Transforming Medical Data Access: The Role and Challenges of Recent Language Models in SQL Query Automation
Abstract
:1. Introduction
2. Literature Overview
2.1. Classical Approaches and Foundational Works in Text-to-SQL
2.2. Large Language Models (LLMs) in NLP
2.3. Advances in Text-to-SQL for Healthcare
2.4. Retrieval-Augmented Generation (RAG) Techniques
2.5. Challenges and Future Directions
3. Research Methodology
3.1. Dataset and Query Preparation
3.2. Language Models Under Test
3.3. Prompt Structure
- Job description: A clear and concise description of the role of the model, focusing on its task of converting user queries into SQL commands specifically designed for use in medical databases. This contextual setting helps the model align its outputs with the intended domain-specific requirements.
- Instructions: Detailed guidelines that outline the constraints and expectations for SQL generation. These included using SQLite-compatible functions, avoiding placeholders or overly complex expressions, and prioritizing simple, readable SQL solutions. Such explicit instructions reduce ambiguity and improve the relevance of the generated outputs.
- Schema information: A comprehensive database schema description, including table structures, field names, data types, and their purposes. This information equips the model with the necessary context to generate syntactically correct and semantically meaningful SQL queries.
- 1-shot example query: A single example demonstrating a natural language query and its corresponding SQL output. This example serves as a reference point to condition the model’s behavior and guides its interpretation of subsequent inputs, particularly in aligning its outputs with the expected format and style.
- Data preview: Sample rows from key tables in the database, providing additional context about the data stored within. This contextual data allows the model to understand the relationships between tables and fields better, improving its ability to generate precise and contextually appropriate queries.
3.4. Measurements
- Syntactic validity: The generated SQL query must be syntactically correct and executable in an SQLite environment without errors.
- Semantic correctness: The query must return results that match the expected output for the given natural language query.
- Alignment with constraints: The query must adhere to the specified constraints, such as avoiding placeholders or using SQLite-compatible functions.
- Efficiency: While not the primary criterion, the query should avoid unnecessary complexity and redundancy where possible.
- Input tokens: The number of tokens in the input prompt, including the query, schema information, instructions, and other contextual details.
- Output tokens: The number of tokens generated by the model as part of the SQL output.
- Pricing structure: The pricing structure specific to each model is typically defined as a cost per input and output token.
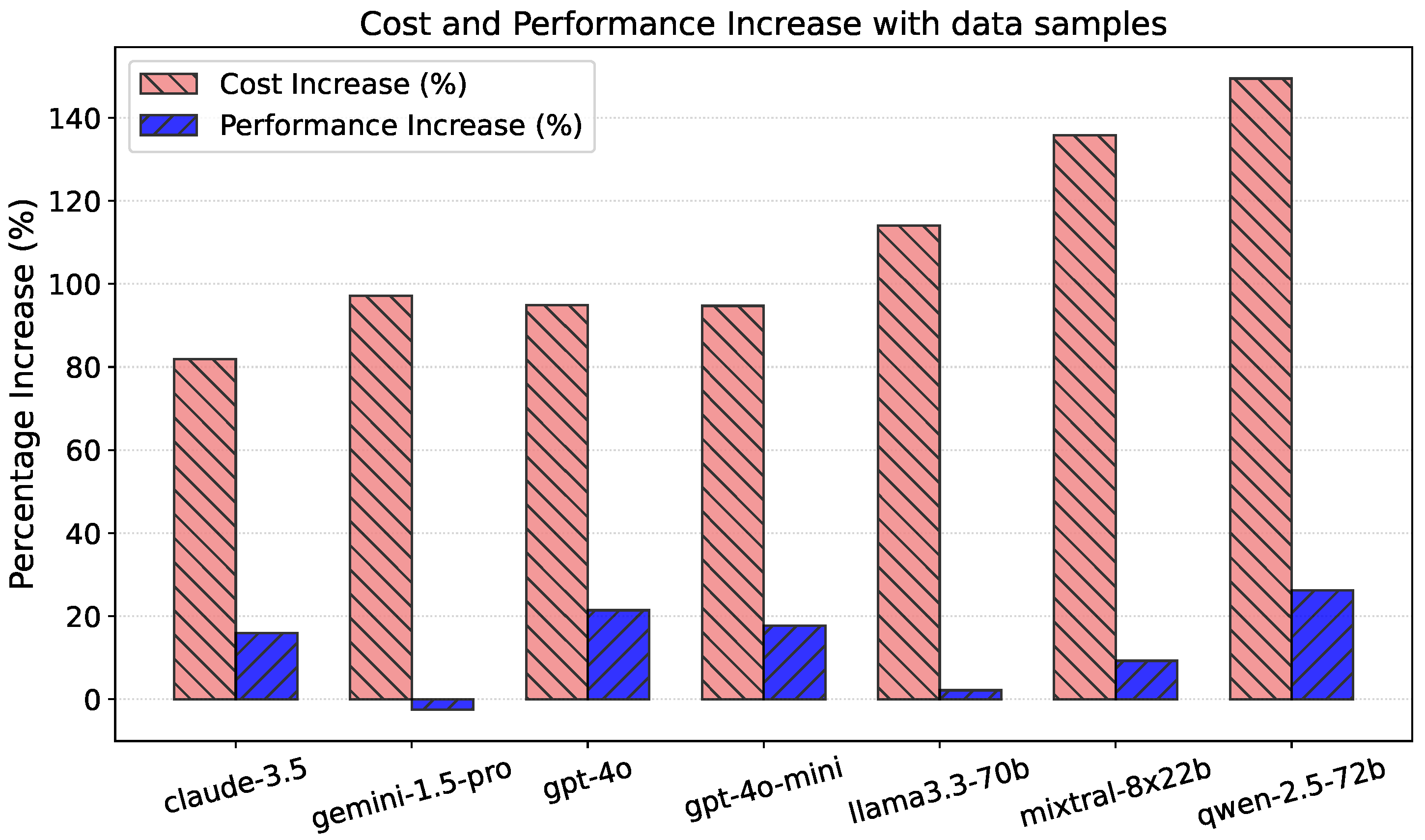
3.5. Ablation Study
- Both data preview and example question–answer are included.
- Only the data preview is included.
- Only the example question–answer is included.
- Neither the data preview nor the example question–answer is included.
4. Results
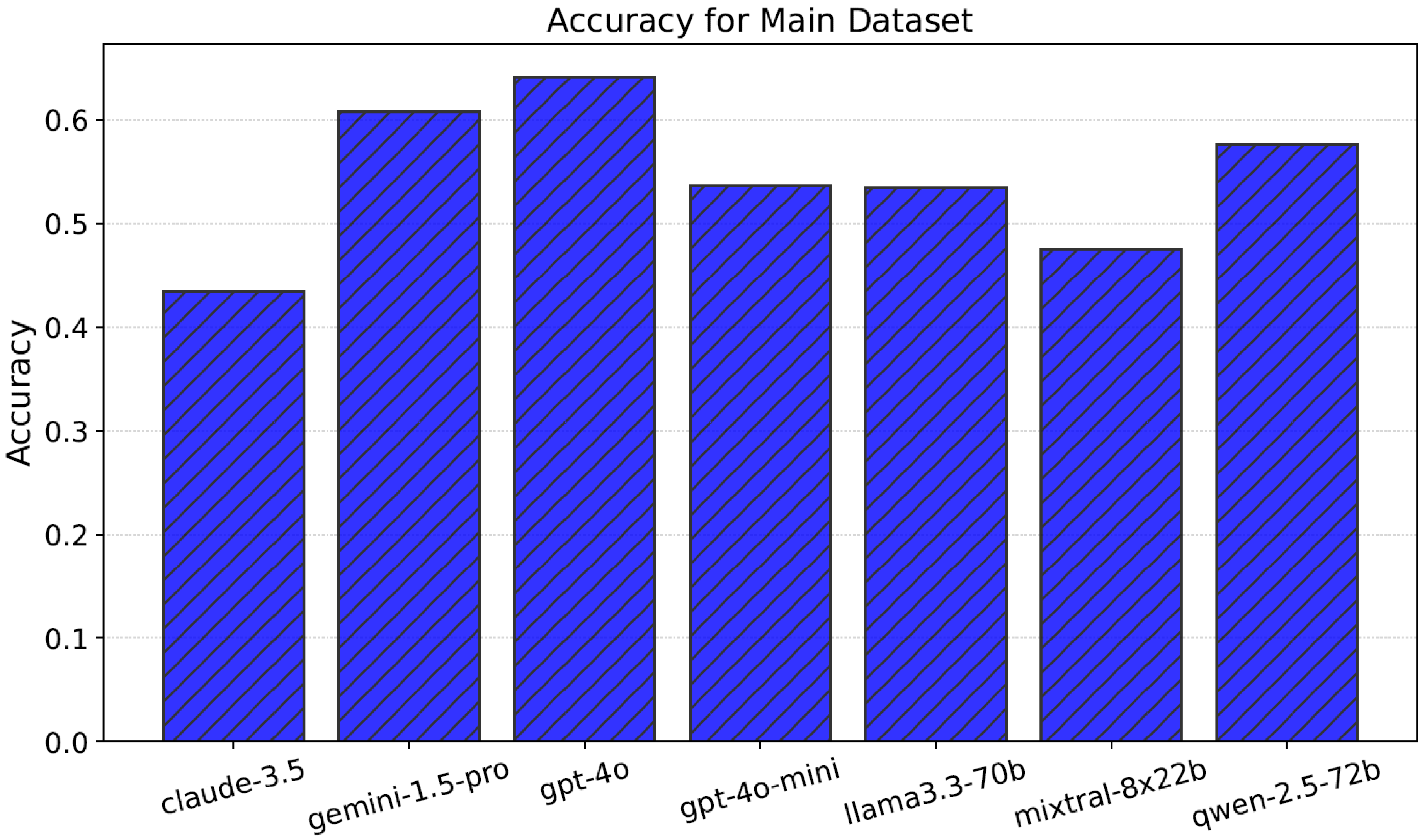
4.1. Performance and Efficiency
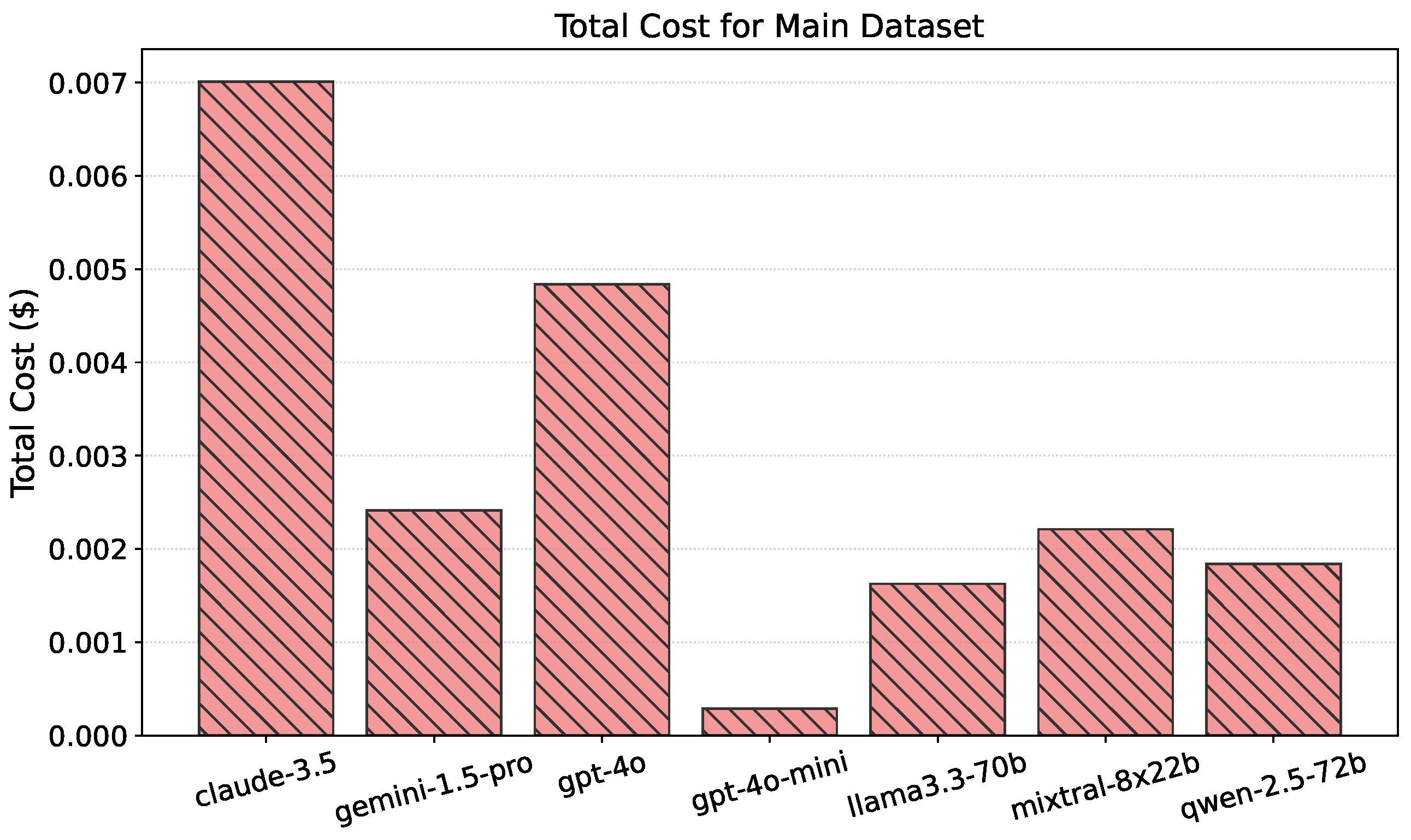
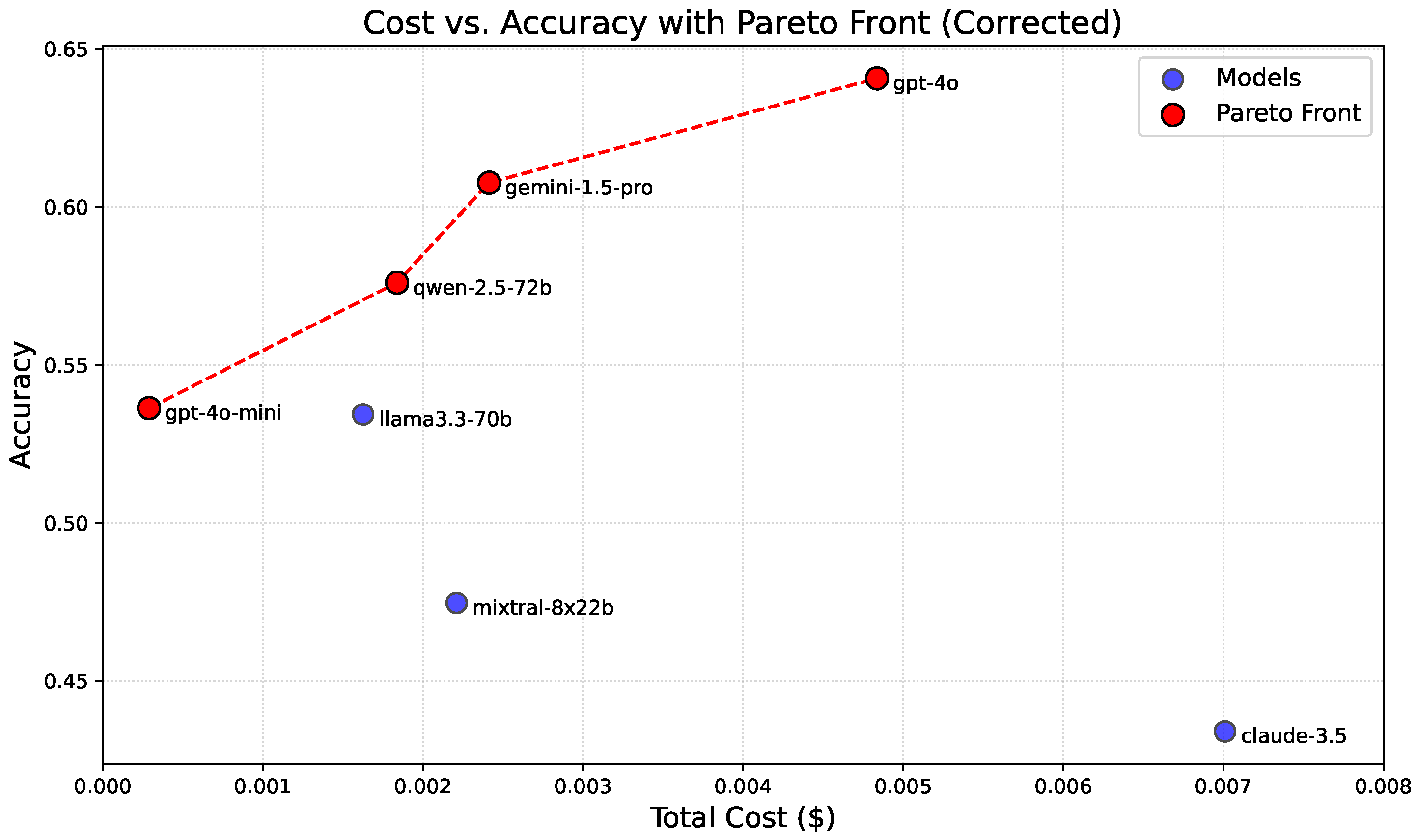
4.2. Cost-Effectiveness and Trade-Offs
4.3. Commercial vs. Open-Source Considerations
4.4. Recommendations
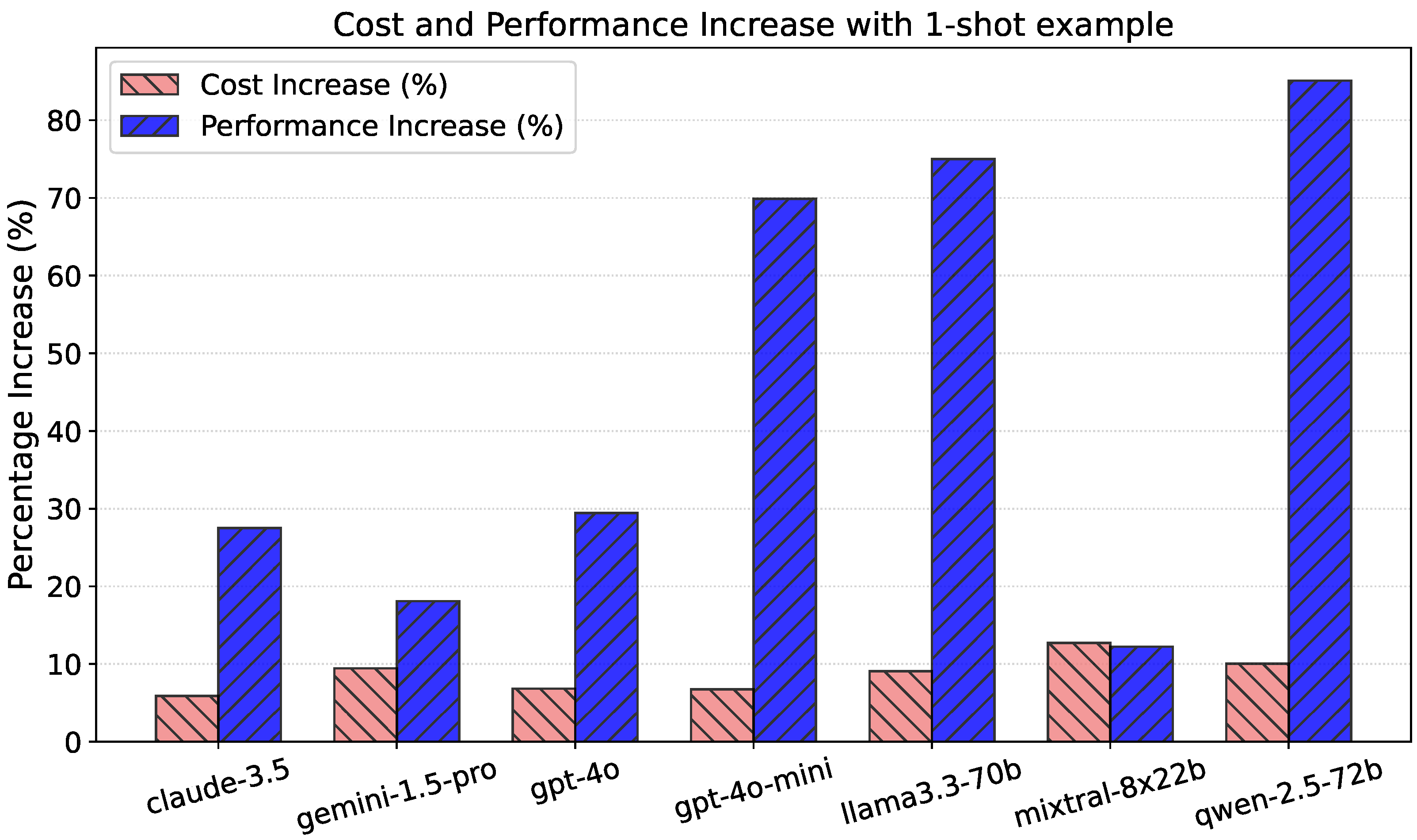
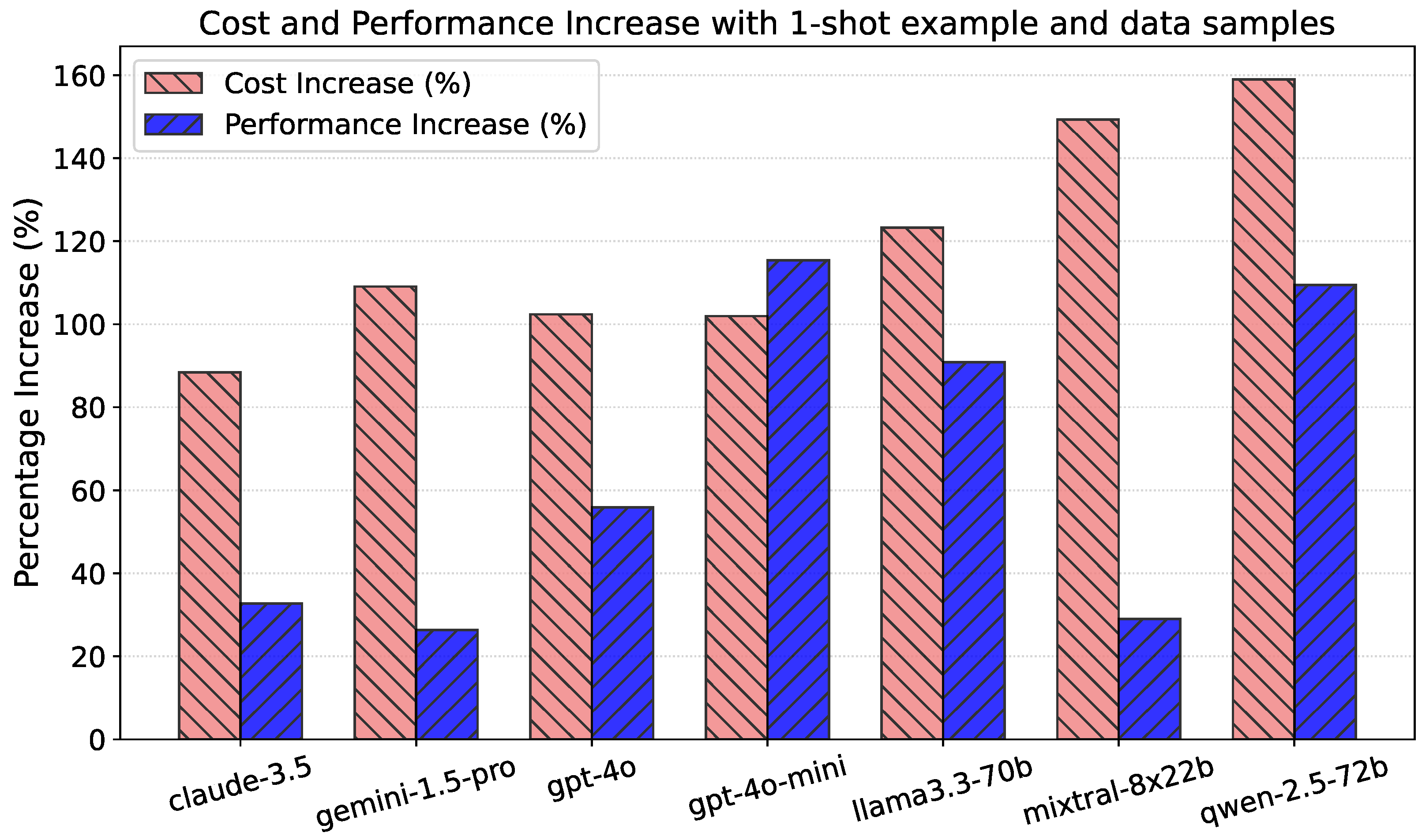
5. Prompt Structure Ablation Study
5.1. Table Data Samples in Prompt
5.2. One-Shot Examples in Prompt
5.3. Full Prompt with Both
5.4. Ablation Study Conclusions
6. Limitations and Future Directions
6.1. Challenges in Medical Name Resolution
6.2. Potential of Retrieval-Augmented Generation (RAG)
6.3. Future Directions
6.4. Threats to Validity
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Prompt Structure
- Generate accurate SQL queries aligned with the user’s intent.
- Return SQL statements without explanations or comments.
- Prefix all selected or counted columns with the table name and wrap them in double quotes.
- Do not use placeholders (e.g., {{table_name}}, {{column_name}}).
- Wrap SQL code in “‘sql blocks.
- Assume an SQLite database and avoid functions unavailable there.
- Prefer simple solutions with minimal function calls or redundant boolean operations.
- Do not use table aliases.
- When matching strings, match them in lowercase using LOWER().
Appendix A.1. Section [Example] for Ablation Study
Appendix A.2. Section [Schema] for Ablation Study
Appendix A.3. Section [Data_Preview] for Ablation Study
Appendix B. Examples of Prompted SQL Queries
Appendix B.1. Case Number: ac54a82fadd7f1ed60b5f097dcccc668
- Llama3.3-70b:SELECT DEMOGRAPHIC."DAYS_STAY", LAB."CATEGORY"FROM DEMOGRAPHICINNER JOIN LAB ON DEMOGRAPHIC."SUBJECT_ID" = LAB."SUBJECT_ID"WHERE DEMOGRAPHIC."SUBJECT_ID" = 18372
- Mixtral-8x22b:SELECT DEMOGRAPHIC."DAYS_STAY", LAB."CATEGORY"FROM DEMOGRAPHICINNER JOIN LAB ON DEMOGRAPHIC."SUBJECT_ID" = LAB."SUBJECT_ID"WHERE DEMOGRAPHIC."SUBJECT_ID" = 18372
- Gemini 1.5-pro:SELECT DEMOGRAPHIC."DAYS_STAY", LAB."CATEGORY"FROM DEMOGRAPHICINNER JOIN LAB ON DEMOGRAPHIC."SUBJECT_ID" = LAB."SUBJECT_ID"WHERE DEMOGRAPHIC."SUBJECT_ID" = 18372
- Qwen-2.5-72B:SELECT "demographic"."DAYS_STAY", "lab"."CATEGORY"FROM "demographic"INNER JOIN "lab" ON "demographic"."HADM_ID" = "lab"."HADM_ID"WHERE "demographic"."SUBJECT_ID" = 18372
- Claude-3.5:SELECT DEMOGRAPHIC."DAYS_STAY", LAB."CATEGORY"FROM DEMOGRAPHICLEFT JOIN LAB ON DEMOGRAPHIC."SUBJECT_ID" = LAB."SUBJECT_ID"WHERE DEMOGRAPHIC."SUBJECT_ID" = 18372GROUP BY DEMOGRAPHIC."DAYS_STAY", LAB."CATEGORY"
- GPT-4o:SELECT DEMOGRAPHIC."DAYS_STAY", LAB."CATEGORY"FROM DEMOGRAPHICINNER JOIN LAB ON DEMOGRAPHIC.HADM_ID = LAB.HADM_IDWHERE DEMOGRAPHIC."SUBJECT_ID" = 18372
- GPT-4o-mini:SELECT DEMOGRAPHIC."DAYS_STAY", LAB."CATEGORY"FROM DEMOGRAPHICLEFT JOIN LAB ON DEMOGRAPHIC."HADM_ID" = LAB."HADM_ID"WHERE DEMOGRAPHIC."SUBJECT_ID" = 18372
Appendix B.2. Case Number: a90ecf2fa6bc7c4af1d8fc44eb09a881
- Llama3.3-70b:SELECT COUNT(DISTINCT DEMOGRAPHIC."SUBJECT_ID")FROM DEMOGRAPHICINNER JOIN DIAGNOSES ON DEMOGRAPHIC."HADM_ID" = DIAGNOSES."HADM_ID"WHERE LOWER(DEMOGRAPHIC."MARITAL_STATUS") = ’married’AND LOWER(DIAGNOSES."SHORT_TITLE") = ’pyelonephritis, unspecified’
- Mixtral-8x22b:SELECT COUNT(DISTINCT DEMOGRAPHIC."SUBJECT_ID")FROM DEMOGRAPHICINNER JOIN DIAGNOSES ON DEMOGRAPHIC."SUBJECT_ID" = DIAGNOSES."SUBJECT_ID"WHERE LOWER(DEMOGRAPHIC."MARITAL_STATUS") = ’married’AND LOWER(DIAGNOSES."SHORT_TITLE") = ’pyelonephritis, unspecified’
- Gemini 1.5-pro:SELECT COUNT(DISTINCT DEMOGRAPHIC."SUBJECT_ID")FROM DEMOGRAPHICINNER JOIN DIAGNOSES ON DEMOGRAPHIC.HADM_ID = DIAGNOSES.HADM_IDWHERE LOWER(DEMOGRAPHIC."MARITAL_STATUS") = ’married’AND LOWER(DIAGNOSES."SHORT_TITLE") = ’pyelonephritis, unspecified’
- Qwen-2.5-72B:SELECT COUNT(DISTINCT "demographic"."SUBJECT_ID")FROM "demographic"INNER JOIN "diagnoses" ON "demographic"."HADM_ID" = "diagnoses"."HADM_ID"WHERE LOWER("demographic"."MARITAL_STATUS") = ’married’AND LOWER("diagnoses"."SHORT_TITLE") = ’pyelonephritis, unspecified’
- Claude-3.5:SELECT COUNT(DISTINCT DEMOGRAPHIC."SUBJECT_ID")FROM DEMOGRAPHICINNER JOIN DIAGNOSES ON DEMOGRAPHIC."HADM_ID" = DIAGNOSES."HADM_ID"WHERE LOWER(DEMOGRAPHIC."MARITAL_STATUS") = ’married’AND LOWER(DIAGNOSES."SHORT_TITLE") = ’pyelonephritis nos’
- GPT-4o:SELECT COUNT(DISTINCT DEMOGRAPHIC."SUBJECT_ID")FROM DEMOGRAPHICINNER JOIN DIAGNOSES ON DEMOGRAPHIC."HADM_ID" = DIAGNOSES."HADM_ID"WHERE LOWER(DEMOGRAPHIC."MARITAL_STATUS") = ’married’AND LOWER(DIAGNOSES."SHORT_TITLE") = ’pyelonephritis, unspecified’
- GPT-4o-mini:SELECT COUNT ( DISTINCT DEMOGRAPHIC."SUBJECT_ID" )FROM DEMOGRAPHICINNER JOIN DIAGNOSES ON DEMOGRAPHIC.HADM_ID = DIAGNOSES.HADM_IDWHERE DEMOGRAPHIC."MARITAL_STATUS" = "MARRIED"AND LOWER(DIAGNOSES."SHORT_TITLE") = "pyelonephritis, unspecified"
References
- Blumenthal, D. Stimulating the adoption of health information technology. N. Engl. J. Med. 2009, 360, 1477–1479. [Google Scholar] [CrossRef] [PubMed]
- Jetley, G.; Zhang, H. Electronic health records in IS research: Quality issues, essential thresholds and remedial actions. Decis. Support Syst. 2019, 126, 113137. [Google Scholar] [CrossRef]
- Huang, K.; Altosaar, J.; Ranganath, R. ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission. arXiv 2020, arXiv:1904.05342. [Google Scholar]
- Alsentzer, E.; Murphy, J.R.; Boag, W.; Weng, W.H.; Jin, D.; Naumann, T.; McDermott, M.B.A. Publicly Available Clinical BERT Embeddings. arXiv 2019, arXiv:1904.03323. [Google Scholar]
- Singhal, K.; Azizi, S.; Tu, T.; Mahdavi, S.S.; Wei, J.; Chung, H.W.; Scales, N.; Tanwani, A.; Cole-Lewis, H.; Pfohl, S.; et al. Large language models encode clinical knowledge. Nature 2023, 620, 172–180. [Google Scholar] [CrossRef]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language models are few-shot learners. arXiv 2020, arXiv:2005.14165. [Google Scholar]
- Ayaz, M.; Pasha, M.F.; Alzahrani, M.Y.; Budiarto, R.; Stiawan, D. The Fast Health Interoperability Resources (FHIR) Standard: Systematic Literature Review of Implementations, Applications, Challenges and Opportunities. JMIR Med. Inform. 2021, 9, e21929, Erratum in JMIR Med. Inform. 2021, 9, e32869. [Google Scholar] [CrossRef]
- Engelhardt, M. Hitching Healthcare to the Chain: An Introduction to Blockchain Technology in the Healthcare Sector. Technol. Innov. Manag. Rev. 2017, 7, 22–34. [Google Scholar] [CrossRef]
- European Union. General Data Protection Regulation (GDPR); European Union: Brussels, Belgium, 2018.
- Shanafelt, T.D.; Dyrbye, L.N.; Sinsky, C.; Hasan, O.; Satele, D.; Sloan, J.; West, C.P. Relationship Between Clerical Burden and Characteristics of the Electronic Environment With Physician Burnout and Professional Satisfaction. Mayo Clin. Proc. 2016, 91, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Frost, M.J.; Tran, J.B.; Khatun, F.; Friberg, I.K.; Rodríguez, D.C. What Does It Take to Be an Effective National Steward of Digital Health Integration for Health Systems Strengthening in Low- and Middle-Income Countries? Glob. Health Sci. Pract. 2018, 6, S18–S28. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Deng, H.; Liu, B.; Hu, A.; Liang, J.; Fan, L.; Zheng, X.; Wang, T.; Lei, J. Systematic Evaluation of Research Progress on Natural Language Processing in Medicine Over the Past 20 Years: Bibliometric Study on PubMed. J. Med. Internet Res. 2020, 22, e16816. [Google Scholar] [CrossRef] [PubMed]
- Au Yeung, J.; Shek, A.; Searle, T.; Kraljevic, Z.; Dinu, V.; Ratas, M.; Al-Agil, M.; Foy, A.; Rafferty, B.; Oliynyk, V.; et al. Natural language processing data services for healthcare providers. BMC Med. Inform. Decis. Mak. 2024, 24, 356. [Google Scholar] [CrossRef] [PubMed]
- Marshan, A.; AlMutairi, A.N.; Ioannou, A.; Bell, D.; Monaghan, A.; Arzoky, M. MedT5SQL: A transformers-based large language model for text-to-SQL conversion in the healthcare domain. Front. Big Data 2024, 7, 1371680. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, Y.; Song, Y.; Wei, V.J.; Tian, Y.; Qi, Y.; Chan, J.H.; Wong, R.C.W.; Yang, H. Natural Language Interfaces for Tabular Data Querying and Visualization: A Survey. IEEE Trans. Knowl. Data Eng. 2023, 36, 6699–6718. [Google Scholar] [CrossRef]
- Li, F.; Jagadish, H.V. Constructing an Interactive Natural Language Interface for Relational Databases. Proc. VLDB Endow. 2014, 8, 73–84. [Google Scholar] [CrossRef]
- Katsogiannis-Meimarakis, G.; Koutrika, G. A survey on deep learning approaches for text-to-SQL. VLDB J. 2023, 32, 905–936. [Google Scholar] [CrossRef]
- Iacob, R.; Brad, F.; Apostol, E.; Truică, C.O.; Hosu, I.A.; Rebedea, T. Neural Approaches for Natural Language Interfaces to Databases: A Survey. In Proceedings of the 28th International Conference on Computational Linguistics, Barcelona, Spain, 8–13 December 2020. [Google Scholar] [CrossRef]
- Qin, B.; Hui, B.; Wang, L.; Yang, M.; Li, J.; Li, B.; Geng, R.; Cao, R.; Sun, J.; Si, L.; et al. A Survey on Text-to-SQL Parsing: Concepts, Methods, and Future Directions. arXiv 2022, arXiv:2208.13629. [Google Scholar] [CrossRef]
- Jin, Y.; Chandra, M.; Verma, G.; Hu, Y.; De Choudhury, M.; Kumar, S. Better to Ask in English: Cross-Lingual Evaluation of Large Language Models for Healthcare Queries. In Proceedings of the ACM Web Conference 2024, Singapore, 13–17 May 2024; Association for Computing Machinery: New York, NY, USA; pp. 2627–2638. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, Q.; Cui, L.; Liu, Y. Large Language Model Enhanced Text-to-SQL Generation: A Survey. arXiv 2024, arXiv:2410.06011. [Google Scholar]
- Liu, X.; Shen, S.; Li, B.; Ma, P.; Jiang, R.; Zhang, Y.; Fan, J.; Li, G.; Tang, N.; Luo, Y. A Survey of NL2SQL with Large Language Models: Where are we, and where are we going? arXiv 2024, arXiv:2408.05109. [Google Scholar]
- Hong, Z.; Yuan, Z.; Zhang, Q.; Chen, H.; Dong, J.; Huang, F.; Huang, X. Next-Generation Database Interfaces: A Survey of LLM-based Text-to-SQL. arXiv 2024, arXiv:2406.08426. [Google Scholar]
- Yang, R.; Ning, Y.; Keppo, E.; Liu, M.; Hong, C.; Bitterman, D.S.; Ong, J.C.L.; Ting, D.S.W.; Liu, N. Retrieval-augmented generation for generative artificial intelligence in health care. npj Health Syst. 2025, 2, 2. [Google Scholar] [CrossRef]
- Yang, E.; Amar, J.; Lee, J.H.; Kumar, B.; Jia, Y. The Geometry of Queries: Query-Based Innovations in Retrieval-Augmented Generation. arXiv 2024, arXiv:2407.18044. [Google Scholar]
- Holzinger, A.; Langs, G.; Denk, D.; Zatloukal, K.; Müller, H. Causability and Explainability of AI in Medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. Adv. Neural. Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- Meta-AI. LLaMA 3.3-70B. 2024. Available online: https://www.llama.com/ (accessed on 11 December 2024).
- Mistral AI. Mixtral 8x22B: Mixture-of-Experts Model. 2024. Available online: https://mistral.ai/news/mixtral-8x22b (accessed on 11 December 2024).
- Google DeepMind. Gemini Pro. 2024. Available online: https://deepmind.google/technologies/gemini/pro/ (accessed on 11 December 2024).
- Anthropic. Claude 3.5. 2024. Available online: https://www.anthropic.com/news/claude-3-5-sonnet (accessed on 11 December 2024).
- OpenAI. GPT-4o. 2024. Available online: https://openai.com/index/hello-gpt-4o/ (accessed on 11 December 2024).
- OpenAI. GPT-4o-Mini: Advancing Cost-Efficient Intelligence. 2024. Available online: https://openai.com/index/gpt-4o-mini-advancing-cost-efficient-intelligence/ (accessed on 11 December 2024).
- QwenLM. Qwen2.5. 2024. Available online: https://github.com/QwenLM/Qwen2.5 (accessed on 11 December 2024).
| Natural Language Question | SQL Query |
|---|---|
| What are the details of male patients admitted in 2020? | SELECT * FROM demographics WHERE gender=’M’ AND admission_year=2020; |
| List all laboratory tests with abnormal results for patient ID 12345. | SELECT test_name, result FROM lab_tests WHERE patient_id=12345 AND abnormal=1; |
| Show prescriptions of antibiotics for patients under 18 years old. | SELECT * FROM prescriptions WHERE drug_type=’antibiotic’ AND age<18; |
| Retrieve all diagnoses for patients who underwent surgery in June 2021. | SELECT diagnoses FROM procedures JOIN diagnoses ON procedures.patient_id = diagnoses.patient_id WHERE procedures.date BETWEEN "1 June 2021" AND "30 June 2021" AND procedures.type=’surgery’; |
| Model (Commercial) | Parameter Count | Architecture and Key Features |
|---|---|---|
| LLaMA 3.3-70B (No) | 70 Billion | Transformer decoder; open-source, highly tunable, efficient token usage for large datasets. |
| Mixtral 8x22B (No) | 141 Billion (Ensemble) | Ensemble transformer; designed for scalability, optimized for distributed tasks with multiple smaller models. |
| Gemini-1.5 (Yes) | 1.5 Trillion | Transformer decoder; lightweight and cost-effective, ideal for minimal token usage tasks. |
| Claude 3.5 (Yes) | Proprietary | Transformer decoder; human-aligned, optimized for conversational tasks and intent recognition. |
| GPT-4o (Yes) | Proprietary | Transformer decoder; high accuracy, general-purpose capabilities, suitable for premium use cases. |
| GPT-4o-Mini (Yes) | Proprietary | Transformer decoder; compact design, tuned for budget-sensitive tasks while maintaining strong performance. |
| Qwen-2.5-72B (Yes) | 72 Billion | Transformer decoder; specialized expert models for coding and math; structured data handling. |
| Model | Accuracy (%) | Average Tokens In | Average Tokens Out |
|---|---|---|---|
| Claude-3.5 | 43.4 ± 0.1 | 1986.052 | 70.085 |
| Gemini-1.5-pro | 60.8 ± 0.4 | 1716.036 | 53.653 |
| GPT-4o | 64.1 ± 0.6 | 1710.875 | 55.880 |
| GPT-4o-mini | 53.6 ± 0.3 | 1710.875 | 55.155 |
| Llama3.3-70b | 53.4 ± 0.2 | 1747.020 | 60.461 |
| Mixtral-8x22b | 47.5 ± 0.3 | 2365.230 | 91.056 |
| Qwen-2.5-72b | 57.6 ± 0.0 | 1981.154 | 61.747 |
| Model | Price/1M Input Tokens ($) | Price/1M Output Tokens ($) | Price per Query ($) |
|---|---|---|---|
| Claude-3.5 | 3.00 | 15.00 | 0.00701 |
| Gemini-1.5-pro | 1.25 | 5.00 | 0.00241 |
| GPT-4o | 2.50 | 10.00 | 0.00484 |
| GPT-4o-mini | 0.15 | 0.60 | 0.00029 |
| Llama3.3-70b | 0.90 | 0.90 | 0.00163 |
| Mixtral-8x22b | 0.90 | 0.90 | 0.00221 |
| Qwen-2.5-72b | 0.90 | 0.90 | 0.00184 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanković, N.; Šajina, R.; Lorencin, I. Transforming Medical Data Access: The Role and Challenges of Recent Language Models in SQL Query Automation. Algorithms 2025, 18, 124. https://doi.org/10.3390/a18030124
Tanković N, Šajina R, Lorencin I. Transforming Medical Data Access: The Role and Challenges of Recent Language Models in SQL Query Automation. Algorithms. 2025; 18(3):124. https://doi.org/10.3390/a18030124
Chicago/Turabian StyleTanković, Nikola, Robert Šajina, and Ivan Lorencin. 2025. "Transforming Medical Data Access: The Role and Challenges of Recent Language Models in SQL Query Automation" Algorithms 18, no. 3: 124. https://doi.org/10.3390/a18030124
APA StyleTanković, N., Šajina, R., & Lorencin, I. (2025). Transforming Medical Data Access: The Role and Challenges of Recent Language Models in SQL Query Automation. Algorithms, 18(3), 124. https://doi.org/10.3390/a18030124






