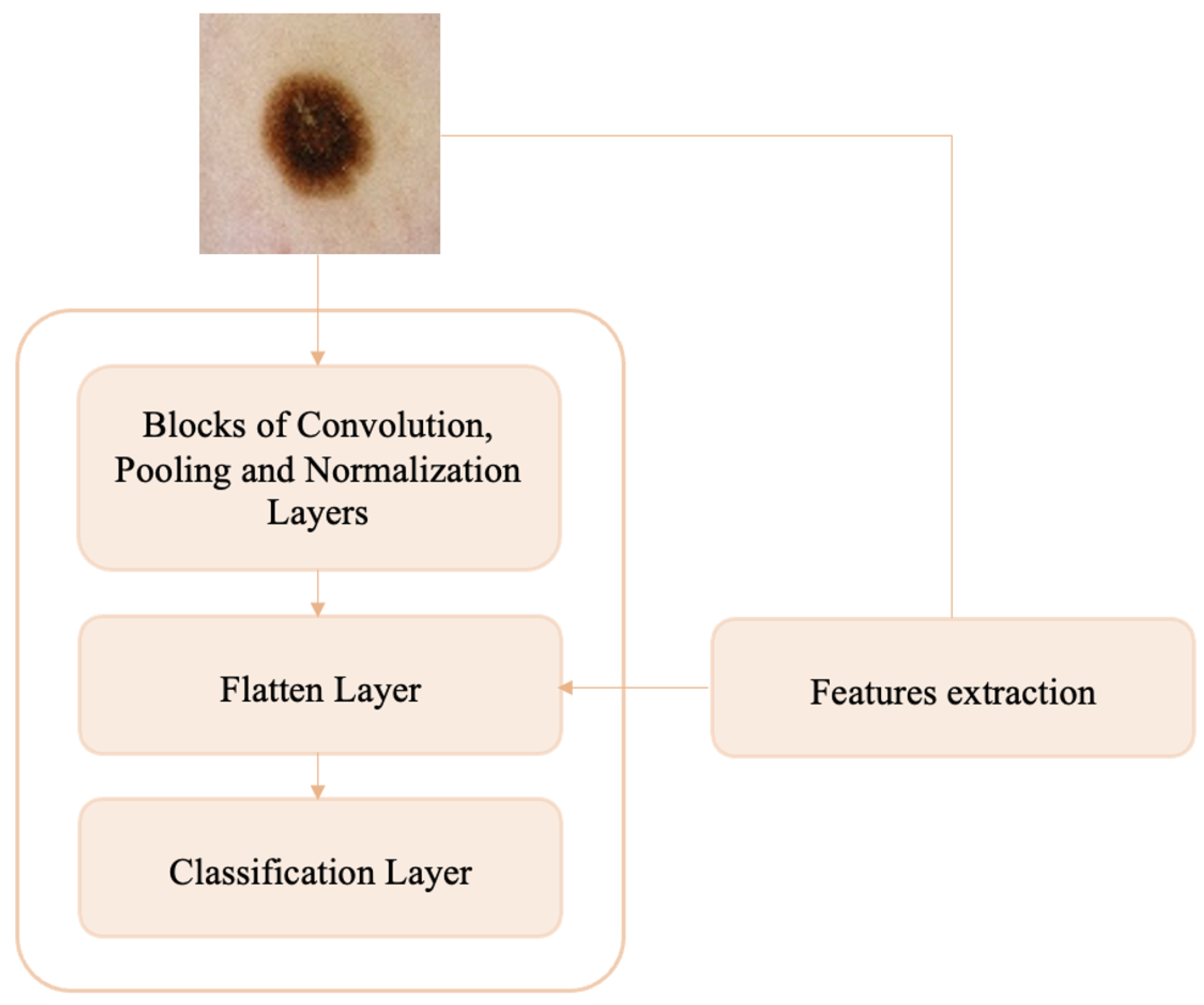
Melanoma detection can be understood as an anomaly detection problem in skin lesions because melanomas represent anomalies (shape, size, color, etc.) in the nevi population. In this paper, we propose an anomaly detection method that involves only the manual and automatic feature extraction and classification steps. Specifically, we develop a custom CNN that, in addition to the dataset of melanoma and nevi images, is trained on additional handcrafted texture features extracted from whole of dermoscopic images. This avoids the segmentation step and could capture some aspects of pathology related to the tissue surrounding the lesions. In addition, no preprocessing is performed on skin lesion images, which reduces the working time. The main contributions of this paper are:
- 1.
A custom convolutional neural network model with added handcrafted features is proposed that classifies melanomas and nevi accurately without the need for image preprocessing and segmentation.
- 2.
On the same test dataset, the performance of the proposed model exceeds that of the most widely used pre-trained models for skin lesion classification (ResNet50, VGG16, DenseNet121, DenseNet169, DenseNet201, and MobileNet).
- 3.
The execution time required by the proposed model to execute the output results is much less than that required by the pre-trained models tested.
- 4.
The proposed model achieves better accuracy on the ISIC 2016 dataset than other existing deep learning models.
- 5.
A brief interpretability analysis of the proposed model is performed.
The paper is organized into the following sections. Related work in the areas of skin lesion classification and anomaly detection is reported in
Section 2.
Section 3 discusses the main challenges and opportunities of skin cancer detection.
Section 4 presents the materials used for the present work, namely public datasets of dermoscopic images.
Section 5 shows the workflow of the proposed methodology, including the extraction of handcrafted features from dermoscopic images and the construction and training of the CNN network for the melanoma–nevi discrimination task.
Section 6 provides details on the settings of the experiments and the creation of the datasets used in training and testing the model. In addition, some ablation experiments and the results obtained for classification are presented. In
Section 7, the results obtained by comparing our model with some transfer learning (TL) approaches commonly used in the field of dermatology and with the literature on the ISIC 2016 dataset and DermIS database are discussed. In addition, a brief interpretability analysis of the proposed model is made. In
Section 8, the conclusions reached are summarized, and some future arrangements are discussed.
6. Results
Implementation details: A macOs computer with an Apple M1 Pro processor and 16 GB RAM was used for training and validation of the proposed model. Models were created with Spyder 5.1.5 and Python 3.9.12 using the Keras library. Scikit-learn, OpenCV, Pandas, and Numpy were used as dependencies, and Tensorflow, which specializes in effective training of deep neural networks by exploiting graphics cards, served as the backend engine. The choice of the Python language was related to its power and accessibility, which have made it the most popular programming language for data science.
Details of datasets used: The model was trained with the ISIC 2018 and ISIC 2019 training datasets and was tested on the ISIC 2016 test dataset to ensure comparison of the results obtained with those reported in other papers. As mentioned above, only melanoma and nevi images were selected from each dataset. In the training datasets, the division between classes shows a strong imbalance in favor of the nevi class, whose images are numerically more than three times as numerous as those of melanomas (a widespread phenomenon in the medical field because it is relatively easy to get normal data, but it is quite difficult to get abnormal data). To arrive at a balance between the two classes under consideration, random downsampling (DS) of the nevi class was performed; specifically, the same number of nevi images were selected from the two training datasets such that their sum was equal to the number of melanoma images. The nevi images were randomly selected from each dataset. The initial and final numbers of melanomas and nevi in the two training datasets are reported in
Table 3.
Table 3.
Number of N and M samples in training datasets.
Table 3.
Number of N and M samples in training datasets.
| Dataset | Before | After |
|---|
| | N | M | Tot | N | M | Tot |
| ISIC 2018 | 6705 | 1113 | 7818 | 2818 | 1113 | 3931 |
| ISIC 2019 | 12,875 | 4522 | 17,397 | 2818 | 4522 | 7340 |
The 10% training data are used as a validation set to provide the impartial process of model training.
Network configuration. After testing several configurations obtained by varying the optimizer between Adam, Adamax, and SGD; the initial learning rate between and ; and the batch size between 32 and 512; the parameters set for the proposed CNN network were:
Classification results: As already anticipated, for the present work, GLCM and LBP texture features were considered for injection into the CNN network to improve its classification performance. Several experiments were conducted to try to find out whether it is convenient to use additional information to that already automatically extracted from the network and, if so, whether it is better to use the two separately or combined. In addition, to understand whether it is necessary to use a complex approach such as a CNN instead of simpler methods, texture features were classified by the most widely used ML methods for skin lesion classification, namely SVM, KNN, and LDA.
- -
SVM: Both the linear kernel (with the values of the inverse regularization parameter C varied between 1, 10, 100, and 1000) and the RBF and polynomial kernels (with all possible combinations between the values of C equal to the previous ones and the values of the gamma parameter equal to 0.001 and 0.0001) were tested. For both GLCM and LBP feature classification, the best results were obtained using the SVM with the RBF kernel and setting the parameters C = 1000 and gamma = 0.001; for GLCM + LBP feature classification, the best result is obtained with a linear SVM and setting the parameter C = 100.
- -
KNN: The number of neighbors was varied between 3 and 10. For both GLCM and LBP feature classification, the best result is obtained with K = 9; while for GLCM + LBP combination, the best result is obtained with K = 10.
- -
LDA: Singular Value Decomposition (SVD) and Least Squares Solution (LSQR) resolution methods were tested, the former of which yielded the best classification performance for all features.
For all ML methods, “best result” means the one that shows the best trade-off between high sensitivity and specificity. The results obtained on the test dataset in the different situations are shown in
Table 4.
The ML methods show similar sensitivity values to those obtained with the various CNN networks, but they have lower specificity, accuracy, and AUC. Looking at the results obtained with CNNs, it is evident from the results that none of the situations is clearly superior to the others, but the best is the one in which LBP texture features are injected into the proposed CNN network. For these latter four situations, ROC curve plots are shown in
Figure 3.
One might also ask whether the LBP features extracted from the images are all relevant or whether a selection of the most significant ones should be performed. In this regard, two alternative scenarios were simulated in which Principal Component Analysis (PCA) was applied to reduce the dimensionality of the features to 1/3 and 1/2 of their original sizes. The results obtained by training the proposed CNN with the addition of these reduced features are shown in
Table 5 along with those obtained by considering all the original features.
In the PCA(1/3) case, sensitivity increases compared with the situation where feature dimensionality is not reduced, but specificity decreases greatly. In contrast, in the PCA(1/2) case, the results are lower than the CNN LBP situation for all metrics. From the results obtained, it can be inferred that it is not necessary to perform dimensionality reduction of LBP texture features since they are all important for classification of skin lesions.
In addition, the present work aims to investigate whether a preliminary image preprocessing step is necessary for the addressed anomaly detection task. In this regard, a comparison is made between the performance obtained by training the custom CNN LBP with the original images and with images preprocessed using the main techniques used in the field of dermatology, as is discussed briefly below.
Gaussian filter (GF) is a linear smoothing filter that operates as a kind of low-pass filter [
70]. It is a widely used preprocessing method in image processing and computer vision to attenuate noise. In the present work, the standard deviation for the Gaussian kernel (
) is set to 1, 3, and 5; the smoothing operation is performed identically in all directions, resulting in a filtering action independent of the orientation of the structures in the image.
Histogram Equalization (HE) is the most widely used global method for calibrating contrast in images and is based on the idea of reassigning pixel intensity values to make the intensity distribution maximally uniform [
71].
Color Normalization (CN) is a technique widely used in computer vision to compensate for illumination variations in images. The method chosen for the present work is based on the
technique of color constancy [
72] that ensures that the perceived color of the image remains the same under different illumination conditions in order to facilitate the classification algorithm.
Various combinations of the techniques just described were also tested: GF + HE, GF + CN, and HE + CN. In such combinations, the Gaussian filter was set with
, which has been shown to yield the best classification results. The results of applying the various preprocessing methods to a skin lesion image are shown in
Figure 4.
A comparison of the classification results obtained by training the neural network with original and preprocessed images is shown in
Table 6.
It can be seen from the results that: (1) the Gaussian filter with and leads to high specificity values but very low sensitivity; (2) color normalization (CNN CN), Gaussian filtering combined with histogram equalization (CNN GF + HE), and the combination of histogram equalization and color normalization (CNN HE + CN) allows for the obtainment of higher specificity values—and therefore higher accuracy—than that obtained without preprocessing; however, in all these situations, the sensitivity, which reflects the accuracy in correctly classifying melanomas and is therefore a parameter of utmost importance for the specific application (anomaly detection in the medical field), is much lower than that obtained with CNN w. p.; (3) application of the Gaussian filter with leads to classification results similar to those obtained by training the network with the original images, but lower; (4) the best trade-off between sensitivity, specificity and accuracy seems to be achieved by the CNN network trained using the original images.
Once the best solution for melanoma detection was identified—which was obtained by adding the texture information captured by the LBP operator from the unprocessed images to the CNN network—we wanted to check whether the selection of the training dataset made to balance the two classes was the best one. In this regard, the proposed balancing situation—the results of which have already been shown in
Table 4—is compared with the following two: (1) the imbalance between the classes is not balanced, and therefore, the training set includes all nevi and melanoma images (scenario named M/Ntot). In this case, the number of nevi is more than three times the number of melanomas; (2) to make the number of melanomas approach the total number of nevi, data augmentation is applied on the melanoma images until the number of melanomas is tripled (scenario named DA Mx3). The operations performed to increase the number of images in the training dataset are: rotation (range = 20), width shift (range = 0.1), height shift (range = 0.1), shear (range = 0.3), zoom (range = 0.3), horizontal flip, and fill mode = ’nearest’. The results obtained by training the network with these new datasets and the comparison without using DA are shown in
Table 7.
Paradoxically, by not balancing the classes and thus having one-third as many melanomas as nevi, the specificity is much higher than the sensitivity; on the other hand, as the number of melanomas increases, the ability of the network to recognize nevi also increases, but the network has zero sensitivity, indicating that no melanomas were classified correctly. The results obtained, although it may not seem like it, are “lucky” results; in fact, they could have been even worse considering the fact that during training, the performance of the models on the validation dataset was anything but consistent in both scenarios previously described.
Figure 5 plots the accuracy curves obtained during training in the M/Ntot
Figure 5a and DA Mx3
Figure 5b situations.
Having verified that the best classification results for melanoma detection are obtained by adding the texture information captured using the LBP operator to the CNN network and dividing the two classes equally without applying DA techniques, we refer to this situation below by talking about the “proposed work”. Then, the accuracy and loss plots of the training and validation sets related to the proposed work are shown in
Figure 6.
The model shows an excellent learning rate, as the training accuracy increases with the number of epochs (
Figure 6a), and the validation accuracy, although it tends to decrease at first, stabilizes not far from the training curve. Both training loss and validation loss decrease to the point of stability (
Figure 6b). Training reaches convergence after about 50 epochs. The small gap between the training and validation curves shows that the model is not affected by overfitting.
The results shown by the validation curves are very different from those obtained on the test set (see
Table 7). This could stand to mean that: (1) because the ISIC 2019 dataset is more numerous than the ISIC 2018 dataset, the network learned more about the characteristics of this dataset, and that (2) the 10% of the training dataset intended to constitute the validation set (obtained by means of the
validation split function in Keras) contains many more ISIC 2019 images than ISIC 2018 images, thus making the results obtained on the validation and test sets not coincide. However, good generalization capabilities across similar datasets would be expected from a model showing these curves; in fact, although they were collected in different years and intended for different challenges, the ISIC 2018 and ISIC 2019 datasets both contain dermoscopic images. It is beyond the scope of this paper to investigate possible differences in the acquisition or preprocessing of the images that make up the two datasets, which might be a good reason to balance the training dataset not only from the point of view of classes (as many melanomas as nevi) but also from the point of view of the source from which the images come (as many ISIC 2018 images as ISIC 2019 images).
Section 7 reports and discusses the results obtained from comparing the proposed model with the most widely used pre-trained models for skin lesion classification: VGG16, ResNet50, MobileNet, DenseNet121, DenseNet169, and DenseNet201. In addition, the performances of the proposed network on the ISIC 2016 dataset and on the DermIS database are compared with other state-of-the-art methods from the literature.
7. Discussion
The purpose of the proposed work is an in-depth study of the analysis and classification of dermoscopic images. In fact, we want to understand whether the hard work of preprocessing images of skin lesions—aimed at the elimination of hairs, marker marks, air bubbles, etc.—is really essential for the classification of melanomas vs. nevi and whether useful information can be drawn not only from the lesions under examination but also from the area of skin surrounding them. In fact, imagining that the region of skin surrounding the lesions has also undergone some change that is, however, not yet visible to the naked eye, not taking it into consideration would miss important and potentially useful information for classification/diagnosis purposes. Thus, the proposed method is aimed at examining these aspects and involves training a custom CNN with images that have not undergone any kind of preprocessing to improve their appearance and adding texture information related to whole images (lesion + surrounding skin). This method is compared with the most widely used pre-trained networks in the field of dermatology and other state-of-the-art methods using ISIC 2016 as a test set.
7.1. Comparison with Common Pre-Trained Networks
Although public dermatology datasets are large enough to successfully train even complex custom neural networks, the use of pre-trained networks— typically on the ImageNet dataset—is widespread. In the dermatology field, the most widely used are: VGG16, ResNet50, MobileNEt, DenseNet121, DenseNet169, and DenseNet201. A comparison of the proposed model is performed with all these networks to see if it is the best one for the task at hand. The results of the comparison between the proposed network and the tested pre-trained models are shown in
Table 8.
Pre-trained models have millions of pre-trained parameters and only a small fraction of trainable parameters, whereas in a custom CNN, since there is no prior knowledge, all parameters are trainable. Information already learned from pre-trained networks can be very useful in classifying new data, but it is not always the best solution, especially when the pre-trained and newly trained data do not have similar characteristics. From the results, it can be seen that the proposed model, in addition to being better able to distinguish melanomas and nevi than the other networks, takes much less time for each epoch of training because of the simplicity of its architecture. This is no small detail considering that many epochs are planned for the training phase.
7.2. Comparison with the Literature
The different existing works regarding the melanoma vs. nevi classification task and using ISIC 2016 as the test dataset, described in [
19], are reported below and are finally compared with the proposed model. In [
73], the authors propose three different scenarios for using the VGG16 network. First, they train the network from scratch, obtaining the least accurate results. Then, they apply the TL method, which turns out to be better than the first method but suffers from the phenomenon of overfitting. Finally, they apply fine-tuning, obtaining the best results. The authors of [
30] propose a framework that performs image preprocessing and performs the classification task using a hybrid CNN consisting of three feature extraction blocks whose results are merged to provide the final output. In [
74], an approach is proposed in which shape, color, and texture features are extracted from previously segmented skin lesion images and then concatenated to features automatically extracted from a custom CNN.
The comparison between the performance achieved by the proposed model and that of works reported in the literature is summarized in
Table 9.
On the 2016 ISIC dataset, the performance of the model is very high and reflects the accuracy and loss curves shown in
Section 6 (
Figure 6). Both in terms of accuracy and in terms of sensitivity, specificity, and AUC score, the proposed model outperforms the models reported in the literature. Whether this result is due to the fact that, in contrast to [
30], the images were not preprocessed; or to the fact that, in contrast to [
74], the texture features were extracted from the entire image and not only from the segmented lesion; or again, that both may have contributed to the result is not known for sure.
To show the robustness of the proposed work, an external validation was conducted using the online public database Dermatology Information System (DermIS) [
75]. This dataset, which contains 1000 dermoscopic images, of which 500 are benign and 500 are malignant, is the largest public dataset after the ISIC archive. Containing a limited number of data, the DermIS dataset is integrated with other public or private datasets, and classification results are reported globally but not for the split datasets, thus preventing comparison with the literature. A paper was identified that reports classification results on the DermIS dataset not combined with other datasets, the comparison of which with the proposed method is shown in
Table 10.
Although the sensitivity of the model proposed in [
76] is higher than that obtained in the present work, the specificity is much lower. This leads to a good ability to classify melanomas correctly, but there is a high rate of false positives, which, although less severe than false negatives, lead to increased costs for patients, who will have to undergo unnecessary additional visits and diagnostic tests. The proposed model once again shows good results compared with the literature.
7.3. Analysis of Interpretability and Limitations of the Proposed Model
With the interpretability analysis of neural networks, it is possible to have a visual explanation of the decisions made by the so-called “black-box”. For such analysis, the Grad-CAM algorithm is used in the present work. This algorithm uses the gradient information (global mean) flowing into the last convolutional layer of the CNN to assign importance values (weights) to each neuron for a particular decision of interest [
77]. The convolutional layers naturally retain spatial information that is lost in the fully connected layers; therefore, the last convolutional layers can be expected to have the best trade-off between high-level semantics and detailed spatial information. Neurons in these layers search for class-specific semantic information in the image (e.g., parts of objects). The heat map of the image that results from applying this algorithm highlights the features of the image. The Grad-CAM algorithm is applied on the last convolutional layer of the proposed custom CNN, that is, before the injection of texture features. Some results obtained are shown in
Figure 7.
As can be seen, the network focuses mainly on the pixels of the skin lesion; from here emerges the contribution of adding texture features extracted not only from the lesion but also from what surrounds it, which is not automatically considered by the neural network and which could be useful for the correct classification of the lesion. However, injecting into the network additional features extracted from images that have not undergone any preprocessing of cropping out parts that are not relevant to the diagnosis may lead the network to attribute to one of the two classes a specific feature that is not related to the pathology at all. These are spurious correlations, and in the present case, these correlations involve dark image contours due to the image acquisition process and associated with the class of melanoma (
Figure 8).
Such images are classified correctly, but interpretability analysis shows that to arrive at the correct result, even before feature injection, the network focuses on regions of the image that are not diagnostically important. If the network already sees these details as possible important features for assigning the N or M label to the images, it is possible that the additional feature vector is going to contribute in the same direction by emphasizing the spurious correlation. One would then need to conduct an analysis on the outcome of the classification after appropriately cropping the images so as to eliminate the black edge disturbance while retaining as much of the surrounding tissue as can be saved.
Another important aspect to consider is that the proposed model is among the methodologies that can classify images of single skin lesions but not total body images. The single-lesion approach allows the application of some dermatologic rules, such as the ABCDE rule, but does not allow consideration of the global picture of patients’ skin lesions (as opposed to dermatologic examinations in which macro-screening is performed). In fact, with this approach, it is not possible to assess the presence of the so-called “ugly duckling”: that is, a nevus that is different from a subject’s other nevi and is therefore suspect [
78]. The underlying concept is that most normal lesions resemble each other, whereas melanomas differ in size, shape, and color just like they are ugly ducklings (abnormal data in the context of anomaly detection). The total body photography (TBP) technique allows photographs of the entire body or portions of the body (wide-field approaches) to be acquired, allowing lesions to be mapped and the entire skin surface of the patient to be monitored [
79]. This technique was combined with DL techniques using 2D [
80,
81,
82] and 3D [
83] images and showed excellent classification performance. The advantages of using this technique in computer-assisted diagnosis systems relate to the possibility of getting an overview of patients’ skin lesions, the possibility of implementing teledermatology since there is no specific acquisition instrumentation but images can be taken using traditional cameras, and finally, the possibility of assessing the appearance of new skin lesions during follow-up. There are limitations, however, in that, compared with the single-lesion method, the total-body approach implies reduced image quality (even using high-resolution cameras, the image quality of skin lesions is lower than that obtained with dermoscopy), and in addition, the absence of public databases makes comparison with the literature impossible.
8. Conclusions and Future Work
In the present work, the identification of cutaneous melanoma is intended as an anomaly detection problem for which a simple custom CNN is proposed, in which additional texture information from dermoscopic images of skin lesions is included to aid in its increased classification performance for the melanoma vs. nevi task. The goal, in addition to creating a network that is accurate in melanoma detection (anomaly detection), is to test whether such a network, trained on unprocessed images from which features are extracted in full, can achieve high performance. In fact, although lesion segmentation allows the ABCD rules commonly used by dermatologists for the examination of skin lesions to be applied, it is not necessarily the case that the features of the tissue surrounding the lesions do not contain important information overlooked in clinical diagnosis. Several experiments were conducted to (1) demonstrate that simple ML models are not suitable for the purpose of this work; (2) show how there is no need to operate dimensionality reduction of extracted features, which are found to be totally important; (3) demonstrate that preprocessing of dermoscopic images is not strictly necessary; and (4) in the present case, the data augmentation technique does not improve classification performance but even worsens it. The proposed network, trained on the ISIC 2018 and ISIC 2019 datasets, shows better results than the most commonly used pre-trained networks in dermatology, especially with regard to computational cost. Moreover, on the ISIC 2016 dataset, the proposed model achieves very high performance that is higher than that reported in the literature. On the DermIS database, the proposed model also performs well. This could mean that image preprocessing is not so necessary but also that image segmentation from which additional information is then extracted tends to leave out important aspects of the images not necessarily related to the lesion. An interpretability analysis is then conducted, which shows that additional texture information related to the unsegmented whole images may add to the network classification—which instead dwells on the skin lesion—but also that these features could contribute to increased spurious correlations. Therefore, probably a preliminary step of partial elimination of any disturbances, such as black image contours, could lead to improved classification results. Future work will go in this direction.