Abstract
Hybrid kinetic models, linking structured models of cell (nano-scale) metabolic processes to the dynamics of macroscopic variables of the bioreactor, are proven to lead to more precise predictions of all key-species dynamics under variable operating conditions, being of an exceptional importance in engineering evaluations to in-silico (math-model-based) determine the optimal operating mode of a fed-batch bioreactor (FBR). Even if such extended dynamic models require more experimental and computational efforts, their use has proven to be advantageous. The approached probative example refers to the simulation of the dynamics of some key species of the central carbon metabolism (CCM) of a modified E. coli cell, linked to the state variables of a FBR used for the tryptophan (TRP) production. By using several optimization algorithms, and an original application of the Pareto-optimal front technique, this paper compares various operating alternatives by using multiple control variables, aiming to maximize TRP production, with minimum substrate consumption. The used E. coli strain was modified to drastically amplify the glucose (GLC) uptake into the cell.
1. Introduction
Biosyntheses spare today a lot of attention to any mean or device improving bioprocess yields. Eventually, they are able to replace the complex chemical processes, which are energetically intensive and generate lot of toxic wastes [1,2]. In a production chain of producing fine-chemicals, different key-operations are managed and optimized (i.e., cultivation, purification, filtration, capture, polishing steps, bioreactor operation, etc.). The fermentative processes conducted in bioreactors with microbial cultures, or the enzymatic reactors [1], are currently used to produce a large variety of valuable molecules [3,4,5], by integrating genetic and engineering methods [6,7]. Bioreactors are constructed and operated in multiple alternative forms, as reviewed in the literature [1,5,8].
In spite of their larger volumes, continuously mixed aerated (oxygenated) tank reactors, operated in BR (batch), or FBR (fed-batch) modes, are the most used, because they ensure a high oxygen transfer and a rigorous temperature/pH control, as is also the case here for the tryptophan (TRP) production.
This work is focused on the engineering aspect, seeking the production maximization of a FBR used for the TRP synthesis (i.e., maximize the molecule production in a minimum amount of time, while minimizing the consumption of raw materials), by using in silico engineering techniques [5,9,10].
From the engineering point of view, in addition to the production optimization, there are several essential problems to be addressed. (1) Derivation of an enough adequate bioprocess kinetic model from on-/off-line measurements during the batch preliminary experiments. (2) The in-silico evaluation of optimal operating policies of a given FBR based on an available bioprocess kinetic model. This step can be applied in two ways: (2a) off-line (or ‘run-to-run’), by using an (extended/deterministic) adequate kinetic model previously identified from the collected experimental data (this paper, and other reports [10,11,12,13]); (2b) on-line, by using a simplified, often empirical mathematical model to obtain a state-parameter estimator based on the on-line recorded data (such as the classical Kalman filter) [13,14,15,16,17,18,19].
To be consistent, the in-silico derived optimal operation mode of a bioreactor should be based on a simulation model that must include most of the process key variables. The use of a deterministic dynamic model, based on the process mechanism, and on macroscopic (measurable) state-variables, as the case here, is preferred due to the physical significance of the terms/parameters, which make possible their validation vs. experimental and literature data, even if repeated model updating is often necessary due to the high variability of the bioprocess. Typical optimization objective functions were reviewed, as stated by Engasser [20].
In spite of its low productivity, BRs are commonly used for slow bioprocesses, because they are highly flexible and easy to operate in various alternative forms [5], detailed as follows: (i) simple BR, when substrate(s), a biocatalyst, and additives are initially loaded in the recommended amounts (concentrations) (Figure 1) [5,21,22]. The BR (single- or multi-objective) optimization determines the batch time and the substrate/biocatalyst initial load [14,23]; (ii) a BR-to-BR optimization, which includes a model-updating intermediate step (‘tendency modelling’ [24]) to evaluate the optimal load of the next BR [4,9,12,14,25,26,27,28]; (iii) a serial sequence of BRs (SeqBR) [28]. The SeqBR includes a series of BRs of quasi-equal volumes. The content of every BR is transferred to the next, by adjusting the reactants and/or biocatalyst concentrations of the latter. Each BR initial load adjustment is determined in silico to ensure the optimal operation of the whole ensemble [4,25]. (iv) BRP, that is, a BR with reactants and/or a biocatalyst added during the batch process in a pulse-like addition of equal/uneven solution volumes, with the frequency determined correspondingly [5]; (v) a semi-batch (that is, fed-batch) reactor (SBR or FBR) (Figure 1), with an optimally varied feeding strategy of the biocatalyst/substrate(s) [5,10,15,27,29,30,31]. Usually, FBRs report better performances compared to other batch operating alternatives. However, they are more difficult to operate. “That is because they need previously prepared stocks of cell-cultures, and substrate(s), of different concentrations (a-priori in-silico determined), to be fed for every ‘time-arc’ of the batch (that is a batch-time division in which the feeding composition is constant; self-understood, the feeding of time-‘arcs’ usually differ between them). This is the price paid for achieving FBR best performances [5,29,32,33]”. Other constructive solutions can be adopted [5,34].
The current (default) approach to solve the model-based design, optimization and control problems of industrial biological reactors is the use of unstructured kinetic models of Monod type (for cell culture reactors) or of Michaelis-Menten type (if only enzymatic reactions are retained) that ignores detailed representations of cell processes. The applied engineering rules are similar to those used by the (bio)chemical engineering and inspired from the nonlinear system control theory [3,15,30,35,36,37,38,39,40]. However, by accounting for only key process variables (biomass, substrate and product concentrations), these global (apparent) models do not properly reflect the metabolic changes, being unsuitable to accurately predict the cell response to environmental perturbations by means of (self-) regulated cell metabolism (e.g., this paper, and Maria and Luta [41]).
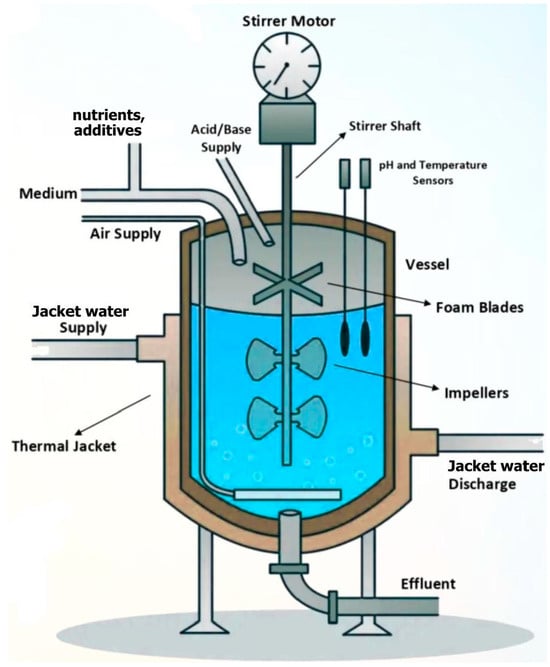
Figure 1.
An oversimplified scheme of a BR or a FBR used to conduct biological processes. In the BR operating mode, the substrate(s), biocatalyst, and additives are only initially loaded in recommended amounts (concentrations). In the FBR operating mode, the substrate(s)/biocatalyst, and additives (nutrients, pH-control substances, etc.) are continuously fed, following a certain (optimal) strategy, to be determined [5]. The FBR presents a similar construction to a BR, and a similar modelling hypothesis. However, unlike BR, the reactants and/or biocatalyst are added during the batch, following a time-step-wise variable (optimal) policy, to be determined off-line [5], or on-line [18].
Figure 1.
An oversimplified scheme of a BR or a FBR used to conduct biological processes. In the BR operating mode, the substrate(s), biocatalyst, and additives are only initially loaded in recommended amounts (concentrations). In the FBR operating mode, the substrate(s)/biocatalyst, and additives (nutrients, pH-control substances, etc.) are continuously fed, following a certain (optimal) strategy, to be determined [5]. The FBR presents a similar construction to a BR, and a similar modelling hypothesis. However, unlike BR, the reactants and/or biocatalyst are added during the batch, following a time-step-wise variable (optimal) policy, to be determined off-line [5], or on-line [18].
An alternative compromise is to use hybrid structured modular dynamic (kinetic) models (HSMDMs) [35] that combine unstructured with structured process characteristics to generate more precise predictions [35,41,42]. The purpose of kinetic HSMDMs is to interconnect process variables (individual or lumped) belonging to at least two hierarchical levels of model details. The resulting composite model is able to simulate the bioreactor dynamics simultaneously at various levels of interest. Thus, the dynamics of the bioreactor macroscopic state variables (i.e., species present in the liquid bulk) are simulated concomitantly with those of the nano-scale key variables, describing the cell metabolic processes of interest together with the core of the central carbon metabolism (CCM), because the macro-/nano-scale species are closely linked, as long as some cell metabolites are imported/excreted from/in the bioreactor bulk. Even if such an extended kinetic model, including complex cell metabolic pathways, requires more experimental and computational efforts to be built-up and identified from structured kinetic data, the resulting hybrid (bi-level) dynamic model presents major and remarkable advantages, as pointed out by Maria [35]:
- (i)
- This model allows for in silico engineering analyses (bioreactor design, off-line optimization) of a higher accuracy, and of a higher degree of detail (the number of considered state variables) compared to the unstructured (empirical, global) models. For instance, this model could better predict the optimal time-step-wise variable feeding strategy of a FBR to maximize its productivity. This numerical analysis is carried out in this paper.
- (ii)
- The extended hybrid HSMDM model can also be used for bioinformatics purposes, by evaluating the influence of the bioreactor operating conditions (control variables) on the dynamics of cell key-species and metabolic fluxes involved in the synthesis of target metabolites. Examples includes conditions for occurrence of glycolytic oscillations [43], or the oscillations in the TRP-operon expression [44,45], or conditions leading to a balanced cell growth (quasi-steady-state QSS, i.e., homeostasis). All these in-silico simulations can direct the design of genetically modified micro-organisms (GMO) with desirable ‘motifs’. Examples are given in the literature [11,46,47];
- (iii)
- The extended hybrid HSMDM models can also be used to obtain lumped (reduced) dynamic models of the process useful for rapid engineering calculations, by employing specific model reduction rules and a check in local operating domains {see the works [48,49] for the nonlinear models cases, or [50,51] for the linear models cases}. As a result, the bioprocess complexity may be described by a succession of local reduced models enfolded on the real process.
- (iv)
- As proved by several case studies, the hybrid bi-level structured models allow more robust extrapolation of the bioprocess behaviour. As an example, Maria and Luta [41] optimized the mercury uptake by modified E. coli cells in a FBR; Maria et al. [52] optimized the succinate production by modified E. coli in a batch reactor; other examples are given on FBR optimization for the monoclonal antibodies (mAbs) production [5,53].
- (v)
- The degree of precision and detail of such a hybrid HSMDM model is remarkable, as proved by some examples, as followings:
- (v-a)
- In the present case study, the HSMDM is able to predict the dynamics of [11 (cell species) + 4 (bulk species)] vs. only [3 (bulk species)] by a classical un-structured FBR model, while covering a wider range of control variables, and for various E. coli cell strains.
- (v-b)
- In another example from the literature [52], the HSMDM is able to predict the metabolic fluxes dynamics of [72 (cell species), involved in 95 reactions + 1 (bulk species, i.e., the biomass)] vs. only [1 (bulk species)] by a classical macroscopic BR model, while covering a wider range of control variables, and GMO E. coli strains.
- (v-c)
- In the case study by Maria [35,41], such an HSMDM is able to accurately predict the dynamics of [26 (cell species) + 3 (bulk species)] vs. only [3 (bulk species)] by a classical macroscopic FBR model, while covering a wider range of substrate input loads, using cloned E. coli cells with various amounts of mercury-plasmids.
In fact, as exemplified in this paper, to offer more precise and detailed support for the engineering evaluations, the HSMDMs should also include the cell metabolic processes responsible for the synthesis of the target metabolite (herein referred to as TRP) in addition to the reactor macro-scale state variables. As a mandatory feature, the cell model should also include the core of the central carbon metabolism (CCM), like the extended version reviewed by Maria et al. [52,54]. This is because the enzymatic reactions in the CCM are interconnected, while the intermediates are starting points in the reaction chains producing all the cell metabolites, understood as the “target” metabolites of practical interest. In spite of concerted efforts to derive a sufficiently accurate/extended dynamic model of the CCM, a reduced model is usually preferred [52,55,56,57,58,59]. This is because, for the extended CCM kinetic models (e.g., 95 enzymatic reactions with 72 metabolites in the model of Edwards and Palsson [60], discussed by Maria et al. [52,54]), it is too difficult to make estimations from the few and often un-structured data, and they are also too complex to be used in quick engineering evaluations. For these reasons, the reduced CCM adequate model, constructed by Maria [11] was adopted in the present paper, being schematically presented in (Figure 2). It accounts for five reaction-pathway interconnected modules (briefly mentioned in Section 3.1, and in the caption of Figure 2), which are as follows: (i) the phosphotransferase system (PTS, in a ‘wild’ strain; that is, the consecutive reactions from GLC(external) to F6P); (ii) glycolysis [54] (the consecutive reactions from GLC(external) to PYR); (iii) the TRP synthesis (the gray area) [44]; (iv) the synthesis of adenosin co-metabolites ATP, ADP, and AMP, as part of the ATP recovery system (the pink rectangle); and (v) the lumped form of the TCA cycle. The connection of the TRP synthesis to glycolysis is realized through the PEP glycolytic node [11,44,45].
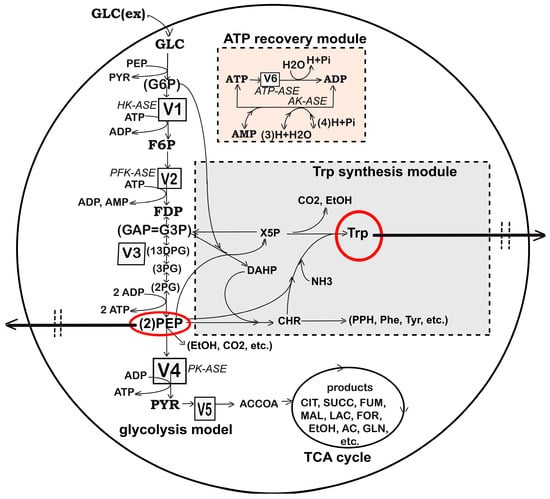
Figure 2.
Simplified structured reaction pathway in E. coli used by Maria [11] to derive the TRP-synthesis modular kinetic model used in this paper. The following modules are included: (i) the phosphotransferase system (PTS, in a ‘wild’ strain; that is, the consecutive reactions from GLC(external) to F6P); (ii) glycolysis [54], (the consecutive reactions from GLC(external) to PYR); that is, the CCM model module [a]; (iii) the TRP synthesis (the grey area) [44]; that is, the CCM model module [c]; (iv) The synthesis of adenosin co-metabolites ATP, ADP, and AMP, as part of the ATP recovery system (the pink rectangle); that is, the CCM model module [b]; and (v) the lumped form of the TCA cycle. The connection of the TRP synthesis to glycolysis is realized through the PEP glycolytic node [11,44,45]. Notations: GLC(ex) = glucose in the cell environment. Species abbreviations are given in the abbreviations list. Species in parenthesis are not explicitly included in the glycolysis model. Italic letters denote the enzymes. Squares include the notations of enzymatic reactions V1–V6 included in the glycolysis model (Tables 2 and 3). Adapted from Maria [32,43], with the courtesy of CABEQ Jl, and completed according to the Maria [11] kinetic model.
Figure 2.
Simplified structured reaction pathway in E. coli used by Maria [11] to derive the TRP-synthesis modular kinetic model used in this paper. The following modules are included: (i) the phosphotransferase system (PTS, in a ‘wild’ strain; that is, the consecutive reactions from GLC(external) to F6P); (ii) glycolysis [54], (the consecutive reactions from GLC(external) to PYR); that is, the CCM model module [a]; (iii) the TRP synthesis (the grey area) [44]; that is, the CCM model module [c]; (iv) The synthesis of adenosin co-metabolites ATP, ADP, and AMP, as part of the ATP recovery system (the pink rectangle); that is, the CCM model module [b]; and (v) the lumped form of the TCA cycle. The connection of the TRP synthesis to glycolysis is realized through the PEP glycolytic node [11,44,45]. Notations: GLC(ex) = glucose in the cell environment. Species abbreviations are given in the abbreviations list. Species in parenthesis are not explicitly included in the glycolysis model. Italic letters denote the enzymes. Squares include the notations of enzymatic reactions V1–V6 included in the glycolysis model (Tables 2 and 3). Adapted from Maria [32,43], with the courtesy of CABEQ Jl, and completed according to the Maria [11] kinetic model.
Examples include a large number of simple/extended models [11,44,52,54,55,56,57,58,59,60,61,62,63,64], and complex simulation platforms, such as the JWS platform of Olivier and Snoep [65]; the MPS platform of Seressiotis and Bailey [66]. The E-cell [67,68], or the V-cell [69] complex models, accounting for thousands of species and reactions, display extended capabilities to predict the dynamics of the cell metabolism under various conditions, based on EcoCyc, KEGG, Prodoric, Brenda and other bio-omics databanks (from a review by Maria [35]).
A special focus was given to the accurate modelling of the glycolysis dynamics and its self-regulation [43,45,54], because most of the glycolysis intermediates are starting nodes (precursors) for the internal production of most of the cell metabolites (e.g., amino-acids, nucleotides, etc.) [35]. Even if such extended structured models are currently used only for research purposes, as they are difficult to identify, it is a question of time until they will be adapted for industrial/engineering purposes in the form of reduced structured hybrid models. The case study presented and discussed here proves this trend.
At this point, we must underline that the cell metabolism is highly sophisticated, involving O(103–4) components, O(103–4) transcription factors (TF-s), activators, inhibitors, and at least one order of magnitude higher number of (bio)chemical reactions, all ensuring the fast adaptation of the cell to the changing environment through complex genetic regulatory circuits (GRC-s) [35]. The cell is highly responsive to the environmental stimuli and highly evolvable, carried out by self-changing its genome/proteome and metabolism, which are the stoichiometry and the reaction rates (fluxes) of the enzymatic reactions, to achieve an optimized and balanced growth using minimum resources (nutrients/substrates).
The development of extended CCM dynamic models on a deterministic basis to adequately simulate in detail the cell metabolism self-regulation, cell growth, and replication for such an astronomical cell metabolism complexity is practically impossible, due to the lack of structured/comprehensive information and computational limitations. A review of some trials is presented by several authors [35,54,70,71].
By using an adequate extended HSMDM (bi-level) from the literature (built-up by Maria [11,54]), the in silico analysis of this paper aims to evaluate and compare the performances of various alternatives to optimally operate an analyzed FBR by using the modified E. coli T5 strain suspended culture provided by Chen [72]. The results are also compared with the experimental data and the non-optimal constant feeding operation of Chen [72], and Chassagnole et al [55]. The characteristics of this strain were reflected in the rate constants estimated by Maria [11].
2. Modified E. coli Strain and Experimental FBR
2.1. Modified E. coli T5 Strain
Although the production of TRP by engineered E. coli has been extensively studied, the complex regulations of the biosynthetic pathways make achieving a high product yield still very challenging. A metabolic flux analysis of Chen et al.’s work [46,47] (Figure 2) suggests that replacement of the ‘wild’ PTS glucose uptake system can improve the TRP yield from GLC. Finally, they obtained a promising T5 strain by using genetic modifications of the TRP producer ‘wild’ strain S028. Basically, they removed the PTS import system, replacing it with a more effective one based on the galactose permease/glucokinase (GalP/Glk) uptake system, by modulating the gene expression of GalP/Glk. The resulting T5 strain showed an increase in the specific TRP production rate in a non-optimal FBR by 53% compared to the initial strain [47], and by 70% for an optimally operated FBR [32]. More details on E. coli mutants presenting alternative routes for GLC uptake are given by various authors [47,72,73,74,75].
2.2. The Experimental Bioreactor and Data Acquisition
To estimate the rate constants of the extended HSMDM (bi-level) for the studied TRP synthesis using the modified E. coli T5 strain, Maria [11] used the experimental kinetic data of Chen [72], obtained in a lab-scale FBR (see the Acknowledgements). The completely automated 10 L capacity FBR in (Table 1) includes a large number of facilities (the automatic addition of nutrients, antibiotics, and pH-control compounds to ensure the optimal growth of the biomass; a reduced scheme is given in Figure 1), all described in detail by Chen [72]. By using this FBR, Chen [72] conducted a ‘nominal’ operation to obtain the kinetic data necessary to derive a kinetic model of the bioprocess. This non-optimal operation meant an addition of a constant feed flow rate and GLC solution during the whole batch time of 63 h.
Samples have been taken from the FBR during the batch with a certain frequency (every 2 to 5 h), by determining the concentration dynamics of four key species from the bulk-phase of interest (GLC, TRP, PYR, biomass-X), with each species time-trajectory including 17 quasi-uniformly distributed recorded points. These data are represented as blue points, corresponding to recorded PYR in (Figure 6), recorded [GLC] in (Figure 8), the recorded [X] in (Figure 9c,d), and the recorded [TRP] in (Figure 10).
Concerning the analytical techniques used to derive such measurements, the reader can refer to the work of Chen [72].

Table 1.
The nominal operating conditions of the FBR (a constant feed flow rate of a GLC solution of constant concentration) used by Chen [72] (Fig. 1) to collect kinetic data for the TRP synthesis by using a suspended culture of genetically modified E. coli cells (T5 strain). SP = set point.
Table 1.
The nominal operating conditions of the FBR (a constant feed flow rate of a GLC solution of constant concentration) used by Chen [72] (Fig. 1) to collect kinetic data for the TRP synthesis by using a suspended culture of genetically modified E. coli cells (T5 strain). SP = set point.
| The FBR Initial Conditions | ||
|---|---|---|
| Parameter | Nominal (Initial) Value | Remarks |
| Biomass initial concentration () | 0.16 (gDW/L) Experimental data of Chen [72] (Figure 9c,d) | courtesy of Chen [72] |
| Batch time (tf) | 3780 min (63 h) | Chen [72] |
| Cell content dilution rate (μ), (1/min) | 1.25 × 10−5–0.015 | Estimated [55] 0.0017 |
| Feed flow rate (FL) | 0.015 L/h (nominal SP, constant) | maintained quasi-constant and non-optimal by Chen [72] |
| 0.01–0.111 L/h | optimally varied in this paper, but not exceeding an imposed 7 L liquid volume at the batch end. | |
| Bioreactor dilution rate (D) | D = FL/VL | Usually adjusted to be close to μ, to avoid the biomass washout [55,76] |
| Bioreactor liquid initial volume (VL,0) | 0.5 L | Maximum 10 L |
| Glucose feeding solution concentration | 3330.5 mM (nominal SP, constant) | maintained constant and non-optimal by Chen [72] |
| 1–5 M | optimized variable, or constant policy in this paper within imposed limits | |
| Glucose solution properties | Solubility in water 5 M (25 °C), 7 M (30 °C), 10.2 M (40 °C) | Bishop [77] |
| 1000 cps (4.5 M, 30 °C), compared to 1094 cps (molasses, 38 °C) | Laos and Harak [78] [Wikipedia—molasse viscosity centi-poise (cps), 2024] | |
| Initial glucose concentration in the bioreactor at (t = 0). | 194.53 mM | Experimental data of Chen [72] (Figure 8) |
| To be optimized for a FBR | Optimized in this paper within imposed limits | |
| Temperature/pH | 37 °C/6.8 | Chen [72] |
| Bioreactor capacity [max()], and facilities | 10 L, automatic control of pH, DO, temperature | Chen [72] |
| Biomass density () | 565.5 gDW·(L cytosol)−1 | Chassagnole et al. [55] |
| Initial concentrations for the glycolytic cell species (in mM) | = 0.6003 = 0.2729 cATP(t = 0) = 2.6729 = 2.6706 = 4.27 [AMDTP]total = 5.82 | measured by Chassagnole et al. [55] |
| Initial concentrations for the TRP-synthesis operon species (in μM) | (t = 0) = 0.01 (t = 0) = 3.32 (nM) (quasi-constant) (t = 0) = 0.01 (t = 0) = 928 (nM) | measured by Bhartiya et al. [79] |
| (t = 0) = 0.164 | this paper; data from Chen [72] | |
3. Bioprocess and Bioreactor Dynamic Model
3.1. The Structured Hybrid Kinetic Model
In this paper, an HSMDM has been adopted from the literature [11,35,44]. Such dynamic models were proven to successfully solve difficult bioengineering problems more accurately and with a higher degree of detail. In such a complex model, the cell-scale part (including nano-level state variables) is linked to the biological reactor macro-scale state variables. Thus, both the model prediction quality and its validity range are improved [35,41,42].
This structured modular HSMDM has been chosen for several reasons: (a) It includes modules characterizing the dynamics of the cell pathways (that is, the glycolysis, ATP-recovery system, TRP-operon expression), and of the biomass growth, involved in the TRP-synthesis. (b) This HSMDM was experimentally identified, and checked the extensive experiments conducted by several authors, for the glycolysis [11,54,55,71] and for the TRP synthesis [11,47,72]. The rate constants of this model have been estimated by Maria [11] using the experimental data of Chen [72]. (c) The relevant plots of (Figures 6, 8, 9 and 10) prove the very good performance of this hybrid model.
Even if such a complex/extended dynamic model requires more experimental and computational efforts to be built-up and identified from structured kinetic data, the resulting hybrid (bi-level) dynamic model presents major and remarkable advantages, well pointed out by the case studies presented by Maria [35]. Among these, we mention only the increased degree of detail of the predictions of such an HSMDM model, and the more robust extrapolations of the bioprocess behaviour [35,41].
With these compelling reasons in mind, an HSMDM from the literature was adopted in the present engineering analysis.
A significant number of simplified kinetic models with lumped terms (species and/or reactions) have been proposed in the literature regarding TRP-synthesis complex regulation process (in a review by Maria et al. [44]). Unlike other models, the adopted HSMDM is able to predict, under certain operating conditions, the oscillation occurrence of the glycolysis, and of the TRP synthesis (not the case here) [43,44,45,79,80,81]. Oscillations in the TRP synthesis are produced due to the concomitant activation and high-order repression of the TRP-operon expression, together with a nonlinear demand for the end product, facilitating the cyclic nature of its expressions. The cell growth and dilution rates [related to the cell cycle, and the liquid residence time in a (semi-)continuous bioreactor] strongly influence the TRP system stability, as proved in silico by Maria [35,45]. The glycolytic oscillation occurrence mechanism is extensively discussed by Maria et al. [43,54].
The hybrid kinetic model ultimately adopted is that of Maria [11], built-up and estimated by using the kinetic data of Chen [72] (Figures 6, 8, 9 and 10). The necessary kinetic data were collected by Chen [72] in a FBR, operated under the non-optimal conditions given in Table 1, using the modified T5 strain of E. coli. This complex structured kinetic model, presented in (Table 2, Table 3 and Table 4), is a deterministic one. Its core is the glycolysis model constructed by Maria [54], validated using literature data. The rate constants of this core model were further used as a starting point in the final estimation step of the complete HSMDM of the FBR used here, and by Maria [11] to simulate the TRP synthesis.
As explained in Section 1, to offer more precise and detailed support for the engineering evaluations, the HSMDMs should also include the cell metabolic processes responsible for the synthesis of the target metabolite (herein referred to as TRP) in addition to the reactor macro-scale state variables.
As a mandatory feature, the cell model should also include the core of the central carbon metabolism (CCM), such as the extended versions reviewed by Maria et al. [52,54]. This is because the enzymatic reactions in the CCM are interconnected, while the intermediates are starting points in the reaction chains producing all the cell metabolites, understood as the “target” metabolites of practical interest. In spite of concerted efforts to derive a sufficiently accurate/extended dynamic model of the CCM, a reduced model is usually preferred [52,54,55,56,57,58,59]. This is because, for the extended CCM kinetic models (e.g., 95 enzymatic reactions with 72 metabolites in the model of Edwards and Palsson [60], discussed by Maria et al. [52,54]), it is too difficult to make estimations from the few and often un-structured data, and they are too complex to be used in quick engineering evaluations. For these reasons, the reduced adequate CCM model, constructed by Maria [11] was adopted in the present paper, being schematically presented in (Figure 2). It accounts for five reaction-pathway interconnected modules (briefly mentioned in Section 3.1, and in the caption of Figure 2), that is: (i) the phosphotransferase system (PTS, in a ‘wild’ strain; that is, the consecutive reactions from GLC(external) to F6P), or another GLC uptake system, like those based on the galactose permease/glucokinase (GalP/Glk) in this paper; (ii) glycolysis [54], (the consecutive reactions from GLC(external) to PYR); that is, the CCM model module [a]; (iii) the TRP synthesis (the grey area) [44]; that is, the CCM model module [c]; (iv) the synthesis of adenosin co-metabolites ATP, ADP, and AMP, as part of the ATP recovery system (the pink rectangle); that is, the CCM model module [b]; and (v) the lumped form of the TCA cycle. The connection of the TRP synthesis to glycolysis is realized through the PEP glycolytic node [11,44,45].
To conclude, the HSMDM includes only the key species of the following four linked reaction modules responsible for the TRP synthesis (Figure 2), using information from the literature as a starting point, with the rate constants taken from the literature [11,45]. The first three modules concern the inner cell reactions, while the fourth concerns the biomass [X] growth dynamics in the FBR bulk, that is:
Module [a]—glycolysis with a natural (‘wild’) GLC-uptake system (PTS), or with a modified GLC-uptake system (GalP/Glk in the modified E. coli strain of this paper).
Module [b]—ATP-recovery system.
Module [c]—TRP-operon expression.
Module [X]—biomass growth dynamics in the FBR bulk.
The considered species’ mass balance of HSMDM is given in (Table 2, Table 3 and Table 4). The dynamic model is hybrid (bi-level) because it connects the macro-state variable of the FBR (biomass X, GLC, TRP, PYR) with the considered cell nano-level variables (GLC, F6P, FDP, PEP, PYR, ATP, in Table 2 and Table 3) for the glycolysis, and (TRP, OR, OT, MRNA) for the TRP-operon expression (in Table 4). A detailed description of these modules is given by Maria [11]. Only some observations are presented here, to understand how the interconnected modules of the modular HSMDM works.

Table 2.
The HSMDM of the studied TRP synthesis in the FBR of Table 1. The model combines the mass balance of the cell glycolytic key species, and of the FBR control variables (GLC, FL) for its time-step-wise variable feeding strategy. See Table 3 for the – flux expressions. Adapted from Maria [11,44,45,54].
Table 2.
The HSMDM of the studied TRP synthesis in the FBR of Table 1. The model combines the mass balance of the cell glycolytic key species, and of the FBR control variables (GLC, FL) for its time-step-wise variable feeding strategy. See Table 3 for the – flux expressions. Adapted from Maria [11,44,45,54].
| Species’ Mass Balance | Auxiliary Relationships, and Estimated Rate Constants |
|---|---|
| Glucose: ; ; with V1 of (Table 3); = control variables to be optimized; j = 1, …, Ndiv (equal time-arcs); (t = 0) is given in (Table 1) for the nominal SP of the FBR of Chen [72] For the optimal FBR with adopted Ndiv = 5, the feeding strategy is given by Equation (6). See also Footnote (a). |
|
| Species inside the cell: | |
| Liquid volume dynamics: ; in (Table 1); j = 1, …, Ndiv (equal time-arcs) |
|
| Biomass dynamics: ; ; ; in (Table 1) |
Rate constants [, aX, bX, NX] estimated by Maria [11] are: = 1.05 × 10−4 (1/min·mM); aX = 10.19; bX = 1.8036 × 10−2 (1/min); NX = 7.334 × 10−2 |
Footnote: (a) For the adopted Ndiv = 5, the j = 1, …, Ndiv time-arcs’ approx. switching points are as follows: T1 = 12.5 h; T2 = 25 h; T3 = 37.5 h; T4 = 50 h; = 63 h. The time-step-wise feed flow rates are to be determined together with the other control variables (that is, ) to ensure the FBR optimal operation. (b) The initial concentrations of the cell species (F6P, FDP, PEP, PYR, ATP) and of the biomass are given in (Table 1).
Module [X]. “The cell culture is considered to be homogeneous, and introduced as a lump X in the FBR model of (Table 2). A Contois model [85], modified by considering a power-law inhibition with the 1-st order growing biomass at the denominator, was proved to be the most adequate vs. the experimental data of Chen [72] (Figure 9c,d). This module [X] is connected to the cell processes, by influencing the GLC dynamics in the bulk phase through the X-growth rate (Table 2) that, in turn, influences the GLC import flux V1 into the cell” (Table 3).
Modules [a] (glycolysis), and [b] (ATP-recovery system). The reduced reaction pathway of the glycolysis {module [a]}, and of the ATP-recovery system {module [b]} are given in (Figure 2), while their reaction rate expressions and parameters are given in (Table 2 and Table 3).
This model, constructed by Maria [11], has been developed based on information from the literature [44,45,54,55,71], by changing the V1 reaction rate expression to reflect the modified GLC import system of the modified E. coli T5 strain. Thus, the dynamics of the species belonging to the three inter-connected modules {[a], [b], and [X]} can be simulated concomitantly, according to the reduced reaction pathway of (Figure 2).
In fact, the glycolysis is the core module of the CCM. In the adopted HSMDM, the glycolysis module [a] (presented in Figure 2), is the reduced model of that constructed by Maria [54]. An adequate modelling of the glycolysis dynamics is important, because the intermediates provide entry/exit points to/from glycolysis. For example, PEP is the starting point for the synthesis of essential amino-acids (AA), such as tryptophan, cysteine, arginine, serine, etc. [44,55,84,86].
Due to the tremendous importance of the glycolysis in simulating the cell CCM, intense efforts have been made both in its experimental study, and in modeling the dynamics of this process specifically in bacteria. The large number of glycolysis reduced or extended kinetic models proposed in the literature [45,54,87] present a complexity ranging from 18–30 species, included in 48–52 reactions, with a total of 24–300 or more rate constants [54]. Most of these models are however too complex to be easily identified from (often) few available kinetic data, and too complex to be further used for engineering calculations. Beside, with few exceptions [45,54], most of them can not satisfactorily reproduce the glycolytic oscillations occurrence on a mechanistic basis [43,45].

Table 3.
Reaction rate expressions V1-V6 of the HSMDM model of (Table 2), describing the dynamics of the cellular glycolytic species according to the kinetic model by Maria [11,54], and by Chassagnole et al. [55]. In the present study, this glycolysis kinetic model was modified by replacing the PTS system (V1 flux) for the GLC uptake in the “wild” E. coli with those of the mutant T5 E. Coli strain tested in this paper, and by Maria [11]. The model rate constants were estimated by Maria [11] to fit the experimental data of Chen [72] presented in (Table 1 and Figures 6, 8, 9 and 10). Species abbreviations are given in the abbreviation list.
Table 3.
Reaction rate expressions V1-V6 of the HSMDM model of (Table 2), describing the dynamics of the cellular glycolytic species according to the kinetic model by Maria [11,54], and by Chassagnole et al. [55]. In the present study, this glycolysis kinetic model was modified by replacing the PTS system (V1 flux) for the GLC uptake in the “wild” E. coli with those of the mutant T5 E. Coli strain tested in this paper, and by Maria [11]. The model rate constants were estimated by Maria [11] to fit the experimental data of Chen [72] presented in (Table 1 and Figures 6, 8, 9 and 10). Species abbreviations are given in the abbreviation list.
| Reactions | Rate Expressions | Estimated Rate Constants (Units in mM, min) |
|---|---|---|
| GLC import system glc + pep → f6p + pyr pyr + atp → pep + adp + h glc + atp → f6p + adp +h | Modification for the T5 strain . | = 1.0902 (mM/min) = 3581.8 (mM) = 0 = 0 |
| f6p + atp → fdp + adp + h | . | = 1.0437 = 2 = 0.062028 (mM/min) = 6.16423 (mM) = 25 μM = 60 μM |
| fdp + 2 adp (+ 2 nad + 2 p) ↔ 2 pep + 2 atp (+ 2 nadh + 2 h + 2 h2o) | . | = 4602.3 (1/min) = 31.917 (1/min) = 0.05 = 3 |
| pep + adp + h → pyr + atp | . | = 1.331879 = 4 = 0.1333655 (mM/min) = 1.146443 (mM) = 0.2 (mM) = 9.3 (mM) |
| pyr → products (accoa, cit, succ, lac, etoh, ac, …) | . | = 9913.5 (1/min) = 395.525 (mM) = 2.6814 |
| atp → adp + h | . | = 552.38 (1/min) |
| 2 adp ↔ atp + amp | . | = 1 |
| (i) Observations by Termonia and Ross [82,83] indicated experimental evidence of a very fast reversible reaction catalyzed by AKase, with the equilibrium being reached quickly. (ii) The k6 constant takes values according to the micro-organism phenotype (related to the gene encoding the enzyme ATPase that catalyzes this reaction). (iii) = constant [82,83]; (iv) results from solving the thermodynamic equilibrium relationship: , that is: . | ||

Table 4.
Species’ mass balance in the TRP-operon expression kinetic model of Bhartiya et al. [79], modified by Maria [11,44], with the following modifications: (i) PEP (from glycolysis) is the substrate of TRP synthesis, and the node coupling this synthesis with the glycolysis module; (ii) a novel model for the TRP-synthesis inhibition was proposed and identified from experiments. The model rate constants are estimated by Maria [11] based on the experimental data of Chen [72] (Table 1 and Figures 6, 8, 9 and 10) collected from a FBR using a modified E. Coli T5 strain. The initial estimate of the rate constants is those of Maria [11]. Species abbreviations are given in the abbreviation list.
Table 4.
Species’ mass balance in the TRP-operon expression kinetic model of Bhartiya et al. [79], modified by Maria [11,44], with the following modifications: (i) PEP (from glycolysis) is the substrate of TRP synthesis, and the node coupling this synthesis with the glycolysis module; (ii) a novel model for the TRP-synthesis inhibition was proposed and identified from experiments. The model rate constants are estimated by Maria [11] based on the experimental data of Chen [72] (Table 1 and Figures 6, 8, 9 and 10) collected from a FBR using a modified E. Coli T5 strain. The initial estimate of the rate constants is those of Maria [11]. Species abbreviations are given in the abbreviation list.
| Rate Expression | Kinetic Model Parameters (Units in mM, μM, min) |
|---|---|
; . | = 59.062, 1/(min·mM) = 0.5443, 1/min = 17.796, 1/min = 14.094, 1/min = 1.157, 1/min = 3.53, μM = 1.92 = 0.04, μM (see Footnote c) |
| ; = 0; . (see Footnotes a and b) | = −0.32 = 0.36365, 1/min = 3.9923 = 0.017153, 1/min = 0.071515 |
Footnote: (a) The adopted modification for the TRP-synthesis inhibition replaces the C3 variable of the Bhartiya et al. [79] model (not displayed here) with a modified Contois model [85], including a power–law inhibition with TRP growth at the denominator. (b) The nitrogen source in the TRP synthesis is considered in excess and included in the constant. In order to connect this module to the to the glycolysis kinetic model, the PEP species dynamics, generated by the glycolysis was explicitly included in the TRP-synthesis rate as a substrate. (c) The initial concentrations of the TRP-operon species (OR, MRNA, E, TRP) are given in Table 1.
The adopted glycolysis kinetic model constructed by Maria [45,54], even in a reduced form, by accounting only for nine key species in 6 lumped reactions, including 17 easily identifiable rate constants belonging to V1–V6 metabolic fluxes (Figure 2, and Table 2 and Table 3), has been experimentally validated. Additionally, this glycolysis model was proven to adequately reproduce the cell glycolysis under steady-state, oscillatory, or transient conditions depending on several factors, which are as follows [43,44,45]: (i) the defined glucose concentration level/dynamics in the bioreactor, (ii) the total A(MDT)P cell energy resources, and (iii) the cell phenotype characteristics related to the activity of enzymes involved in the ATP utilization and recovery system {denoted here as module [b]}. When including the glycolysis model in the HSMDM, the GLC import system (V1) corresponds to the basic PTS system for the ‘wild’ strain [54,55], or presents a form modified by Maria [11] to match the T5 E. coli strain kinetic data of Chen [72]. According to the experimental data, the produced TRP (see module [c] below) is excreted (Figure 2) through a process described by Chen [72]. The PYR key-metabolite concentration in the cell is regulated through complex mechanisms [88,89], with the excess being excreted, as experimentally proven by Chen [72].
As revealed by the reactions figured in the pink square of (Figure 2), the efficiency and the dynamics of the ATP recovery system {that is module [b] of the present model} is essential for the reaction rates of the whole CCM, as long as ATP plays a catalytic-chemical energy provider role. As underlined by Maria et al. [43,44,45], among the involved parameters, an essential factor is the k6 reaction rate (determined by the ATP-ase characteristics in Figure 2) and included in the glycolysis model of (Table 2 and Table 3). The involved enzymes characteristics are directly related to the cell phenotype (that is cell genomics) controlling the [AMDTP] total energy resources level. To not complicate the simulations, the [AMDTP] level was kept unchanged in the present analysis at an average value given in (Table 1), as suggested by Chassagnole et al. [55]. The adopted model for the ATP-ADP-AMP dynamics (V1–V6, and equilibrium relationships) in (Figure 2—the pink square, and Table 2 and Table 3) proposed by Maria [11,45,54], and by Maria et al. [43] was proved to fairly represent the dynamics and the thermodynamics of the cell modules [a,b].”
Module [c] TRP synthesis: Modelling the TRP synthesis on a deterministic (mechanism-based) approach is difficult because this cellular process is known as being, under certain conditions, a QSS, or an oscillatory one [44,45,79,80,81,90].
However, to avoid extended models, which pose difficulties in their estimation and use, most of the reduced dynamic models from the literature do not distinguish the ‘process’ components from ‘regulatory’ components, and lumped reactions/species are considered instead, with the regulatory performance being included via adjustable model parameters and terms. Kinetic models trying to reproduce the TRP-operon expression self-regulation [80,81] are too extended for our engineering evaluations purposes. Due to the process complexity, some modelling approaches [61] focus more on determining correlations between flux distribution, flux control, and the optimized enzyme activity distribution. However, by employing a kinetic model that is too reduced, such models are not able to be linked to the dynamics of the CCM modules.
For these reasons, in the present analysis, simulations of the TRP synthesis (Figure 2) were performed by using the reduced kinetic model constructed by Maria et al. [11,44]. The mass balances of the key species of the TRP-operon expression are given in (Table 4). This dynamic model is derived from those of Bhartiya et al. [79], but it explicitly connects the TRP module to the glycolysis module [a] through the PEP precursor-sharing node (in Figure 2). Consequently, PEP is included as a substrate in the TRP module [c] mass balance in (Table 4). More details on this module construction are given by Maria and Renea [32]. In fact, as remarked by Li et al. [73], the PEP precursor is the limiting factor for the TRP synthesis. For such a reason, intense efforts have been made to increase its production by glycolysis intensification. This can be realized by optimizing the FBR operating strategy (as is carried out in this paper), and/or by using the modified E. coli T5 strain culture of Chen et al. [47,72] (also carried out in this paper).
The operation of the FBR. A simple ideal model of FBR was adopted to describe the key-species dynamics during the batch at a macroscopic level (GLC, X, TRP, and PYR in bulk), but linked to the cell species dynamics [ that is, (GLC, F6P, FDP, PEP, PYR, ATP) for the glycolysis, and (OR, MRNA, E, TRP) for the TRP-operon expression] (Table 2, Table 3 and Table 4) [11].
In total, the developed HSMDM includes 49 rate constants to be estimated from the experimental kinetic curves of four observed species (GLC, TRP, PYR, X), with each species time-trajectory including 17 quasi-uniformly distributed recorded points (Figures 6, 8, 9 and 10). This estimation problem is equivalent to the high-difficulty nonlinear programming problem (NLP) of [11,48] due to its dimension, and the high nonlinearity of the model and constraints.
Details about the used sequential estimation approach to avoid unfeasible local estimates are given by Maria [11]. The estimated rate constants using the experimental data of Chen [72], and the glycolysis initial estimation of Maria [54] are given in (Table 2, Table 3 and Table 4). A standard weighted least-square estimator (WLSQ) was employed, as it was the most suitable statistical estimator for this case study, due to the different standard deviations and units of the observed species [11,48].
As proven by the plotted results given in (Figures 6, 8, 9 and 10), the species time-trajectories (the blue curves) predicted by the HSMDM fit the experimental data fairly well (the blue circles). Consequently, it can be concluded that the HSMDM of the FBR, adopted above, performs well, and it is suitable for further engineering calculations. The estimated rate constants were validated by comparison with the literature data [11,43,44,54].
A comparison, presented by Maria [11], of the model-estimated rate constants for the modified T5 E. coli strain using the FBR experimental data presented by Chen [72] with those of the same model, but estimated for the ‘wild’ E. coli strain by Chassagnole et al. [55], (not presented here), revealed that most of the estimated rate constants present similar values. However, due to the mentioned modifications of the kinetic model for the used E. coli T5 strain of this paper, some important differences are reported for the following: (i) the rate expression and parameters of the GLC import system (V1 in Table 2 and Table 3), (ii) the biomass growing dynamics (Table 2), and (iii) the TRP-synthesis module [c] rate constants and expressions (Table 4).
The bioreactor ideal model (Table 2, and Equations (1)–(4)) main hypotheses are the following [34]: (i) isothermal, iso-pH, iso-DO operation; (ii) it is self-evident that nutrients, additives, antibiotics, and pH-control compounds are added initially and during FBR operation to ensure the optimal grow of the biomass [72]; (iii) aeration in excess (even with pure oxygen [72]) over the batch to ensure an optimal biomass maintenance, and to contribute to the liquid homogeneity; (iv) perfectly mixed liquid phase (with no concentration gradients), of a volume increasing according to the liquid feed flow rate constant or time-varying strategy; (v) the limits of the liquid feed flow rate ( in Equation (12)) are adjusted so that they do not exceed the maximum bioreactor capacity () in Equation (12); (vi) negligible mass resistance to the transport of oxygen and compounds into the liquid and biomass flocks; (vii) GLC-substrate is initially added in the bioreactor and during the batch according to an optimal feeding mode to be determined; (viii) the feed flow rate during the batch is varied according to an optimal feeding strategy to be determined for every ‘time-arc’ index ‘j’ in Equation (5) (that is an interval in which the batch time is divided).
From a mathematical point of view, in a general form, the FBR HSMDM (Table 2, Table 3 and Table 4) translates to a set of 12 differential mass balances (ODE set) written for the key species of the FBR, as follows:
Species in the bulk phase:
“i” denotes species present in the FBR bulk (GLC, X, TRP, PYR); “j” denotes the FBR feeding time-arcs; j = 1, …, Ndiv.
Key-species inside cells:
In the Equations (1) and (2), index “i”denotes the species inside cells; that is, (GLC, F6P, FDP, PEP, PYR, ATP) for the glycolysis, and (OR, MRNA, E, TRP) for the TRP-operon expression.
Biomass in the bulk phase:
Liquid volume dynamics:
In the Equation (4), index “j” denotes the FBR feeding time-arcs; j = 1, …, Ndiv.
In Equation (1), refers to the concentration of the species i in the feeding solution, constant over the time interval j (j = 1, …, Ndiv). In the present case only, GLC is fed in the FBR during the batch. The reaction rate ri expressions, together with the associated rate constants and other details, are given in (Table 2, Table 3 and Table 4). In Equations (1)–(3), c = vector of species concentrations; = initial value of c (at time t = 0) given in (Table 1); = vector of the model rate constants. The reactor content dilution in Equation (1), determined by the increasing VL in Equation (4), is due to the continuously added FL term.
In Equation (1), GLC and FL are the control variables. The optimal can be constant, or it can be variable. In the last case, it has to be determined for every time-step-wise interval, over j = 1, …, Ndiv time-arcs. For instance, for the adopted Ndiv = 5, the j = 1, …, and Ndiv time-arc switching points given in Equation (5) are the following: T1 = /Ndiv (12.5 h); T2 = 2 × T1 (25 h); T3 = 3 × T1 (37.5 h); T4 = 4 × T1 (50 h); = 63 h. More specifically:
Feed flow rate strategy in the time-step-wise operation of the FBR:
Similarly, if a variable feeding strategy was chosen, for the adopted Ndiv = 5 equal time-arcs, the GLC solution concentration feeding strategy in a time-step-wise operation of the FBR is the following:
Ways to intensify the TRP production in the FBR: As revealed by the literature [11,43,44,72], the cell internal and external factors determine whether the TRP production increases strongly, as follows: (1) “The glycolysis intensity (GLC uptake flux, and the average levels of the glycolytic species), transmitted to the TRP-synthesis module [c] via the shared PEP intermediate, and (2) The glycolysis dynamics (QSS, or oscillatory behaviour). On the other hand, the glycolysis intensity is controlled by several cell internal and external factors, well reviewed by Maria [43,45];” (3) The optimal operation type of the FBR—while the factors of (1) and (2) can be influenced by modifying the E. coli characteristics, by developing a GMO [47,72], factor (3) can be solved by applying an in silico, off-line engineering analysis (detailed in this paper).
4. Fed-Batch Bioreactor Optimization Problem
4.1. Preliminary Considerations
The experimentally validated FBR HSMDM (Table 2, Table 3 and Table 4) adopted in this paper achieves a reasonable compromise between its level of detail (the number of accounted intra-cellular species and reaction pathways, included in the lumped key modules [a–c], Figure 2), and its predictive power for the bulk-phase species of interest (that is, X growth, GLC depletion, excreted TRP, and PYR, depicted in Table 2, Table 3 and Table 4). The adequacy of this model vs. the experimental data (Figures 6, 8, 9 and 10) [11] gives it enough credentials to make predictions of a satisfactory quality within the investigated experimental domain.
The optimal FBR operation alternatives derived in this paper are more complex than the simple operation presented by Chen [72], characterized by a constant non-optimal feeding over the FBR batch (Table 1), and the simulated species blue curves in Figures 6–10).
In the present paper, other FBR feeding alternatives will be explored. Thus, (i) the batch time is divided in Ndiv (equal lengths time-arcs), and (ii) the control variables are kept constant over every time-arc only at the optimal values determined from solving an optimization problem (i.e., the maximization of the TRP production). The time intervals of equal lengths Δt = tf /Ndiv are obtained by dividing the batch time tf into Ndiv parts tj−1 ≤ t ≤ tj, where tj = jΔt are switching points (where the reactor input is continuous and differentiable). The time intervals for the present case study with an adopted Ndiv = 5 are shown in Footnote (a) of (Table 2). Additionally, even if a FBR with a constant feeding is chosen, due to its simple operation, the constant values of the control variables will be determined by applying a NLP optimizer, or a Pareto-optimal front technique.
So as not to complicate the engineering calculations, the main assumption in Equations (5) and (6) is the following: On each time-step-wise arc, index j = 1, …, Ndiv, the control variables and are kept constant. Of course, the values on each time-arc do not have to be necessarily equal to each other.
To summarize, in the FBR case with a variable feeding, according to Equations (1)–(6), for every j = 1, …, Ndiv time-‘arcs’, the following control variables are to be determined:
- The time-step-wise continuously added liquid feed flow rate FL,j (j = 0, …, Ndiv − 1); FL,0 = FL (t = 0) of the substrate solution;
- The inlet concentrations [GLC]inlet,j (j = 0, …, Ndiv − 1) for each of the added solutions of FL,j. Usually, the fed FL,j present different concentrations, to be determined together with the initial concentration; that is, [GLC]0 = [GLC](t = 0) = [GLC]inlet,0.
The FBR time-‘arcs’ are defined before Equation (5). For an adopted Ndiv = 5 time-‘arcs’, the FBR optimal operation is determined in two alternatives:
FBR operation alternative (A): A constant feeding along the entire batch, as follows:
This case corresponds to the nominal FBR non-optimal operating conditions of Chen [72]; the control variables and being kept constant on each time-arc at the non-optimal values given in (Table 1).
Of course, a better idea for this alternative (also examined in the present study) is to optimize the initial values and (two unknowns) by applying an NLP procedure (with a single objective function), or a Pareto-optimal front technique (with multiple objectives), with both procedures in the presence of multiple constraints (Section 4.2.4). Details on the used NLP optimization algorithm are given in Section 4.2.5, while on the Pareto front in Section 4.2.3.
FBR operation alternative (B): A time-step-wise variable uneven feeding along the batch (studied in this paper) is achieved by determining the following control variables: , and of , over the j = 0,1,…(Ndiv − 1) time-‘arcs’ of the batch (for Ndiv = 5). These variables result from reaching the optimum of an objective function (the maximum of TRP production). In this NLP optimization alternative, there are 4× Ndiv unknowns in total; that is, 4 × 5 = 20 searching values for the adopted Ndiv = 5. Multi-objective FBR optimization is possible, which is also examined in this paper. See Maria and Crisan [29] as an example.
4.2. Optimization Problem Formulation
4.2.1. The Selection of Control Variables for the FBR with Variable Feeding
By analyzing the FBR model of (Table 2), the natural option is to choose the easy-to-handle macroscopic ones as the control variables. Here, according to the model form (Section 3.1), the following two variables were chosen, both related to the reactor feeding:
- (a)
- The substrate (j = 1, …, Ndiv) whose concentration plays a major role in the cell glycolysis intensification and TRP production;
- (b)
- The liquid feed flow rate (j = 1, …, Ndiv) with the GLC solution, which is directly linked to the GLC feeding, and responsible for the reactor content dilution.
In the FBR time-step-wise variable feeding policy (alternative B, above), each control variable is kept constant over each time-arc (index j). Of course, they are not necessarily equal between different time-arcs. Thus, for Ndiv = 5, there are 5 × 2 = 10 unknowns displayed in Equation (7), to be determined by optimization under certain constraints.
4.2.2. Single Objective Function (Ω) Optimization (NLP)
By considering the mentioned control variables in Equation (7), the optimization of the FBR with a variable feeding consists of simultaneously determining its optimal initial load with its feeding strategy for every time interval (‘arc’) during the batch, leading to the maximization of [TRP] production, outlined as follows:
The [TRP](t) dynamics in Equation (10) are evaluated by solving the FBR ODE dynamic model [Equations (1)–(6)] over the whole batch time (t) ∈ [0, tf].
The choice of the number of time-‘arcs’ (Ndiv): The FBR operating strategy with a variable feeding (alternative B) implies a time-step-wise variable feeding over an adopted (Ndiv = 5), with equal time-arcs that cover the whole batch time. Each time-arc ‘j’ (j = 1, …, Ndiv) is characterized by optimal levels of the feed flow rate , and of the . It is self-evident that, over a time interval, the control variables are kept constant. Of course, the optimal values on various intervals may differ from each other. A brief survey of the FBR optimization literature [5,18,26] reveals that a small number (Ndiv) < 10 is commonly used due to its multiple advantages, extensively pointed out by Maria [5,32]. Additionally, FBR operation, using a large Ndiv time-arc, can present special operating problems when including PAT (Process Analytical Technology) tools [91].
Thus, to not complicate the computational analysis, (Ndiv) = 5 was adopted, with equal time-arcs covering the batch time = 100 h.
4.2.3. Multi-Objective Optimization by Using the Pareto-Optimal Front with an Optimal but Constant FBR Feeding
When more than one objective function is simultaneously considered, the optimization problem is more difficult to solve. For multi-objective optimization, several alternatives can be utilized [92,93]. One elegant option is to obtain the set of Pareto-optimal solutions, also called a Pareto front, for the case of at least two adverse objectives [94]. A Pareto solution is one where any improvement in one objective can only take place at the cost of the other objective. In fact, the Pareto curve is the geometrical place of all the points (problem solutions) that realize the best compromise between the two considered objective functions. The choice of a solution (a point) from this Pareto curve is subjective, usually dependent on other criteria not included in the Pareto optimization.
For the present case study of an FBR operated with a constant optimal feeding (in terms of feed flow rate FL and inlet [GLC]), several opposing objectives can be considered at the batch end, such as maximum TRP production, minimum substrate [GLC] consumption, minimum initial necessary viable biomass [X]o, minimum feed flow rate FL (to avoid overly diluting the bioreactor content), and others. Of course, the Pareto-optimal fronts can be obtained by using any pair of these opposite objective functions. In the considered case study, the following four opposite objectives are considered two-by-two, based on their adverse effect in the process model, as follows:
Due to the limited efficiency of the applied Pareto optimizer (Section 4.2.5), the rough (slightly oscillating) Pareto curves obtained are difficult to interpret. To better point out their monotony, approximate Pareto curves will be generated by using the cubic spline procedure of Matlab™ (MathWorks, Natick, MA, USA) with a suitable smoothing constant.
4.2.4. Optimization Problem Constraints
The optimization problems from Equations (10) and (11) are subjected to the following multiple constraints, as follows:
- (a)
- (b)
- (c)
- To limit the substrate excessive consumption, by also accounting for the limited reactor volume, feasible searching ranges are imposed on the control/decision variables, namely [95,96]:
- (d)
- Physical meaning of the searching variables:
- (e)
- Physical meaning of the state variables:
ci(t) ≥ 0 (i = 1, …, number of species in the model)
- (f)
- Limit the maximum cell resources in AMDTP [11,55]
[ATP](t) < Total [AMDTP] of (Table 1);
With [ATP](t) obtained from solving the FBR models of Equations (1)–(6)
With [ATP](t) obtained from solving the FBR models of Equations (1)–(6)
As an observation, the imposed control variables ranges are related not only to the implementation facilities, but also to the economic reasons, meaning minimum substrate consumption, the reduced dilution of the reactor content, and an effective bioreactor control.
4.2.5. The Applied Numerical Solvers
The time-evolution of the model species (inside cell, or in the bulk-phase) is obtained by solving the FBR dynamic model Equations (1)–(6) with the initial condition of Cj,0 = Cj (t = 0) of (Table 1) for the inside cell species, excepting the bulk [GLC]0, FL,0 to be determined from the FBR optimization, as indicated by Equations (7) and (9)–(11). The imposed batch time tf, and the optimal medium conditions are those of (Table 1). The dynamic model solution was obtained with a high accuracy, by using the high-order stiff integrator (ODE15S) of the Matlab™, with suitable quadrature parameters to keep the integration error very low.
Because of the highly nonlinear FBR model Equations (1)–(6), the nonlinear optimization objective Equation (10), and the nonlinear problem constraints Equations (12)–(15) the formulated problem Equations (7)–(10) translates into a nonlinear optimization problem (NLP) with a multimodal objective function and a non-convex searching domain. To obtain the global feasible solution with enough precision, the multi-modal optimization solver MMA of Maria [48,97] has been used, as being proved in previous works to be more effective compared to the common (commercial) algorithms. The computational time was reasonably short (minutes) using a common PC, thus offering a quick implementation of the obtained FBR optimal operating mode. The NLP solution was checked by using several (randomly generated) starting points for the rate-constants”.
The rough Pareto-optimal fronts have been generated by using the dedicated algorithm (GAMULTIOBJ) of the Matlab™ math computational package. Due to the limited efficiency of the used optimizer, the obtained rough Pareto curves are difficult to interpret. To better point out their monotony, approximate Pareto curves have been generated by using the cubic smoothing spline procedure of Matlab™ (function CSAPS), with a suitable smoothing constant.
5. Optimization Results and Discussion
The results obtained by solving the NLP optimization problem, and by deriving the Pareto-optimal fronts, are presented in the following forms:
- -
- -
- -
- -
- A comparison of all FBR operating alternatives in terms of TRP production and raw material consumption is given in Table 5, even if the batch time of the Chassagnole et al. [55] experiment is much shorter.
It is worth mentioning that the FBR operation in Table 5 is considered in several alternative forms: (i) a time-step-wise variable feeding (Equations (1)–(6), and Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10), (ii) a constant non-optimal feeding (literature results); or (iii) a constant but optimal feeding (this paper, the NLP-optimal, or the Pareto-optimal) (Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10).
In Table 5, the substrate (GLC) consumption for the FBR case was evaluated with the following relationship:

Table 5.
Efficiency of the modified E. coli T5 strain in GLC uptake, and TRP synthesis in the tested FBR. of (Table 1). Initial liquid volume, VL,0 = 0.5 L. Only the FBR of Chassagnole [55] (first row) uses the “wild” E. coli strain.
Table 5.
Efficiency of the modified E. coli T5 strain in GLC uptake, and TRP synthesis in the tested FBR. of (Table 1). Initial liquid volume, VL,0 = 0.5 L. Only the FBR of Chassagnole [55] (first row) uses the “wild” E. coli strain.
| Bioreactor Operation | Substrate Consumption, and Biomass Production (A) | Max [TRP], (mM) | Final VL (L) | ||||
|---|---|---|---|---|---|---|---|
| Type | Ndiv | GLC, (Moles) (Eq. (16)) | max [X] (gDW/L) (Predicted) | % | |||
| FBR Constant non-optimal feeding of Chassagnole [55]; see Maria [44] ‘wild strain’ | 1 | Nominal feeding (B, E, F) | 0.06 (wild strain) | 10 (tf = 100 min) | 4.1 (worst alternative) | 0.55 | |
| [GLC]in | 200 | ||||||
| D | 3 × 10−4–10−2 | ||||||
| FL | 3 × 10−3 L/min | ||||||
| [Xv]o | 8.7 | ||||||
| FBR Constant non-optimal feeding, Chen [72] ‘modified strain’ (C) | 1 | Nominal feeding (E) | 3.15 (T5 strain) | 25 (tf = 63 h) | 188 (poor alternative) | 0.94 | |
| [GLC]in | 3330.5 | ||||||
| FL | 0.015 | ||||||
| [Xv]o | 8.7 | ||||||
| FBR Constant, but NLP optimal feeding, (this paper), ‘modified strain’ (C) | 1 | Optimal inlet conc., Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10 (E) | 10.99 (T5 strain) | 600 (tf = 63 h) | 219 (poor alternative, high consumption, high final volume) | 6.49 | |
| [GLC]in | 1694 | ||||||
| FL | 0.103 | ||||||
| [Xv]o | 8.7 | ||||||
| FBR NLP optimal variable feeding, (this paper) ‘modified strain’ (E) | 5 | Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10 (D, E) | 4.53 (T5 strain) | 595 (tf = 63 h) | 237 (fairly good) | 2.7 | |
| [GLC]in | variable | ||||||
| FL | variable Figure 9 | ||||||
| [Xv]o | 8.7 | ||||||
| FBR Constant, but Pareto-optimal feeding, (this paper) ‘modified strain’ (C) | 1 | Optimal inlet conc., Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10 (E) | 2.14 (T5 strain) | 600 (tf = 63 h) | 266 (best alternative) | 2.64 | |
| [GLC]in | 1000.5 | ||||||
| FL | 0.042 | ||||||
| [Xv]o | 8.7 | ||||||
Footnotes: (A). The displayed digits comes from the numerical simulations. (B). The checked FBR set-points of Maria [44]. (C). The FBR operation with a constant-over-time feeding for all the control variables implies that (1-blue, 3-red, and 4-green curves in Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10): ; ; the only two variables to be optimized being the initial (inlet) values and , under the constraints Equations (12)–(14). See the resulting FBR optimal operating mode in Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10. (D). The FBR optimal time-step-wise variable feeding policy is obtained by using the control variable limits of Equation (12). In this case of variable feeding, the control variables FLj and [GLC]inlet,j; j = 0, 1, …(Ndiv − 1) of Equations (5)–(7) and Table 2, follow an uneven optimization policy (that is, 10 unknowns for Ndiv = 5). Details on the optimization rule used are given in Section 4.2.5. The optimal control variables policy is given in Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10. (E). The units are as follows: [GLC], mM; FL, L/h; [Xv], (gDW/L). (F). Results were simulated using the Chassagnole et al. [55] model and the data available on the JWS platform of Olivier and Snoep [65].
By analyzing the resulting FBR optimal operating modes (plots 2–4 in Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10, and Table 5) compared to those of the sub-optimal (nominal) operation of Chen [72] (plot 1 in the same figures), several observations can be derived, as follows:
- The HSMDM adopted in this paper fairly fits the experimental data of Chen [72], being adequate enough to derive engineering evaluations of the FBR optimal operation.
- Even if the FBR is operated under the nominal, non-optimal conditions of (Table 1), the modified E. coli T5 strain of Chen [72] reported a much higher (45×) TRP production compared to the wild strain of Chassagnole et al [55]. Such a result is explained by a much higher GLC uptake rate in the modified E. coli T5 strain, where the PTS import system of GLC was replaced by a more effective one based on the galactose permease/glucokinase (GalP/Glk), by modulating the gene expression of GalP/Glk [47,72].
- The efficiency in the TRP production of the optimally operated FBR with a variable feeding is significantly higher (237 mM) compared to the same FBR, but with a constant NLP optimal feeding (219 mM), even if the same modified E. coli T5 strain was employed in both cases.
- The FBR optimal NLP operation with a variable feeding reported a similar dilution of the reactor content (2.7 L final volume), compared to the constant feeding in the best Pareto-optimal alternative (2.64 L final volume), as revealed by (Table 5, Figure 9b). Also, the high biomass growth is similar in both cases.On the other hand, as expected, a higher TRP productivity requires a higher substrate consumption, as is the case when using the modified E. coli T5 strain presented by Chen [72] (3.15 moles GLC), compared to the wild type of Chassagnole et al. [55] (0.06 moles GLC). As revealed by Table 5, the GLC consumption is influenced by the FBR operating mode, even if the same cell strain is used. Not surprisingly, the optimally operated FBR requires less substrate consumption. Thus, the same low GLC consumption was reported by the constant Pareto-optimal feeding policy (2.14 moles), and by the variable NLP-optimal feeding (4.53 moles), compared to the NLP-optimal but constant feeding (10.99 moles).
- The comparative analysis of the glycolytic species dynamics in (Figure 6) reveals close trajectories (even quasi-identical for the F6P and FDP species), with any accumulation tendency, for both non-optimal (curve 1), or optimal (curves 2–4) FBR operation. By contrast, the intermediate PEP and the ATP species tend to be formed in higher amounts, but then they are quickly consumed in the subsequent TRP synthesis, or in the glycolysis reactions, ultimately tending to reach their QSS. Such an intensive GLC import during the optimal FBR variable feeding operation (curve 2), and the GLC successive transformation over the glycolysis pathway, and Trp-operon expression is reflected by a higher ATP consumption compared to the non-optimal or sub-optimal FBR operation. The PYR metabolite is consumed in the TCA cycle, and the excess is excreted in the bulk phase (fairly predicted by our kinetic model, matching the experimental data). The better GLC use for an optimal FBR operation is also proven by the more produced and consumed secondary metabolite PYR (curve 2), and by a smaller QSS concentration of the PEP intermediate, quickly transformed in the final product TRP.
- The comparative analysis of the Trp-operon expression species dynamics in (Figure 7) reveals very close trajectories, except for the excreted TRP, for both the non-optimal (curves 1), or optimal (curves 2–4) FBR operation. Such a result can be explained by the operon expression mechanism, involving a tight control via its inhibition terms presented in (Table 4).
- The comparative plots of the GLC concentration dynamics in the FBR bulk phase are presented in (Figure 8). They indicate similar decreasing trajectories for all the investigated FBR operating alternatives. However, in the optimal operation cases (curves 2–4), the GLC decline in the bulk phase is more pronounced compared to the non-optimal operating case (curves 1). The higher GLC consumption in these last optimal operating cases can be explained by the higher TRP productivity. The unevenness in curve 2 is linked to the variable feeding with GLC of the optimally operated FBR (see the feeding plots in the top part of the figure).
- The HSMDM prediction of the biomass dynamics in the bulk phase (curve 1 in Figure 9c,d) fits the experimental data of Chen very well [72], thus proving its adequacy. The comparative plots of the biomass dynamics in the FBR bulk phase in all operating alternatives are presented in (Figure 9c). They reveal similar significantly increasing trajectories. In the best optimal operation cases, the biomass growth is more intense, due to a significantly higher GLC uptake, and a better GLC use during the batch, thus offering more favourable biomass growth conditions. Their maximum increase was limited during the bioreactor optimization in Equation (12), to avoid hydrodynamic problems.
- The TRP concentration dynamics in the bulk phase are comparatively plotted in (Figure 10) for all the investigated optimal operating alternatives of the FBR, and compared to the nominal (non-optimal) operation, with the constant feeding of (Table 1) of Chen [72] [that is, the experimental blue curve 1, and the experimental data (●, blue)]. As can be observed, the applied HSMDM of this paper fits the experimental data fairly well. From all the optimal FBR operation alternatives (curves 2–4), only the Pareto-optimal operation with a constant feeding exhibits a slightly better TRP productivity (curve 4).
- The rough classification given in Table 5 for the different FBR operating alternatives (i.e., poor, worst, good, best) is based on the TRP production maximization, on the substrate (GLC) minimum consumption, and on the minimum increase in the liquid volume.
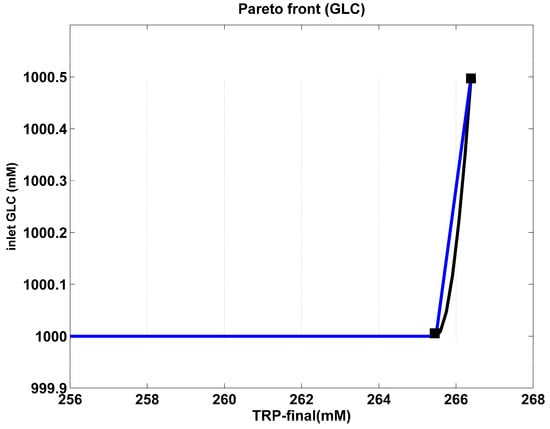
Figure 3.
The Pareto-optimal front for the analyzed FBR of Table 1 in terms of two opposite objectives, namely maximum TRP production vs. minimum substrate (GLC) consumption. The blue curve is the “rough” Pareto-optimal front. The black curve illustrates the approximated Pareto-optimal front by using the cubic spline procedure of Matlab™ (function “CSAPS”), with a smoothing constant of 10−20. For this problem, detailed in Equation (11), a solution was obtained by imposing the control variable limits given in Equation (12). The end point was chosen as being the most favourable solution of this optimization problem. However, the break point can be chosen as well, according to the suggestions of Dan-Maria [94].
Figure 3.
The Pareto-optimal front for the analyzed FBR of Table 1 in terms of two opposite objectives, namely maximum TRP production vs. minimum substrate (GLC) consumption. The blue curve is the “rough” Pareto-optimal front. The black curve illustrates the approximated Pareto-optimal front by using the cubic spline procedure of Matlab™ (function “CSAPS”), with a smoothing constant of 10−20. For this problem, detailed in Equation (11), a solution was obtained by imposing the control variable limits given in Equation (12). The end point was chosen as being the most favourable solution of this optimization problem. However, the break point can be chosen as well, according to the suggestions of Dan-Maria [94].
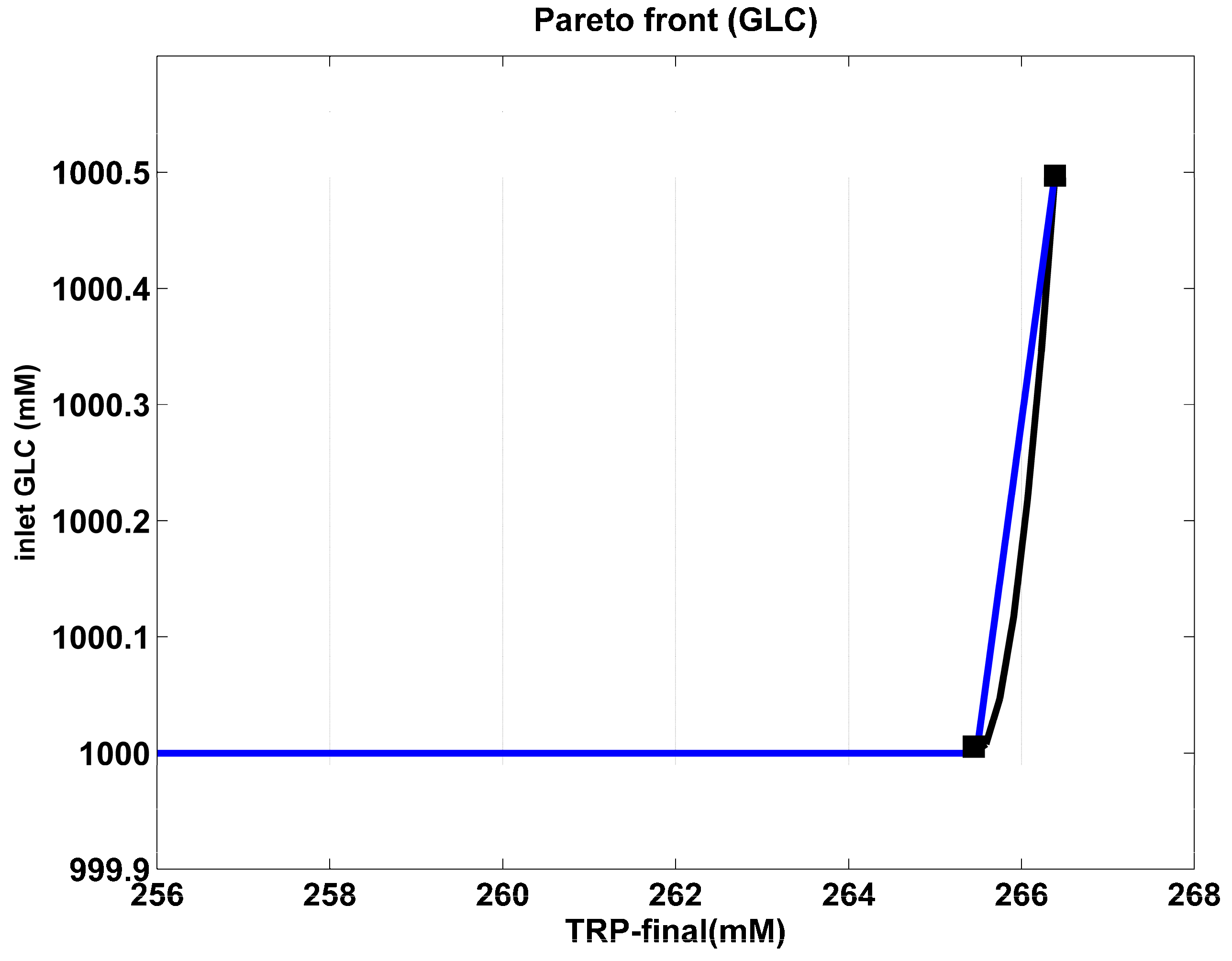
Figure 4.
The Pareto-optimal operating mode of the FBR in terms of required constant feed flow rate, for various amounts of maximum TRP produced. The blue curve is the “rough” Pareto-optimal front. The black curve illustrates the approximated Pareto-optimal front by using the cubic spline procedure of Matlab™ (function “CSAPS”), with a smoothing constant of 10−20. The marked points correspond to those of the Pareto-optimal curve of Figure 4.
Figure 4.
The Pareto-optimal operating mode of the FBR in terms of required constant feed flow rate, for various amounts of maximum TRP produced. The blue curve is the “rough” Pareto-optimal front. The black curve illustrates the approximated Pareto-optimal front by using the cubic spline procedure of Matlab™ (function “CSAPS”), with a smoothing constant of 10−20. The marked points correspond to those of the Pareto-optimal curve of Figure 4.
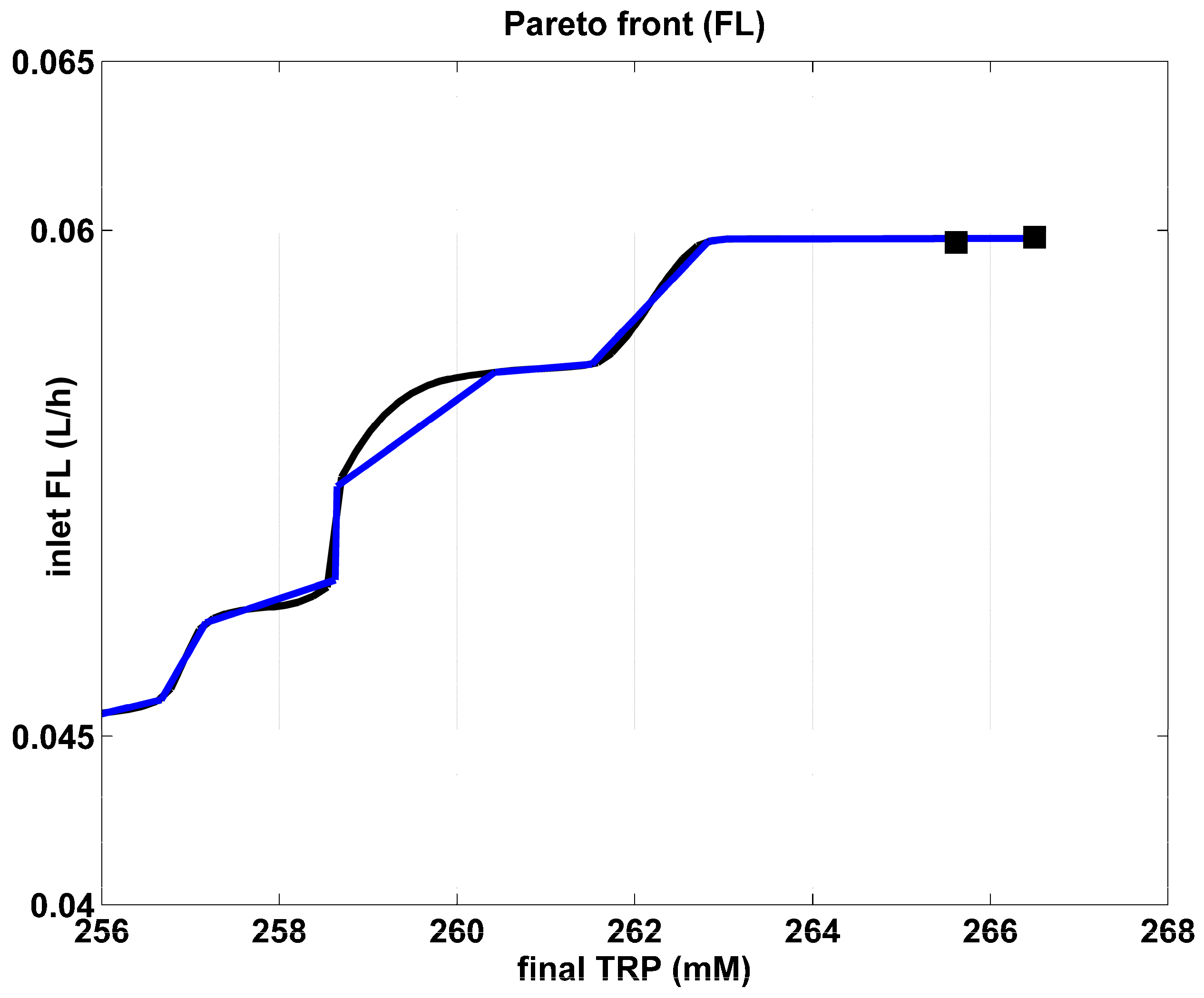
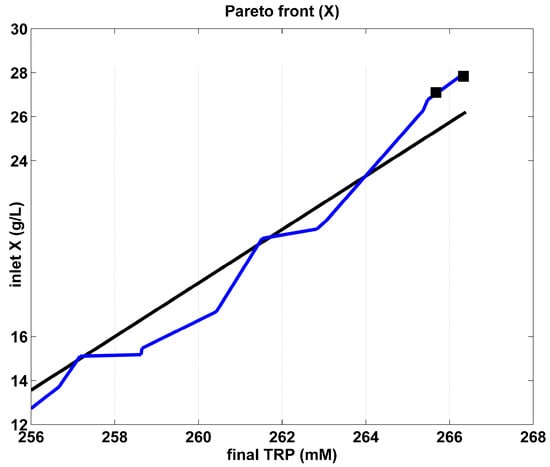
Figure 5.
The Pareto-optimal operating mode of the FBR in terms of the required initial biomass, for various amount of maximum TRP produced. The blue curve is the “rough” Pareto-optimal front. The black curve illustrates the approximated Pareto-optimal front by using the cubic spline procedure of Matlab™ (function “CSAPS”), with a smoothing constant 10−20. The marked points correspond to those of the Pareto-optimal curve of Figure 3.
Figure 5.
The Pareto-optimal operating mode of the FBR in terms of the required initial biomass, for various amount of maximum TRP produced. The blue curve is the “rough” Pareto-optimal front. The black curve illustrates the approximated Pareto-optimal front by using the cubic spline procedure of Matlab™ (function “CSAPS”), with a smoothing constant 10−20. The marked points correspond to those of the Pareto-optimal curve of Figure 3.
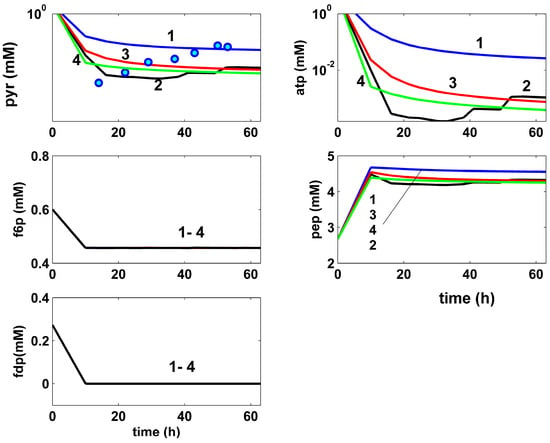
Figure 6.
Model-based simulated trajectories ( ) for the glycolytic key species (PYR, F6P, FDP, ATP, PEP) in the modified E. coli T5 strain for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the glycolytic key species (PYR, F6P, FDP, ATP, PEP) in the modified E. coli T5 strain for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
 ) for the glycolytic key species (PYR, F6P, FDP, ATP, PEP) in the modified E. coli T5 strain for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the glycolytic key species (PYR, F6P, FDP, ATP, PEP) in the modified E. coli T5 strain for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
Figure 6.
Model-based simulated trajectories ( ) for the glycolytic key species (PYR, F6P, FDP, ATP, PEP) in the modified E. coli T5 strain for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the glycolytic key species (PYR, F6P, FDP, ATP, PEP) in the modified E. coli T5 strain for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
 ) for the glycolytic key species (PYR, F6P, FDP, ATP, PEP) in the modified E. coli T5 strain for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the glycolytic key species (PYR, F6P, FDP, ATP, PEP) in the modified E. coli T5 strain for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
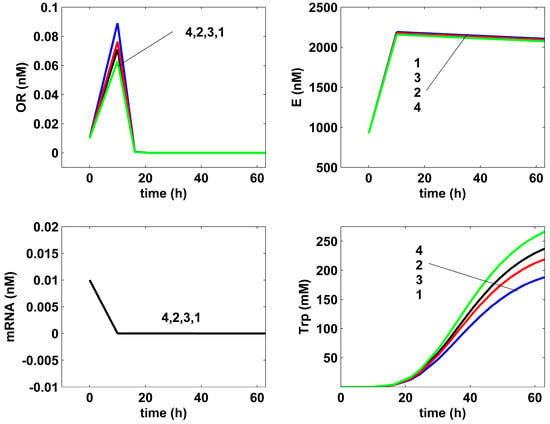
Figure 7.
Model-based simulated trajectories ( ) for the key species involved in the TRP-operon expression module (TRP, OR, MRNA, E) in the modified E. coli T5 strain for the FBR operated in two alternative forms: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the key species involved in the TRP-operon expression module (TRP, OR, MRNA, E) in the modified E. coli T5 strain for the FBR operated in two alternative forms: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
 ) for the key species involved in the TRP-operon expression module (TRP, OR, MRNA, E) in the modified E. coli T5 strain for the FBR operated in two alternative forms: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the key species involved in the TRP-operon expression module (TRP, OR, MRNA, E) in the modified E. coli T5 strain for the FBR operated in two alternative forms: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
Figure 7.
Model-based simulated trajectories ( ) for the key species involved in the TRP-operon expression module (TRP, OR, MRNA, E) in the modified E. coli T5 strain for the FBR operated in two alternative forms: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the key species involved in the TRP-operon expression module (TRP, OR, MRNA, E) in the modified E. coli T5 strain for the FBR operated in two alternative forms: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
 ) for the key species involved in the TRP-operon expression module (TRP, OR, MRNA, E) in the modified E. coli T5 strain for the FBR operated in two alternative forms: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the key species involved in the TRP-operon expression module (TRP, OR, MRNA, E) in the modified E. coli T5 strain for the FBR operated in two alternative forms: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).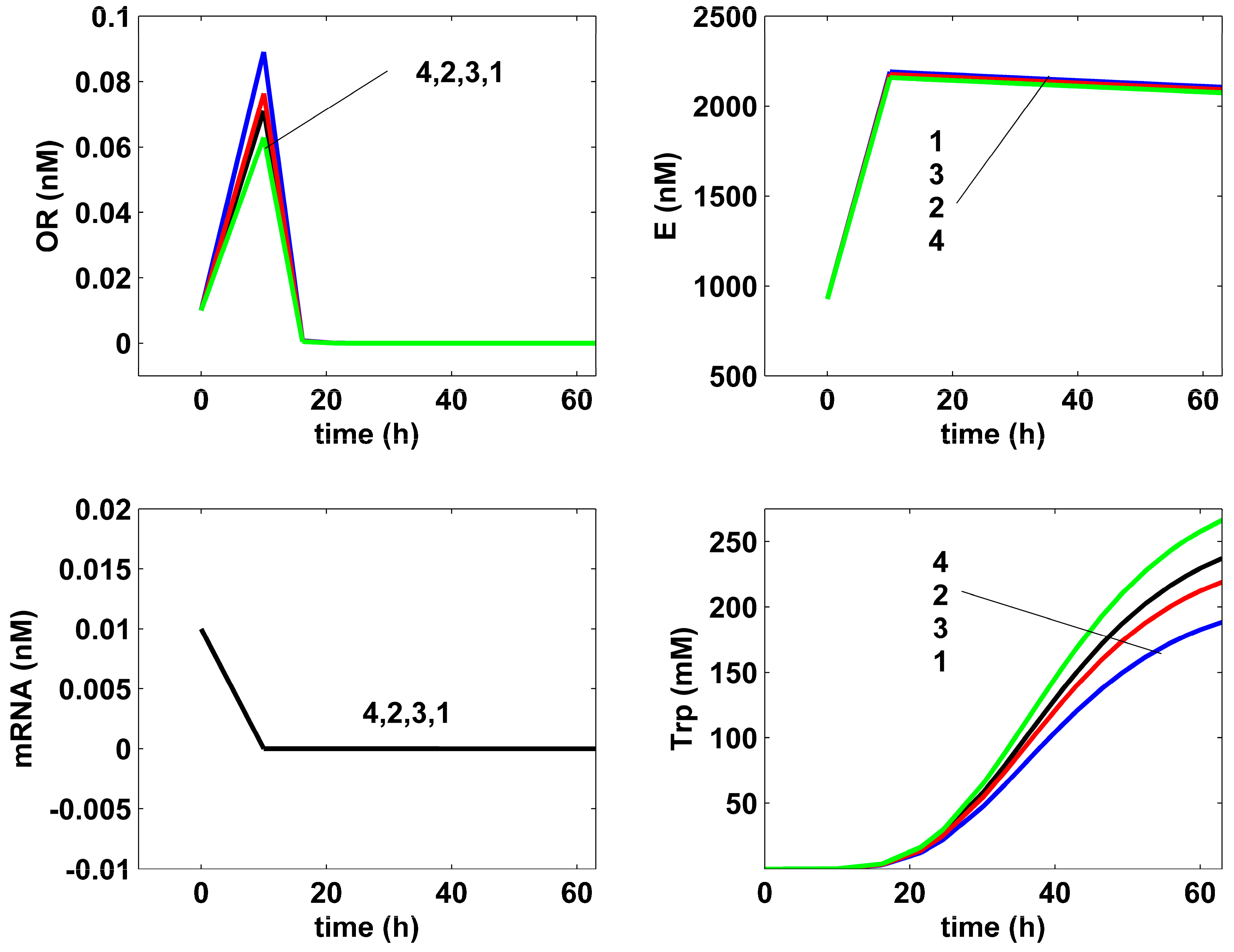
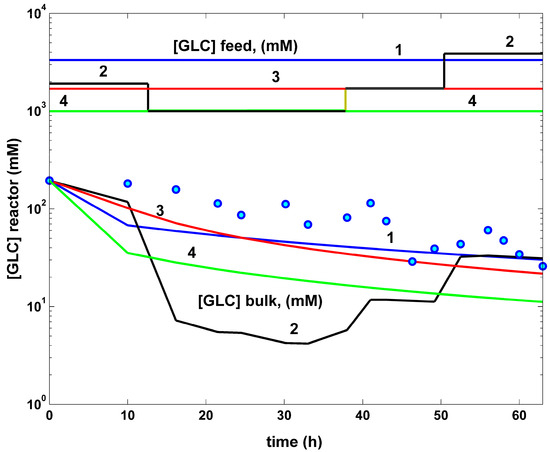
Figure 8.
Top curves: The time-step-wise optimal feeding strategy of GLC in the bioreactor (j = 1, …,5 time-arcs) for FBR operated in different ways, using a modified E. coli T5 strain [32]. (Bottom curves): Model-based simulated trajectories ( ) of glucose (GLC) in the bioreactor bulk, for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL ), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) of glucose (GLC) in the bioreactor bulk, for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL ), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
 ) of glucose (GLC) in the bioreactor bulk, for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL ), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) of glucose (GLC) in the bioreactor bulk, for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL ), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
Figure 8.
Top curves: The time-step-wise optimal feeding strategy of GLC in the bioreactor (j = 1, …,5 time-arcs) for FBR operated in different ways, using a modified E. coli T5 strain [32]. (Bottom curves): Model-based simulated trajectories ( ) of glucose (GLC) in the bioreactor bulk, for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL ), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) of glucose (GLC) in the bioreactor bulk, for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL ), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
 ) of glucose (GLC) in the bioreactor bulk, for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL ), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) of glucose (GLC) in the bioreactor bulk, for the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL ), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).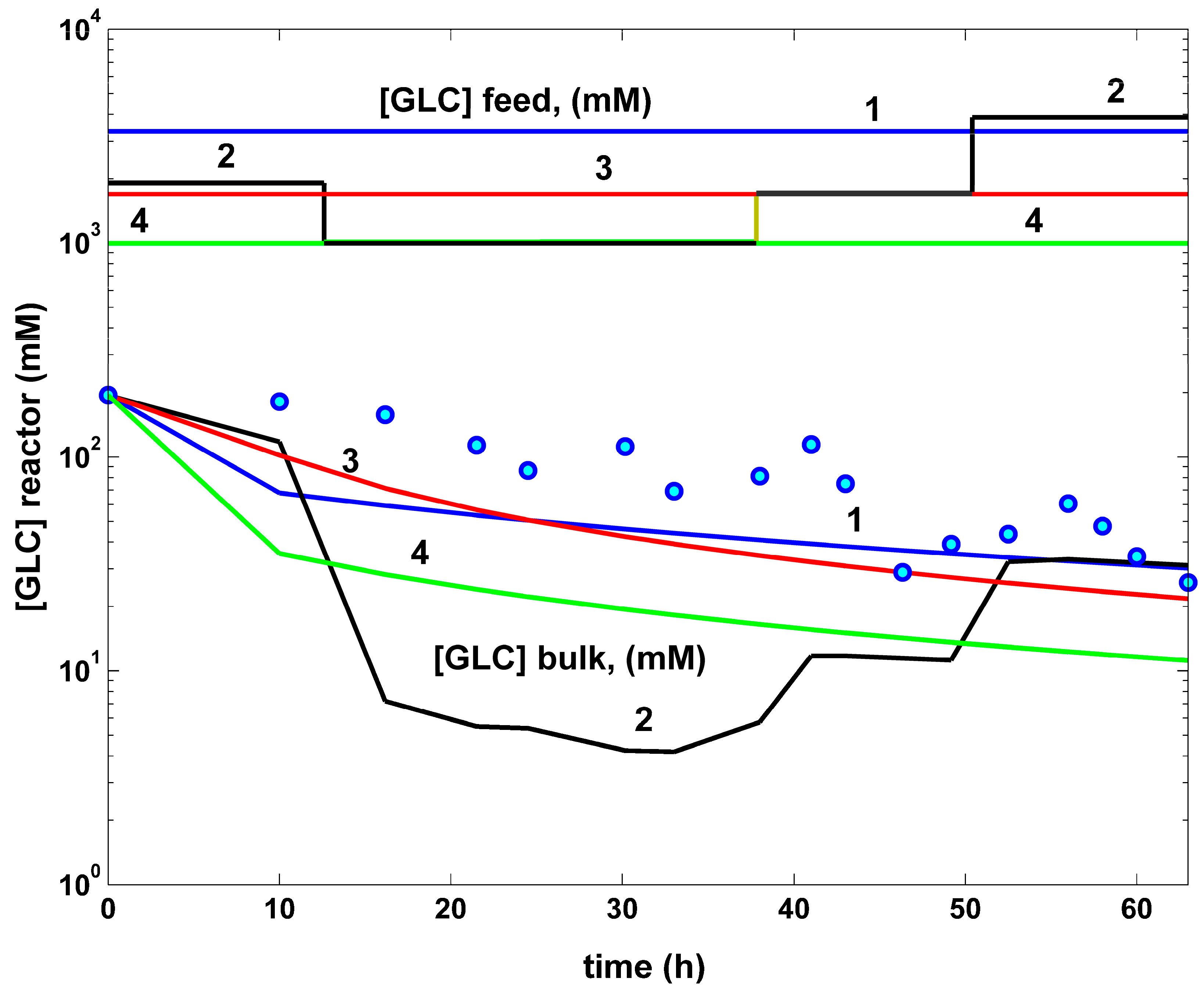
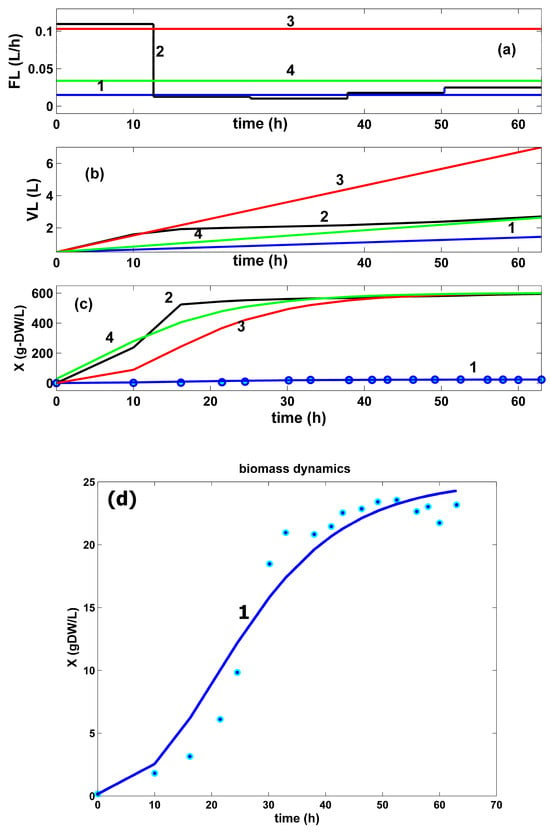
Figure 9.
Top (a): the time-step-wise optimal policy of the feed flow rate (FL), (j = 1, …,5 time-arcs) in the bioreactor ( ) for the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the below legend). Middle (b): the predicted dynamics of the liquid volume (VL) in the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the legend). Down (c): predictions of the viable biomass (X) concentration in the same FBR (using the modified E. coli T5 strain). Down (d): A magnification of the simulated curves, and the experimental data (●, blue) for the viable biomass (X) concentration in the FBR (using the modified E. coli T5 strain) under the nominal, non-optimal operation of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. All these results correspond to the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the below legend). Middle (b): the predicted dynamics of the liquid volume (VL) in the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the legend). Down (c): predictions of the viable biomass (X) concentration in the same FBR (using the modified E. coli T5 strain). Down (d): A magnification of the simulated curves, and the experimental data (●, blue) for the viable biomass (X) concentration in the FBR (using the modified E. coli T5 strain) under the nominal, non-optimal operation of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. All these results correspond to the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
 ) for the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the below legend). Middle (b): the predicted dynamics of the liquid volume (VL) in the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the legend). Down (c): predictions of the viable biomass (X) concentration in the same FBR (using the modified E. coli T5 strain). Down (d): A magnification of the simulated curves, and the experimental data (●, blue) for the viable biomass (X) concentration in the FBR (using the modified E. coli T5 strain) under the nominal, non-optimal operation of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. All these results correspond to the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the below legend). Middle (b): the predicted dynamics of the liquid volume (VL) in the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the legend). Down (c): predictions of the viable biomass (X) concentration in the same FBR (using the modified E. coli T5 strain). Down (d): A magnification of the simulated curves, and the experimental data (●, blue) for the viable biomass (X) concentration in the FBR (using the modified E. coli T5 strain) under the nominal, non-optimal operation of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. All these results correspond to the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
Figure 9.
Top (a): the time-step-wise optimal policy of the feed flow rate (FL), (j = 1, …,5 time-arcs) in the bioreactor ( ) for the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the below legend). Middle (b): the predicted dynamics of the liquid volume (VL) in the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the legend). Down (c): predictions of the viable biomass (X) concentration in the same FBR (using the modified E. coli T5 strain). Down (d): A magnification of the simulated curves, and the experimental data (●, blue) for the viable biomass (X) concentration in the FBR (using the modified E. coli T5 strain) under the nominal, non-optimal operation of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. All these results correspond to the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the below legend). Middle (b): the predicted dynamics of the liquid volume (VL) in the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the legend). Down (c): predictions of the viable biomass (X) concentration in the same FBR (using the modified E. coli T5 strain). Down (d): A magnification of the simulated curves, and the experimental data (●, blue) for the viable biomass (X) concentration in the FBR (using the modified E. coli T5 strain) under the nominal, non-optimal operation of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. All these results correspond to the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
 ) for the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the below legend). Middle (b): the predicted dynamics of the liquid volume (VL) in the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the legend). Down (c): predictions of the viable biomass (X) concentration in the same FBR (using the modified E. coli T5 strain). Down (d): A magnification of the simulated curves, and the experimental data (●, blue) for the viable biomass (X) concentration in the FBR (using the modified E. coli T5 strain) under the nominal, non-optimal operation of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. All these results correspond to the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
) for the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the below legend). Middle (b): the predicted dynamics of the liquid volume (VL) in the FBR (with modified E. coli T5 strain) operated in several alternative forms (see 1–4 in the legend). Down (c): predictions of the viable biomass (X) concentration in the same FBR (using the modified E. coli T5 strain). Down (d): A magnification of the simulated curves, and the experimental data (●, blue) for the viable biomass (X) concentration in the FBR (using the modified E. coli T5 strain) under the nominal, non-optimal operation of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. All these results correspond to the FBR operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).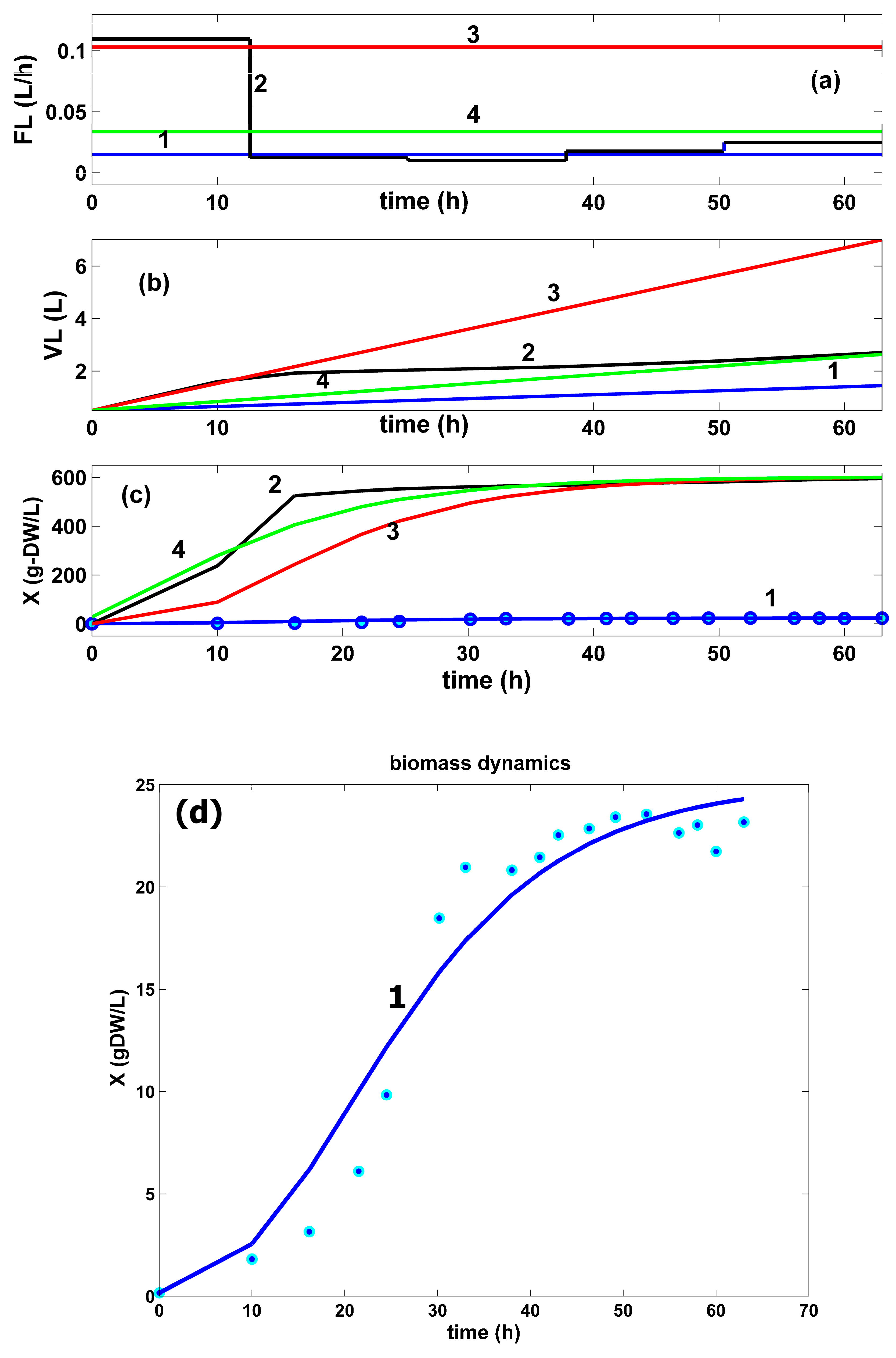
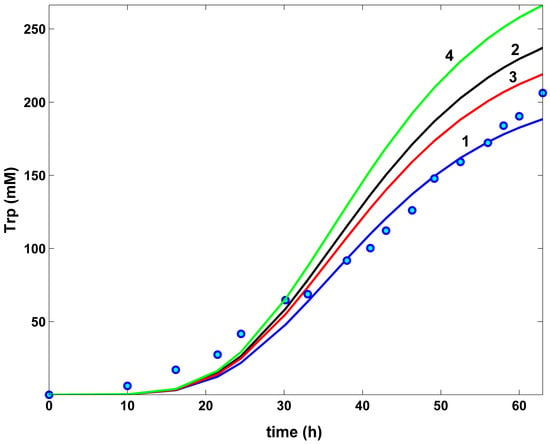
Figure 10.
Model-based predictions of the tryptophan (TRP) concentration dynamics in the analyzed FBR with using the modified E. coli T5 strain, but operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
Figure 10.
Model-based predictions of the tryptophan (TRP) concentration dynamics in the analyzed FBR with using the modified E. coli T5 strain, but operated in several alternative forms, as follows: (1, blue) Curves simulated with the HSMDM (this paper, and Maria [11]) compared to the experimental data (●, blue) of Chen [72] under the nominal, non-optimal operation of the FBR of (Table 1), which involves a constant feed [GLC], and constant feed flow rate FL. (2, black) The optimal operation of the FBR (this paper), with a time-step-wise optimally varied feed [GLC], and feed flow rate, with the control variable limits of Equation (12). (3, red) The optimal operation of the FBR (this paper), with a constant but optimal feed [GLC], and feed flow rate (FL), with the control variable limits of Equation (11). (4, green) The optimal operation of the FBR (this paper), with a constant but Pareto-optimal feed [GLC], and feed flow rate (FL; see Figure 3, Figure 4 and Figure 5), with the control variable limits of Equation (12).
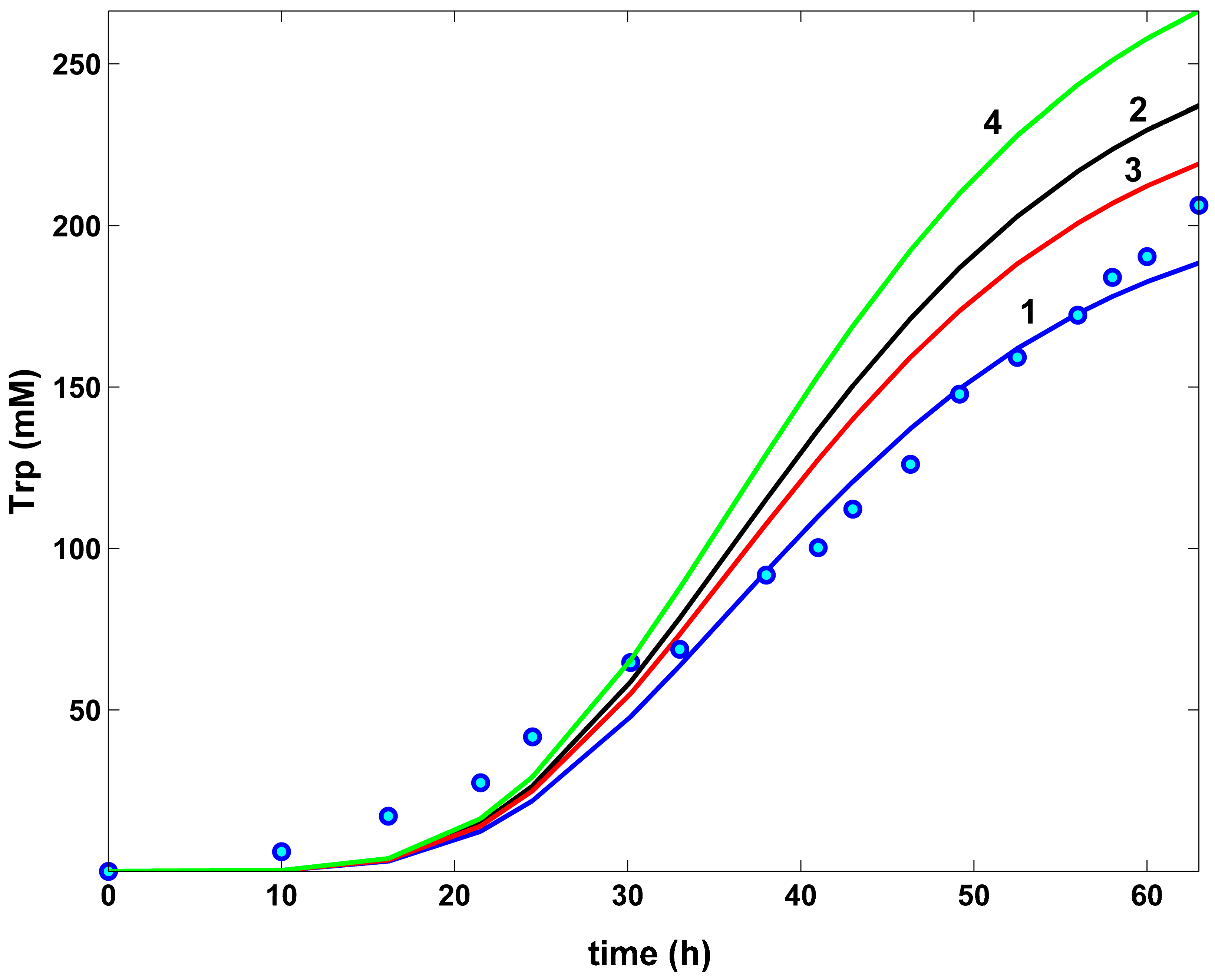
By analyzing the optimization algorithms applied in this paper, and the above (1–10) remarks regarding the obtained optimization results, the present work includes a significant number of novel elements, as follows:
- (a)
- It proves the advantages and feasibility of using the HSMDM in solving difficult biochemical engineering problems, such as the determination of an optimal operation strategy for an industrial bioreactor (FBR).
- (b)
- It proves that an experimentally validated modular HSMDM, of a moderate size, can be used to efficiently solving engineering problems with a much higher degree of precision and detail regarding the species dynamics predictions, compared to the unstructured (global) dynamic models.
- (c)
- It proves that the medium-size, bi-level (hybrid) HSMDM {11 cell metabolites, plus 4 bulk species, in a construction including four interconnected reaction modules, which are: [a] glycolysis, [b] ATP-recovery system, [c] TRP-operon expression, and the biomass [X] growth} adopted in this paper is able to adequately and concomitantly represent the dynamics for both macroscopic state variables of the FBR, and for the cell key species of the CCM related to the TRP synthesis. This hybrid structured model, linking the macro-state variables to the nano cell-scale variables, validated by Maria [11] using the recorded data from the lab-scale FBR over a sufficient batch time (63 h), offers enough credentials for providing predictions of an increased accuracy.
- (d)
- By coupling the structured reduced modular kinetic model of CCM with those for the TRP synthesis (trp-operon expression module) in the E. coli modified cells, this paper performs an in silico analysis, for the first time in the dedicated literature, regarding (i) the way by which the two processes interfere through the PEP glycolysis shared node, (ii) the consequences on the TRP-synthesis yield, and (iii) the way by which the cellular (nano-scale) bioprocess model, directly coupled with the bioreactor macroscopic dynamic model allows for determining the bioreactor’s best operating mode, leading to the TRP biosynthesis maximization.
- (e)
- Compared to the HSMDMs from the literature, the applied (hybrid) HSMDM of this paper, even in a reduced form, is able to simulate the influence of the glycolysis process intensity on the target metabolite (TRP) production, and on the raw material consumption, for the “wild” strain [11,44,98] compared to the modified T5 E. coli strain [11,32].
- (f)
- The HSMDMs allow for an easy evaluation of cellular metabolic fluxes. Such an analysis (not detailed here) quickly opens up the possibility of re-designing some cell fluxes via an in silico re-programming of the cell metabolism for designing GMO with industrial/medical applications [11,32,35,41,43,45,52].
- (g)
- The HSMDM allows for an easy evaluation of TRP-synthesis performance in a FBR depending on the reactor operation mode, but also on characteristics of the E. coli strain used {directly linked to the GLC import system, and to the ATP recovery system efficiency}, both determined by the cell phenotype, which is T5 strain in this paper [11,32,43,44].
- (h)
- Although the production of TRP by engineered E. coli has been extensively studied, “the need of multiple precursors for its synthesis and the complex regulations of the biosynthetic pathways make the achievement of a high product yield still very challenging” [11]. This difficult engineering problem was solved here by the concomitant use of a model-based (in silico) approach, completed with a biological improvement by using the modified T5 E. coli cell culture. Such a concomitant approach is seldom used in the literature.
- (i)
- The derived optimal operating policy of the FBR is given in time intervals (the so-called “time-arcs”) of equal length, of a reduced number, to be easily implemented. The control variables present optimal but constant levels over each time-arc (different between time-arcs) during the FBR operation.
- (j)
- The used biomass culture here refers to a modified E. coli T5 strain. The characteristics of this strain were reflected in the rate constants estimated by Maria [11]. This T5 strain was produced by Chen [72] and Chen et al. [47] to increase TRP production in their bench-scale FBR. They performed genetic modifications of the TRP-producing “wild” strain S028. Basically, “they removed the PTS import-system of GLC, by replacing it with a more effective one based on the galactose permease/glucokinase (GalP/Glk) uptake system, by modulating the gene expression of GalP/Glk. The resulting T5 strain showed an increase in the specific TRP production rate in a non-optimal FBR by 52.93% compared to the initial strain” [47], and by ca. 70% if the FBR used was operated optimally (this paper analysis).
- (k)
- Due to its complex modular structure, the results obtained with the HSMDM of this paper reveal the close link between the cell key metabolites and the FBR operating conditions. Thus, this hybrid model is complex enough to adequately represent the dynamics of the FBR state variables (i.e., the biomass growth, the GLC depletion, and the excreted TRP and PYR in the bulk phase), as well as the dynamics of the cell key species involved in the concerned reaction pathway modules, i.e., (a) glycolysis, (b) ATP recovery system, and (c) TRP-operon expression.
- (l)
- The paper proves that the HSMDM is also suitable to be used in solving multi-objective engineering problems, for instance by using the Pareto-optimal front technique.
6. Conclusions
The extended bi-level (hybrid) HSMDM adopted in this paper was experimentally proved by Maria [11] to adequately represent the dynamics of a studied FBR, for both macroscopic state variables, and for the cell key species of the CCM related to the TRP-synthesis in the FBR, which are: [a] glycolysis, [b] ATP-recovery system, [c] TRP-operon expression, and the biomass [X] growth. The hybrid structured model, linking the macro-state variables to the nano cell-scale variables, was validated by using the recorded data from a lab-scale FBR over an enough long batch time (63 h).
This paper proves how such HSMDMs can be used for engineering calculations if appropriate algorithms are used. The use of a classic NLP single-objective optimization algorithm, or the use of a multi-objective Pareto-optimal front technique, can be both very effective in predicting the optimal operating strategy of a studied FBR, if the problem is correctly formulated in mathematical terms.
The medium-size HSMDMs can offer a greater degree of detail and precision of the predictions compared to the un-structured (global) kinetic models. There are multiple engineering applications, detailed as follows: (a) they allow for the in silico optimization of the target metabolite synthesis (referred to as TRP herein), by optimizing the FBR operation mode; (b) able to predict the cell CCM metabolic fluxes (i.e., homeostatic stationary reaction rates), the HSMDM model can offer suggestions to genetically modify the cell strain [11,32,35,41] to obtain GMOs of desired characteristics (‘motifs’); (c) they can predict the reactor operating conditions when cell oscillatory processes could occur [43,44,45]; (d) they can perform an analysis of the substrate utilization and product synthesis closely related to the reactor operating strategy.
To obtain a high productivity in TRP with an industrial FBR is a difficult problem in the literature. This engineering problem was solved here through the concomitant use of a model-based (in silico) approach, completed with a biological improvement using the modified T5 E. coli cell culture. Such a concomitant approach is seldom used in the literature, but it proved to be very efficient.
The engineering evaluations developed in this paper can be further extended, for instance by deriving a multi-objective optimization of the FBR operating mode, by accounting for not only the product (TRP) maximization, but also minimization of the batch time, and other economic objectives.
The obtained results also proved that, in addition to the cell phenotype (linked to the CCM), the FBR operation modes are the major factors determining the TRP-synthesis efficiency (that is, product maximization, with the minimization of raw material consumption).
As an observation, it could be interesting to investigate, in a future work, how the heterogeneity of the cell culture is reflected in the constants of the kinetic model and in the quality of its predictions [99].
Author Contributions
Conceptualization, G.M.; methodology, G.M.; investigation, G.M. and D.G.; data curation, G.M.; project administration, G.M.; software, G.M.; supervision, G.M.; validation, G.M. and D.G.; writing—original draft, G.M.; writing—review and editing, G.M. and D.G.; formal analysis, D.G. All authors have read and agreed to the published version of the manuscript.
Funding
The authors did not receive support from any organization for the submitted work. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data Availability Statement
Experimental datasets and some of the information used in this study are imported from the literature, and from TU Hamburg, and the source was noted in the text, and in the Acknowledgements section.
Acknowledgments
The authors are deeply grateful to An-Ping Zeng and to their research group (Minliang Chen, Lin Chen, Chengwei Ma) from the Hamburg University of Technology (Institute of Bioprocess and Biosystems Engineering, Germany) for providing, on a free basis, and without any claim of any kind (including no claim of authorship), experimental data from their published works on the TRP synthesis in a fed-batch bioreactor using a genetically modified E. coli (T5 strain) cell culture. I especially want to thank An-Ping Zeng for their constructive comments and discussions on the manuscript.
Conflicts of Interest
The authors confirm that their paper has no conflicts of interest of any kind, and of any nature.
Nomenclature
| , [i] | - | Species ‘i’ concentration |
| - | Biomass concentration | |
| Glucose feeding solution concentration | ||
| Glucose feeding solution concentration over the time-arc ‘J’ | ||
| Glucose initial concentration in the bioreactor | ||
| Glucose concentration in the bulk-phase | ||
| D = FL/VL | - | Bioreactor dilution rate |
| - | Liquid feed flow rate in the bioreactor | |
| k, , , K, n, , , , , etc. - | Reaction rates, and/or equilibrium constants of the kinetic model | |
| - | Species ‘i’ reaction rate | |
| , tf, | - | Time, batch time, cell cycle, respectively |
| – | - | metabolic fluxes in the glycolysis (Table 2 and Table 3, Figure 2). |
| - | liquid volume in the bioreactor | |
| , | - | stoichiometric coefficients |
| Greeks | ||
| α, β, γ, δ | - | Reaction rate constants |
| μ | - | Cell content dilution rate, that is Ln(2)/ |
| Ω | FBR optimization objective function, Equation (10) | |
| - | biomass density | |
| Subscripts | ||
| 0,o | Initial | |
| cell | Referring to the (inside) cell | |
| ext | External to cell (i.e., in the bulk phase) | |
| f | Final | |
| inlet | In the feed | |
| X | Biomass“ | |
| Abbreviations | ||
| 13dpg, pgp | 1,3-diphosphoglycerate | |
| 3pg | 3-phosphoglycerate | |
| 2pg | 2-phosphoglycerate | |
| AA | Amino-acids | |
| Accoa, acetyl-CoA | Acetyl-coenzyme A | |
| AC | Acetate | |
| ADP, adp | Adenosin-diphosphate | |
| AK-ase | Adenylate kinase | |
| ALE | Adaptive laboratory evolution | |
| AMP, amp | Adenosin-monophosphate | |
| ATP, atp | Adenosin-triphosphate | |
| ATP-ase | ATP monophosphatase | |
| CCM | Central carbon metabolism | |
| CIT | Citrate | |
| DO | Dissolved oxygen | |
| DW | Dry mass | |
| E | Enzyme anthranilate synthase in the TRP synthesis model. | |
| ETOH | Ethanol | |
| ext | External to the cell (i.e., in the bulk phase) | |
| FBR | Fed-batch bioreactor | |
| FDP, fdp | Fructose-1,6-biphosphate | |
| F6P, f6p | Fructose-6-phosphate | |
| GalP/Glk | Galactose permease/glucokinase | |
| G3P, g3p, GAP, gap, 3PG, 3pg | Glyceraldehyde-3-phosphate | |
| 2PG, 2pg | 2-phosphoglycerate | |
| G6P, g6p | Glucose-6-phosphate | |
| GLC, glc | Glucose | |
| Glc(ex), [GLC]ext | Glucose in the environment (bulk phase) | |
| GMO | Genetic modified micro-organisms | |
| GRC | Genetic regulatory circuits | |
| HK-ase | Hexokinase | |
| JWS | Silicon Cell project of Olivier and Snoep [65] | |
| LAC, lac | Lactate | |
| MACR | Mechanically agitated continuously aerated reactor | |
| Max (x) | Maxim of (x) | |
| MMA | The adaptive random optimization algorithm of Maria [97] | |
| MRNA | Tryptophan messenger ribonucleic acid during its encoding gene dynamic transcription, and translation | |
| NAD(P)H | Nicotinamide adenine dinucleotide (phosphate) reduced | |
| NLP | Nonlinear programming | |
| ODE | Ordinary differential equations set | |
| OR | The complex between O and R (aporepressor of the TRP gene) | |
| OT | The total TRP operon | |
| P, Pi | Phosphoric acid | |
| PEP, pep | Phosphoenolpyruvate | |
| 13DPG = PGP | 1,3-diphosphoglycerate | |
| PFK-ase | Phosphofructokinase | |
| PK-ase | Pyruvate kinase | |
| PTS | Phosphotransferase, or the phosphoenolpyruvate:glucose phosphotransferase system | |
| PYR, pyr | Pyruvate | |
| QSS | Quasi-steady-state | |
| R5P | Ribose 5-phosphate | |
| mRNA, MRNA | Messenger ribonucleic acid; tryptophan mRNA during its encoding gene dynamic transcription, and translation | |
| SUCC, suc | Succinate | |
| TCA, tca | Tricarboxylic acid cycle | |
| TF | Gene expression transcription factors | |
| TRP, Trp, trp | Tryptophan | |
| X | Biomass | |
| Wt. | Weight | |
| [X] | Concentration of X species | |
References
- Liese, A.; Seelbach, K.; Wandrey, C. Industrial Biotransformations; Wiley-VCH: Weinheim, Germany, 2006. [Google Scholar]
- Moulijn, J.A.; Makkee, M.; van Diepen, A. Chemical Process Technology; Wiley: New York, NY, USA, 2001. [Google Scholar]
- Henson, M.A. Model-based control of biochemical reactors. In The Control Handbook, 2nd ed.; Levine, W., Ed.; Taylor and Francis: New York, NY, USA, 2010. [Google Scholar]
- Dewasme, L.; Cote, F.; Filee, P.; Hantson, A.L.; Wouwer, A.V. Macroscopic dynamic modeling of sequential batch cultures of hybridoma cells: An experimental validation. Bioengineering 2017, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Maria, G. Model-based optimization of a fed-batch bioreactor for mAb production using a hybridoma cell culture. Molecules 2020, 25, 5648. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, K.; Hempel, D.C. From gene to product (editorial). Eng. Life Sci. 2006, 6, 437. [Google Scholar] [CrossRef]
- Hempel, D.C. Development of biotechnological processes by integrating genetic and engineering methods. Eng. Life Sci. 2006, 6, 443–447. [Google Scholar] [CrossRef]
- Nedovic, V.; Willaert, R. Applications of Cell Immobilisation Technology; Springer: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Dewasme, L.; Amribt, Z.; Santos, L.O.; Hantson, A.L.; Bogaerts, P.; Wouwer, A.V. Hybridoma cell culture optimization using nonlinear model predictive control. Proc. Int. Fed. Autom. Control 2013, 46, 60–65. [Google Scholar] [CrossRef]
- Amribt, Z.; Dewasme, L.; Wouwer, A.V.; Bogaerts, P. Optimization and robustness analysis of hybridoma cell fed-batch cultures using the overflow metabolism model. Bioprocess Biosyst. Eng. 2014, 37, 1637–1652. [Google Scholar] [CrossRef]
- Maria, G. A CCM-based modular and hybrid kinetic model to simulate the tryptophan synthesis in a fed-batch bioreactor using modified E. coli cells. Comput. Chem. Eng. 2021, 133, 106628–106635. [Google Scholar] [CrossRef]
- Srinivasan, B.; Primus, C.J.; Bonvin, D.; Ricker, N.L. Run-to-run optimization via control of generalized constraints. Control. Eng. Pract. 2001, 9, 911–919. [Google Scholar] [CrossRef]
- Mendes, R.; Rocha, I.; Pinto, J.P.; Ferreira, E.C.; Rocha, M. Differential evolution for the offline and online optimization of fed-batch fermentation processes. In Advances in Differential Evolution. Studies in Computational Intelligence; Chakraborty, U.K., Ed.; Springer: Berlin, Germany, 2008; pp. 299–317. [Google Scholar]
- Bonvin, D. Real-Time Optimization; MDPI: Basel, Switzerland, 2017. [Google Scholar]
- DiBiasio, D. Introduction to the control of biological reactors. In Chemical Engineering Problems in Biotechnology; Shuler, M.L., Ed.; AIChE: New York, NY, USA, 1989; Volume 1. [Google Scholar]
- Lee, J.; Lee, K.S.; Lee, J.H.; Park, S. An on-line batch span minimization and quality control strategy for batch and semi-batch processes. Control Eng. Pract. 2001, 9, 901–909. [Google Scholar] [CrossRef]
- Ruppen, D.; Bonvin, D.; Rippin, D.W.T. Implementation of adaptive optimal operation for a semi-batch reaction system. Comput. Chem. Eng. 1998, 22, 185–199. [Google Scholar] [CrossRef]
- Loeblein, C.; Perkins, J.; Srinivasan, B.; Bonvin, D. Performance analysis of on-line batch optimization systems. Comput. Chem. Eng. 1997, 21, S867–S872. [Google Scholar] [CrossRef]
- Rao, M.; Qiu, H. Process Control Engineering: A Textbook for Chemical, Mechanical and Electrical Engineers; Gordon and Breach Science Publ.: Amsterdam, The Netherlands, 1993. [Google Scholar]
- Engasser, J.M. Bioreactor engineering: The design and optimization of reactors with living cells. Chem. Eng. Sci. 1988, 43, 1739–1748. [Google Scholar] [CrossRef]
- Wang, C.; Quan, H.; Xu, X. Optimal design of multiproduct batch chemical process using genetic algorithms. Ind. Eng. Chem. Res. 1996, 35, 3560–3566. [Google Scholar] [CrossRef]
- Wang, P. Multi-scale features in recent development of enzymic biocatalyst systems. Appl. Biochem. Biotechnol. 2009, 152, 343–352. [Google Scholar] [CrossRef]
- Ozturk, S.S.; Palsson, B.O. Effect of initial cell density on hybridoma growth, metabolism, and monoclonal antibody production. J. Biotechnol. 1990, 16, 259–278. [Google Scholar] [CrossRef]
- Fotopoulos, J.; Georgakis, C.; Stenger, H.G., Jr. Uncertainty issues in the modeling and optimization of batch reactors with tendency models. Chem. Eng. Sci. 1994, 49, 5533–5547. [Google Scholar] [CrossRef]
- Martinez, E. Batch-to-batch optimization of batch processes using the STATSIMPLEX search method. In Proceedings of the 2nd Mercosur Congress on Chemical Engineering, Rio de Janeiro, Costa Verde, Brazil, 14–18 January 2005. [Google Scholar]
- Lübbert, A.; Jørgensen, S.B. Bioreactor performance: A more scientific approach for practice. J. Biotechnol. 2001, 85, 187–212. [Google Scholar] [CrossRef]
- Binette, J.C.; Srinivasan, B. On the use of nonlinear model predictive control without parameter adaptation for batch processes. Processes 2016, 4, 27. [Google Scholar] [CrossRef]
- Maria, G.; Peptanaru, I.M. Model-based optimization of mannitol production by using a sequence of batch reactors for a coupled bi-enzymatic process—A dynamic approach. Dynamics 2021, 1, 134–154. [Google Scholar] [CrossRef]
- Maria, G.; Crisan, M. Operation of a mechanically agitated semi-continuous multi-enzymatic reactor by using the Pareto-optimal multiple front method. J. Process Control. 2017, 53, 95–105. [Google Scholar] [CrossRef]
- Smets, I.Y.; Claes, J.E.; November, E.J.; Bastin, G.P.; Van Impe, J.F. Optimal adaptive control of (bio)chemical reactors: Past, present and future. J. Process Control 2004, 14, 795–805. [Google Scholar] [CrossRef]
- Franco-Lara, E.; Weuster-Botz, D. Estimation of optimal feeding strategies for fed-batch bioprocesses, Estimation of optimal feeding strategies for fed-batch bioprocesses. Bioprocess Biosyst. Eng. 2005, 28, 71–77. [Google Scholar] [CrossRef][Green Version]
- Maria, G.; Renea, L. Tryptophan production maximization in a fed-batch bioreactor with modified E. coli cells, by optimizing its operating policy based on an extended structured cell kinetic model. Bioengineering 2021, 8, 210. [Google Scholar] [CrossRef] [PubMed]
- Avili, M.G.; Fazaelipoor, M.H.; Jafari, S.A.; Ataei, S.A. Comparison between batch and fed-batch production of rhamnolipid by Pseudomonas aeruginosa. Iran. J. Biotechnol. 2012, 10, 263–269. Available online: https://www.researchgate.net/publication/288966207 (accessed on 21 October 2021).
- Moser, A. Bioprocess Technology-Kinetics and Reactors; Springer: Berlin, Germany, 1988. [Google Scholar]
- Maria, G. Hybrid modular kinetic models linking cell-scale structured CCM reaction pathways to bioreactor macro-scale state variables. In Applications for Solving Bioengineering Problems; Juniper: Irvine, CA, USA, 2023. [Google Scholar]
- Agrawal, P.; Koshy, G.; Ramseier, M. An algorithm for operating a fed-batch fermentator at optimum specific-growth rate. Biotechnol. Bioeng. 1989, 33, 115–125. [Google Scholar] [CrossRef]
- Banga, J.R.; Alonso, A.A.; Singh, P.R. Stochastic optimal control of fed-batch bioreactors. In Proceedings of the AIChE Annual Meeting, San Francisco, CA, USA, 13–18 November 1994. [Google Scholar]
- Doran, P.M. Bioprocess Engineering Principles; Elsevier: Amsterdam, The Netherlands, 1995. [Google Scholar]
- Sarkar, D.; Modak, J.M. Pareto-optimal solutions for multi-objective optimization of fed-batch bioreactors using nondominated sorting genetic algorithm. Chem. Eng. Sci. 2005, 60, 481–492. [Google Scholar] [CrossRef]
- Ashoori, A.; Moshiri, B.; Khaki-Sedigh, A.; Bakhtiari, M.R. Optimal control of a nonlinear fed-batch fermentation process using model predictive approach. J. Process Control. 2009, 19, 1162–1173. [Google Scholar] [CrossRef]
- Maria, G.; Luta, I. Structured cell simulator coupled with a fluidized bed bioreactor model to predict the adaptive mercury uptake by E. coli cells. Comput. Chem. Eng. 2013, 58, 98–115. [Google Scholar] [CrossRef]
- Roubos, J.A. Bioprocess Modeling and Optimization—Fed-Batch Clavulanic Acid Production by Streptomyces clavuligerus. Ph.D. Thesis, TU Delft, Delft, The Netherlands, 2002. [Google Scholar]
- Maria, G.; Mihalachi, M.; Gijiu, C.L. Model-based identification of some conditions leading to glycolytic oscillations in E. coli cells. Chem. Biochem. Eng. Q. 2018, 32, 523–533. [Google Scholar] [CrossRef]
- Maria, G.; Gijiu, C.L.; Maria, C.; Tociu, C. Interference of the oscillating glycolysis with the oscillating tryptophan synthesis in the E. coli cells. Comput. Chem. Eng. 2018, 108, 395–407. [Google Scholar] [CrossRef]
- Maria, G. In-silico determination of some conditions leading to glycolytic oscillations and their interference with some other processes in E. coli cells. Front. Chem. 2020, 8, 526679–526693. [Google Scholar] [CrossRef] [PubMed]
- Chen, L. Rational Metabolic Engineering and Systematic Analysis of Escherichia coli for L-Tryptophan Bioproduction. Ph.D. Thesis, TU Hamburg, Hamburg, Germany, 2016. [Google Scholar]
- Chen, M.; Ma, C.; Chen, L.; Zeng, A.P. Integrated laboratory evolution and rational engineering of GalP/Glk-dependent Escherichia coli for higher yield and productivity of L-tryptophan biosynthesis. Metab. Eng. Commun. 2021, 12, e00167. [Google Scholar] [CrossRef]
- Maria, G. A review of algorithms and trends in kinetic model identification for chemical and biochemical systems. Chem. Biochem. Eng. Q. 2004, 18, 195–222. Available online: http://silverstripe.fkit.hr/cabeq/past-issues/issue/41 (accessed on 22 October 2021).
- Edwards, K.; Edgar, T.F.; Manousiouthakis, V.I. Kinetic model reduction using genetic algorithms. Comput. Chem. Eng. 1998, 22, 239–246. [Google Scholar] [CrossRef]
- Martinez, E.C.; Beltramini, L.J. Lumping upon time-scales: Modeling upon topological factors. Chem. Eng. Sci. 1990, 45, 2103–2108. [Google Scholar] [CrossRef]
- Maria, G. Relations between apparent and intrinsic kinetics of programmable drug release in human plasma. Chem. Eng. Sci. 2005, 60, 1709–1723. [Google Scholar] [CrossRef]
- Maria, G.; Xu, Z.; Sun, J. Multi-objective MINLP optimization used to identify theoretical gene knockout strategies for E. coli cell. Chem. Biochem. Eng. Q. 2011, 25, 403–424. [Google Scholar]
- Dorka, P. Modelling Batch and Fed-Batch Mammalian Cell Cultures for Optimizing MAb Productivity. Master’s Thesis, University of Waterloo, Waterloo, ON, Canada, 2007. [Google Scholar]
- Maria, G. In silico derivation of a reduced kinetic model for stationary or oscillating glycolysis in Escherichia coli bacterium. Chem. Biochem. Eng. Q. 2014, 28, 509–529. [Google Scholar] [CrossRef]
- Chassagnole, C.; Noisommit-Rizzi, N.; Schmid, J.W.; Mauch, K.; Reuss, M. Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol. Bioeng. 2002, 79, 53–73. [Google Scholar] [CrossRef]
- Usuda, Y.; Nishio, Y.; Iwatani, S.; Van Dien, S.J.; Imaizumi, A.; Shimbo, K.; Kageyama, N.; Iwahata, D.; Miyano, H.; Matsui, K. Dynamic modeling of Escherichia coli metabolic and regulatory systems for amino-acid production. J. Biotechnol. 2010, 147, 17–30. [Google Scholar] [CrossRef]
- Kadir, T.A.A.; Mannan, A.A.; Kierzek, A.M.; McFadden, J.; Shimizu, K. Modeling and simulation of the main metabolism in Escherichia coli and its several single-gene knockout mutants with experimental verification. Microb. Cell Factories 2010, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Ceric, S.; Kurtanjek, Z. Model identification, parameter estimation, and dynamic flux analysis of E. coli central metabolism. Chem. Biochem. Eng. Q. 2006, 20, 243–253. Available online: https://hrcak.srce.hr/clanak/7390 (accessed on 10 August 2024).
- Tusek, A.J.; Kurtanjek, Z. Model and global sensitivity analysis of E. coli central metabolism. In Proceedings of the 6th Vienna Conference on Mathematical Modelling MATHMOD, Beč, Austria, 11–13 February 2009; p. 253. [Google Scholar] [CrossRef]
- Edwards, J.S.; Palsson, B.O. The Escherichia coli MG1655 in silico metabolic genotype: Its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. USA 2000, 97, 5528–5533. [Google Scholar] [CrossRef] [PubMed]
- Schmid, J.W.; Mauch, K.; Reuss, M.; Gilles, E.D.; Kremling, A. Metabolic design based on a coupled gene expression—Metabolic network model of tryptophan production in Escherichia coli. Metab. Eng. 2004, 6, 364–377. [Google Scholar] [CrossRef]
- Costa, R.S.; Machado, D.; Rocha, I.; Ferreira, E.C. Large scale dynamic model reconstruction for the central carbon metabolism of Escherichia coli. In Distributed Computing, Artificial Intelligence, Bioinformatics, Soft Computing, and Ambient Assisted Living, Proceedings of the IWANN Conference, Salamanca, Spain, 10–12 June 2009; Part II, LNCS 5518; Springer: Berlin, Germany, 2009; pp. 1079–1083. [Google Scholar]
- Costa, R.S.; Machado, D.; Rocha, I.; Ferreira, E.C. Hybrid dynamic modeling of Escherichia coli central metabolic network combining Michaelis–Menten and approximate kinetic equations. BioSystems 2010, 100, 150–157. [Google Scholar] [CrossRef]
- Machado, D.; Zhuang, K.H.; Sonnenschein, N.; Herrgård, M.J. Current challenges in modeling cellular metabolism. Front. Bioeng. Biotechnol. 2015, 3, 193. [Google Scholar] [CrossRef]
- Olivier, B.G.; Snoep, J.L. Web-based kinetic modelling using JWS Online. Bioinformatics 2004, 20, 2143–2144. [Google Scholar] [CrossRef] [PubMed]
- Seressiotis, A.; Bailey, J.E. MPS: An algorithm and data base for metabolic pathways synthesis. Biotechnol. Lett. 1986, 8, 837–842. [Google Scholar] [CrossRef]
- Tomita, M.; Hashimoto, K.; Takahashi, K.; Shimizu, T.; Matsuzaki, Y.; Miyoshi, F.; Saito, K.; Tanida, S.; Yugi, K.; Venter, J.C. E-Cell: Software environment for whole cell simulation. Bioinformatics 1999, 15, 72–84. [Google Scholar] [CrossRef]
- Tomita, M. Whole-cell simulation: A grand challenge of the 21st century. Trends Biotechnol. 2001, 19, 205–210. [Google Scholar] [CrossRef]
- Slepchenko, B.M.; Schaff, J.C.; Macara, I.; Loew, L.M. Quantitative cell biology with the Virtual Cell. Trends Cell Biol. 2003, 13, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Styczynski, M.P.; Stephanopoulos, G. Overview of computational methods for the inference of gene regulatory networks. Comput. Chem. Eng. 2005, 29, 519–534. [Google Scholar] [CrossRef]
- Visser, D.; Schmid, J.W.; Mauch, K.; Reuss, M.; Heijnen, J.J. Optimal re-design of primarymetabolism in Escherichia coli using linlog kinetics. Metab. Eng. 2004, 6, 378–390. [Google Scholar] [CrossRef]
- Chen, M. Novel Approaches for In Vivo Evolution, Screening and Characterization of Enzymes for Metabolic Engineering of Escherichia coli as Hyper L-Tryptophan Producer. Ph.D. Thesis, TU Hamburg, Hamburg, Germany, 2020. [Google Scholar]
- Li, Z.; Wang, H.; Ding, D.; Liu, Y.; Fang, H.; Chang, Z.; Chen, T.; Zhang, D. Metabolic engineering of Escherichia coli for production of chemicals derived from the shikimate pathway. J. Ind. Microbiol. Biotechnol. 2020, 47, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Li, R.; Liang, Q.; Qi, Q.; Li, Q.; Gu, P. Metabolic engineering for improving L-tryptophan production in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2019, 46, 55–65. [Google Scholar] [CrossRef]
- Carmona, S.B.; Flores, N.; Martínez-Romero, E.; Gosset, G.; Bolívar, F.; Escalante, A. Evolution of an Escherichia coli PTS− strain: A study of reproducibility and dynamics of an adaptive evolutive process. Appl. Microbiol. Biotechnol. 2020, 104, 9309–9325. [Google Scholar] [CrossRef]
- Lei, F.; Jorgensen, S.B. Estimation of kinetic parameters in a structured yeast model using regularization. J. Biotechnol. 2001, 88, 223. [Google Scholar] [CrossRef]
- Bishop, M. An Introduction to Chemistry; Chiral Publ.: Carmel, CA, USA, 2013; Available online: https://preparatorychemistry.com/Bishop_contact.html (accessed on 10 August 2024).
- Laos, K.; Harak, M. The viscosity of supersaturated aqueous glucose, fructose and glucose-fructose solutions. J. Food Phys. 2014, 27, 27–30. [Google Scholar]
- Bhartiya, S.; Chaudhary, N.; Venkatesh, K.V.; Doyle, F.J., III. Multiple feedback loop design in the tryptophan regulatory network of E. coli suggests a paradigm for robust regulation of processes in series. J. R. Soc. Interface 2006, 3, 383–391. [Google Scholar] [CrossRef]
- Xiu, Z.L.; Zeng, A.P.; Deckwer, W.D. Model analysis concerning the effects of growth rate and intracellular tryptophan level on the stability and dynamics of tryptophan biosynthesis in bacteria. J. Biotechnol. 1997, 58, 125–140. [Google Scholar] [CrossRef]
- Xiu, Z.L.; Chang, Z.Y.; Zeng, A.P. Nonlinear dynamics of regulation of bacterial trp operon: Model analysis of integrated effects of repression, feedback inhibition, and attenuation. Biotechnol. Prog. 2002, 18, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Termonia, Y.; Ross, J. Oscillations and control features in glycolysis: Numerical analysis of a comprehensive model. Proc. Natl. Acad. Sci. USA 1981, 78, 2952–2956. [Google Scholar] [CrossRef] [PubMed]
- Termonia, Y.; Ross, J. Oscillations and control features in glycolysis: Analysis of resonance effects. Proc. Natl. Acad. Sci. USA 1981, 78, 3563–3566. [Google Scholar] [CrossRef]
- Stephanopoulos, G.; Simpson, T.W. Flux amplification in complex metabolic networks. Chem. Eng. Sci. 1997, 52, 2607–2627. [Google Scholar] [CrossRef]
- Carlsson, B.; Zambrano, J. Analysis of simple bioreactor models—A comparison between Monod and Contois kinetics. In Proceedings of the IWA Special International Conference: Activated Sludge—100 Years and Counting, Essen, Germany, 12–14 June 2014. [Google Scholar] [CrossRef]
- Calhoun, K.A.; Swartz, J.R. Total amino acid stabilization during cell-free protein synthesis reactions. J. Biotechnol. 2006, 123, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Noor, E.; Eden, E.; Milo, R.; Alon, U. Central Carbon Metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol. Cell 2010, 39, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Kreth, J.; Lengeler, J.W.; Jahreis, K. Characterization of Pyruvate Uptake in Escherichia coli K-12. PLoS ONE 2013, 8, E67125. [Google Scholar] [CrossRef] [PubMed]
- Ruby, E.G.; Nealson, K.H. Pyruvate production and excretion by the luminous marine bacteria. Appl. Environ. Microbiol. 1977, 34, 164–169. Available online: https://aem.asm.org (accessed on 4 May 2021). [CrossRef]
- Castro-Lopez, D.A.; Gonzalez de la Vara, L.E.; Santillán, M.; Martínez-Antonio, A. A molecular dynamic model of tryptophan overproduction in Escherichia coli. Fermentation 2022, 8, 560. [Google Scholar] [CrossRef]
- Bharat, A. Process Analytical Technology (PAT). Master’s Thesis, Padmashree Dr.Vithalrao Vikhe Patil Foundation, College of Pharmacy, Ahmed Nagar, India, 2013. Available online: https://www.slideshare.net/anjalibharat19/process-analytical-tchnology (accessed on 21 October 2021).
- Nagrath, D.; Avila-Elchiver, M.; Berthiaume, F.; Tilles, A.W.; Messac, A.; Yarmush, M.L. Soft constraints-based multiobjective framework for flux balance analysis. Metab Eng. 2010, 12, 429–445. [Google Scholar] [CrossRef]
- Rao, S.S. Engineering Optimization—Theory and Practice; Wiley: New York, NY, USA, 2009; Chapter 14.10. [Google Scholar]
- Dan, A.; Maria, G. Pareto optimal operating solutions for a semibatch reactor based on failure probability indices. Chem. Eng. Technol. 2012, 35, 1098–1103. [Google Scholar] [CrossRef]
- Yoon, Y.J.; Murthy, H.N.; Hahn, E.J.; Paek, K.Y. Biomass Production of Anoectochilus formosanus Hayata in a Bioreactor System. J. Plant Biol. 2007, 50, 573–576. [Google Scholar] [CrossRef]
- Buchmaier, J.; Brunner, C.; Griesbacher, U.; Phan, A.N.; Harvey, A.P.; Krishna Gudiminchi, R.; Nidetzky, B.; Mustera, B. Oscillatory flow bioreactor (OFB) applied in enzymatic hydrolysis at high solid loadings. Chem. Biochem. Eng. Q. 2019, 33, 459–470. [Google Scholar] [CrossRef]
- Maria, G. ARS combination with an evolutionary algorithm for solving MINLP optimization problems. In Modelling, Identification and Control; Hamza, M.H., Ed.; IASTED/ACTA Press: Anaheim, CA, USA, 2003; pp. 112–117. [Google Scholar]
- Maria, G.; Mihalachi, M.; Gijiu, C.L. In silico optimization of a bioreactor with an E. coli culture for tryptophan production by using a structured model coupling the oscillating glycolysis and tryptophan synthesis. Chem. Eng. Res. Des. 2018, 135, 207–221. [Google Scholar] [CrossRef]
- Kindler, O.; Pulkkinen, O.; Cherstvy, A.G.; Metzler, R. Burst statistics in an early biofilm quorum sensing model: The role of spatial colony-growth heterogeneity. Sci. Rep. 2019, 9, 12077. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
