The Variation Driven by Differences between Species and between Sites in Allometric Biomass Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biomass Data
2.2. Methods
2.2.1. The Rationale for the Proposed Methodology
2.2.2. VPC of nested ANOVA
- (a)
- A multilevel model (random intercept and slope model) was fitted to log-log transformed data (for each dataset),where ln(AGB)ijk is the log of AGB for tree i from site j and species k; β0 is the fixed part of the intercept; β1 is the fixed part of the ln(D) slope; β2 is the fixed part of the ln(H) slope; δj0 is the random part of the intercept attributable to differences between sites; δj1 is the random part of the ln(D) slope attributable to differences between sites; δj2 is the random part of the ln(H) slope attributable to differences between sites; γk0 is the random part of the intercept attributable to differences between species; γk1 is the random part of the ln(D) slope attributable to differences species; γk2 is the random part of the ln(H) slope attributable to the differences between species; εijk is the error term of tree i from site j and species k, εijk~N(0,σ2).
- (b)
- A back-transformed nonlinear model was used to calculate the predicted AGB. As the error distribution becomes lognormal in original scale, a back transformation correction factor (exp(σ2/2)) [41,42] was used, where σ is the residual standard error of the model in log-log scale. The back-transformed model that predicts AGB as a function of D and H is:where β0 is the fixed part of the intercept (Equation (1)); β1 is the fixed part of ln(D) slope (Equation (1)); β2 is the fixed part of ln(H) slope (Equation (1)); D and H are the diameter at breast height and tree height; σ2 is the residual variance in Equation (1).
- (c)
- The relative residual for each tree was calculated as,where AGBijk is the observed AGB of tree i, from site j and species k; is the predicted AGB of tree i, from site j and species k (Equation (2)).
2.2.3. Bootstrap Analysis
2.2.4. The effect of Including H as Predictor in Allometric Biomass Models
2.2.5. Analysis of Random Effects
2.2.6. Data Processing
3. Results
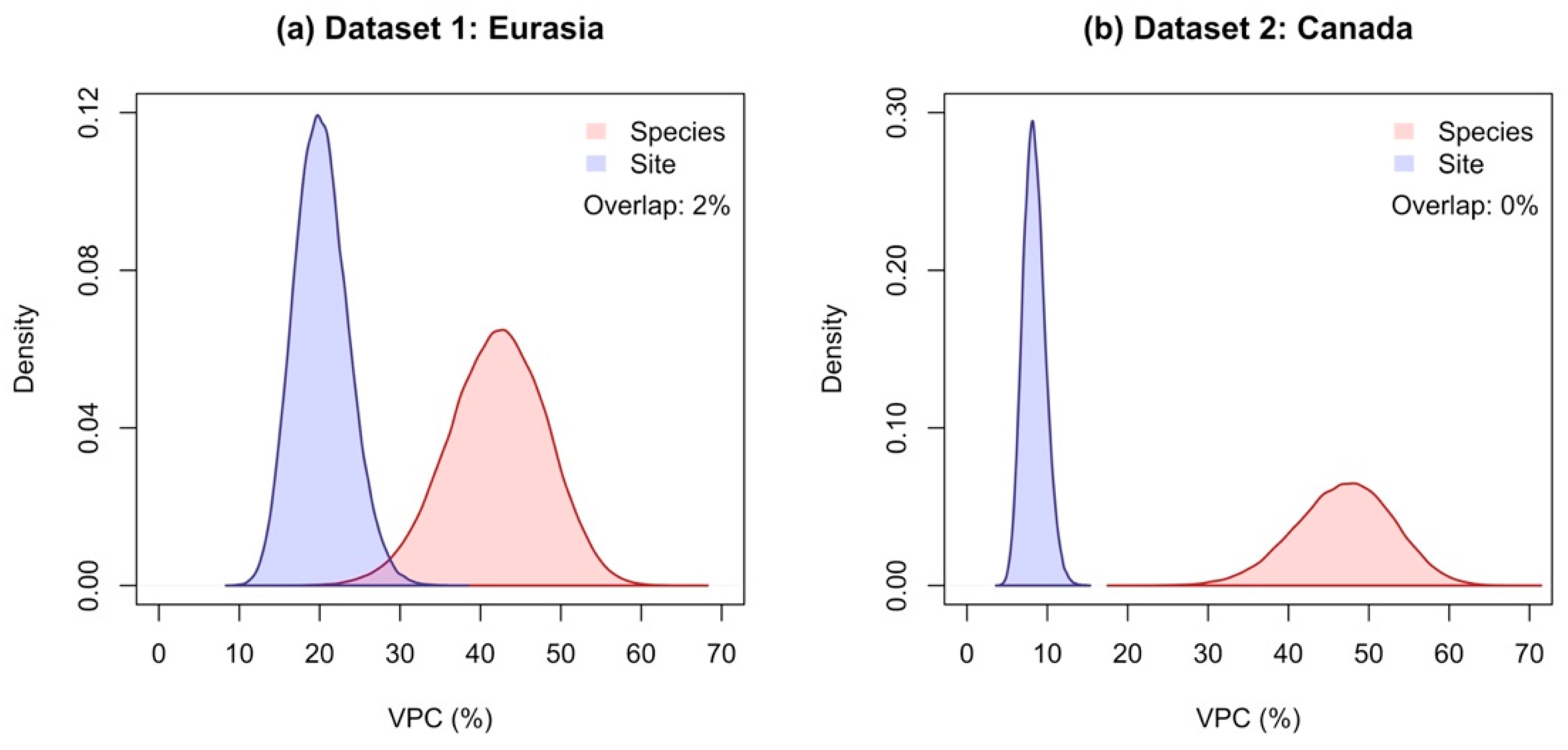
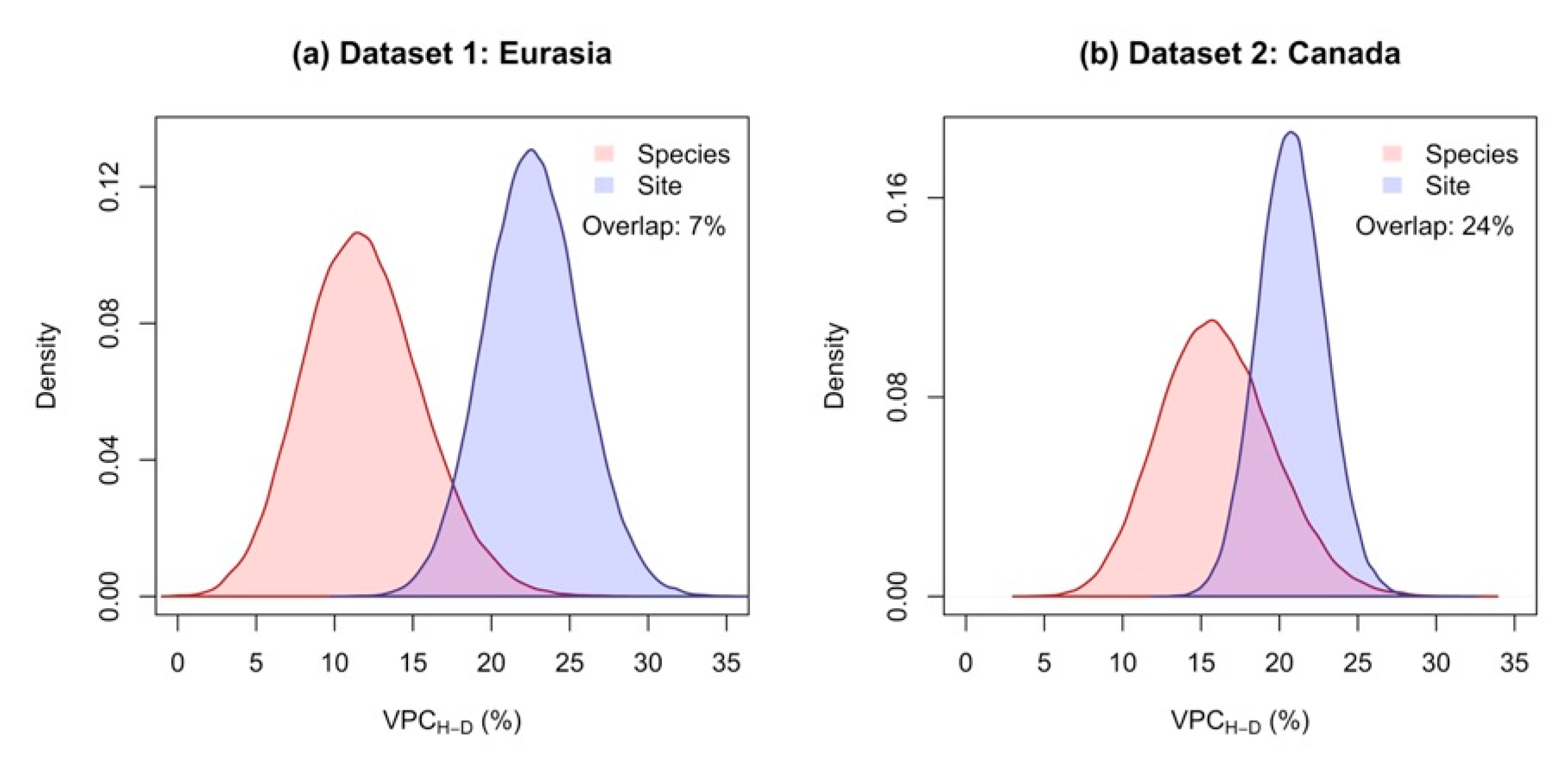
3.1. The Species Explained a Larger Proportion of Variance in Allometric Models Compared to Sites
3.2. Including H as Predictor in Allometric Models Reduced The Proportion of Variance Attributable to Differences between Sites, but Had Marginal Effect on That Attributable to Differences between Species
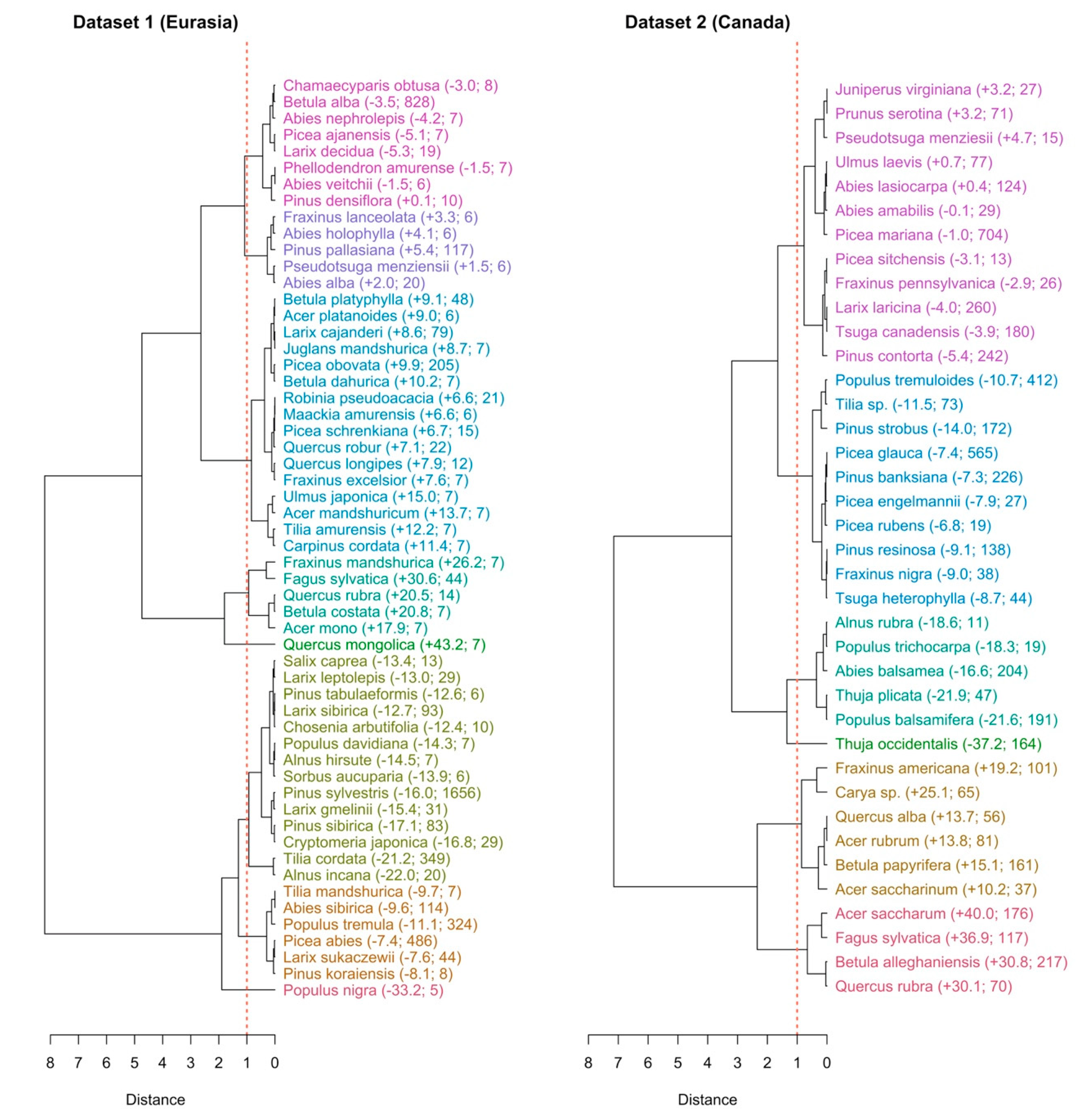
3.3. The Species and Site Random Effects
4. Discussion
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Chave, J.; Andalo, A.C.; Brown, A.S.; Cairns, A.M.A.; Chambers, J.Q.; Eamus, A.D.; Foïster, A.H.; Fromard, A.F.; Higuchi, N.; Kira, A.T.; et al. Tree allometry and improved estimation of carbon stocks and balance in tropical forests. Oecologia 2005, 145, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Chave, J.; Réjou-Méchain, M.; Búrquez, A.; Chidumayo, E.; Colgan, M.S.; Delitti, W.B.C.; Duque, A.; Eid, T.; Fearnside, P.M.; Goodman, R.C.; et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Chang. Biol. 2014, 20, 3177–3190. [Google Scholar] [CrossRef] [PubMed]
- Picard, N.; Saint-André, L.; Henry, M. Manual for Building Tree Volume and Biomass Allometric Equations: From Field Measurement to Prediction; FAO: Rome, Italy; CIRAD: Montpellier, France, 2012. [Google Scholar]
- Huxley, S.J. Problems of Relative Growth, 1st ed.; The Dial Press: New York, NY, USA, 1932. [Google Scholar]
- West, G.B.; Brown, J.H.; Enquist, B.J. A General Model for the Origin of Allometric Scaling Laws in Biology. Science 1997, 276, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Enquist, B.J.; West, G.B.; Charnov, E.L.; Brown, J.H. Allometric scaling of production and life-history variation in vascular plants. Nature 1999, 401, 907–911. [Google Scholar] [CrossRef]
- Pretzsch, H. Species-specific allometric scaling under self-thinning: Evidence from long-term plots in forest stands. Oecologia 2006, 146, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Zianis, D.; Radoglou, K. Comparison between empirical and theoretical biomass allometric models and statistical implications for stem volume predictions. Forestry 2006, 79, 477–487. [Google Scholar] [CrossRef] [Green Version]
- Wang, C. Biomass allometric equations for 10 co-occurring tree species in Chinese temperate forests. For. Ecol. Manag. 2006, 222, 9–16. [Google Scholar] [CrossRef]
- Ducey, M.J. Evergreenness and wood density predict height–diameter scaling in trees of the northeastern United States. For. Ecol. Manag. 2012, 279, 21–26. [Google Scholar] [CrossRef]
- Montagu, K.D.; Düttmer, K.; Barton, C.V.M.; Cowie, A.L. Developing general allometric relationships for regional estimates of carbon sequestration—an example using Eucalyptus pilularis from seven contrasting sites. For. Ecol. Manag. 2005, 204, 115–129. [Google Scholar] [CrossRef]
- Segura, V.; Cilas, C.; Costes, E. Dissecting apple tree architecture into genetic, ontogenetic and environmental effects: Mixed linear modelling of repeated spatial and temporal measures. New Phytol. 2008, 178, 302–314. [Google Scholar] [CrossRef]
- Fukatsu, E.; Nakada, R. The timing of latewood formation determines the genetic variation of wood density in Larix kaempferi. Trees 2018, 32, 1233–1245. [Google Scholar] [CrossRef]
- Barthélémy, D.; Caraglio, Y. Plant Architecture: A Dynamic, Multilevel and Comprehensive Approach to Plant Form, Structure and Ontogeny. Ann. Bot. 2007, 99, 375–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urban, J.; Holušová, K.; Menšík, L.; Čermák, J.; Kantor, P. Tree allometry of Douglas fir and Norway spruce on a nutrient-poor and a nutrient-rich site. Trees 2013, 27, 97–110. [Google Scholar] [CrossRef]
- Archibald, S.; Bond, W.J. Growing tall vs. growing wide: Tree architecture and allometry of Acacia karroo in forest, savanna, and arid environments. Oikos 2003, 102, 3–14. [Google Scholar] [CrossRef]
- Dutcă, I.; Negruţiu, F.; Ioraş, F.; Maher, K.; Blujdea, V.N.B.; Ciuvăţ, L.A.; Negrutiu, F.; Ioras, F.; Maher, K.; Blujdea, V.N.B.; et al. The influence of age, location and soil conditions on the allometry of young Norway spruce ( Picea abies L. Karst.) trees. Not. Bot. Horti Agrobot. Cluj-Napoca 2014, 42, 579–582. [Google Scholar] [CrossRef]
- Poorter, H.; Niklas, K.J.; Reich, P.B.; Oleksyn, J.; Poot, P.; Mommer, L. Biomass allocation to leaves, stems and roots: Meta-analyses of interspecific variation and environmental control. New Phytol. 2012, 193, 30–50. [Google Scholar] [CrossRef]
- Delucia, E.H.; Maherali, H.; Carey, E.V. Climate-driven changes in biomass allocation in pines. Glob. Chang. Biol. 2000, 6, 587–593. [Google Scholar] [CrossRef]
- António, N.; Tomé, M.; Tomé, J.; Soares, P.; Fontes, L. Effect of tree, stand, and site variables on the allometry of Eucalyptus globulus tree biomass. Can. J. For. Res. 2007, 37, 895–906. [Google Scholar] [CrossRef]
- Yang, B.; Xue, W.; Yu, S.; Zhou, J.; Zhang, W. Effects of Stand Age on Biomass Allocation and Allometry of Quercus Acutissima in the Central Loess Plateau of China. Forests 2019, 10, 41. [Google Scholar] [CrossRef]
- Lines, E.R.; Zavala, M.A.; Purves, D.W.; Coomes, D.A. Predictable changes in aboveground allometry of trees along gradients of temperature, aridity and competition. Glob. Ecol. Biogeogr. 2012, 21, 1017–1028. [Google Scholar] [CrossRef]
- Naidu, S.L.; DeLucia, E.H.; Thomas, R.B. Contrasting patterns of biomass allocation in dominant and suppressed loblolly pine. Can. J. For. Res. 1998, 28, 1116–1124. [Google Scholar] [CrossRef]
- Dutcă, I.; Mather, R.; Ioraş, F. Tree biomass allometry during the early growth of Norway spruce (Picea abies) varies between pure stands and mixtures with European beech (Fagus sylvatica). Can. J. For. Res. 2018, 48, 77–84. [Google Scholar] [CrossRef]
- Messier, C.; Mitchell, A.K. Effects of thinning in a 43-year-old Douglas-fir stand on above- and below-ground biomass allocation and leaf structure of understory Gaultheria shallon. For. Ecol. Manag. 1994, 68, 263–271. [Google Scholar] [CrossRef]
- Fayolle, A.; Doucet, J.-L.; Gillet, J.-F.; Bourland, N.; Lejeune, P. Tree allometry in Central Africa: Testing the validity of pantropical multi-species allometric equations for estimating biomass and carbon stocks. For. Ecol. Manag. 2013, 305, 29–37. [Google Scholar] [CrossRef]
- Ngomanda, A.; Engone Obiang, N.L.; Lebamba, J.; Moundounga Mavouroulou, Q.; Gomat, H.; Mankou, G.S.; Loumeto, J.; Midoko Iponga, D.; Kossi Ditsouga, F.; Zinga Koumba, R.; et al. Site-specific versus pantropical allometric equations: Which option to estimate the biomass of a moist central African forest? For. Ecol. Manag. 2014, 312, 1–9. [Google Scholar] [CrossRef]
- Xiang, W.; Zhou, J.; Ouyang, S.; Zhang, S.; Lei, P.; Li, J.; Deng, X.; Fang, X.; Forrester, D.I. Species-specific and general allometric equations for estimating tree biomass components of subtropical forests in southern China. Eur. J. For. Res. 2016, 135, 963–979. [Google Scholar] [CrossRef]
- Paul, K.I.; Roxburgh, S.H.; Ritson, P.; Brooksbank, K.; England, J.R.; Larmour, J.S.; John Raison, R.; Peck, A.; Wildy, D.T.; Sudmeyer, R.A.; et al. Testing allometric equations for prediction of above-ground biomass of mallee eucalypts in southern Australia. For. Ecol. Manag. 2013, 310, 1005–1015. [Google Scholar] [CrossRef]
- Paul, K.I.; Roxburgh, S.H.; Chave, J.; England, J.R.; Zerihun, A.; Specht, A.; Lewis, T.; Bennett, L.T.; Baker, T.G.; Adams, M.A.; et al. Testing the generality of above-ground biomass allometry across plant functional types at the continent scale. Glob. Chang. Biol. 2016, 22, 2106–2124. [Google Scholar] [CrossRef]
- Ishihara, M.I.; Utsugi, H.; Tanouchi, H.; Aiba, M.; Kurokawa, H.; Onoda, Y.; Nagano, M.; Umehara, T.; Ando, M.; Miyata, R.; et al. Efficacy of generic allometric equations for estimating biomass: A test in Japanese natural forests. Ecol. Appl. 2015, 25, 1433–1446. [Google Scholar] [CrossRef]
- Vieilledent, G.; Vaudry, R.; Andriamanohisoa, S.F.D.; Rakotonarivo, O.S.; Randrianasolo, H.Z.; Razafindrabe, H.N.; Bidaud Rakotoarivony, C.; Ebeling, J.; Rasamoelina, A.M.; Rakotoarivony, C.B.; et al. A universal approach to estimate biomass and carbon stock in tropical forests using generic allometric models. Ecol. Appl. 2012, 22, 572–583. [Google Scholar] [CrossRef]
- Rutishauser, E.; Noor′an, F.; Laumonier, Y.; Halperin, J.; Hergoualc’h, K.; Verchot, L. Generic allometric models including height best estimate forest biomass and carbon stocks in Indonesia. For. Ecol. Manag. 2013, 307, 219–225. [Google Scholar] [CrossRef]
- Schepaschenko, D.; Shvidenko, A.; Usoltsev, V.; Lakyda, P.; Luo, Y.; Vasylyshyn, R.; Lakyda, I.; Myklush, Y.; See, L.; McCallum, I.; et al. A dataset of forest biomass structure for Eurasia. Sci. Data 2017, 4, 170070. [Google Scholar] [CrossRef] [PubMed]
- Ung, C.H.; Bernier, P.; Guo, X.J. Canadian national biomass equations: New parameter estimates that include British Columbia data. Can. J. For. Res. 2008, 38, 1123–1132. [Google Scholar] [CrossRef]
- Ung, C.H.; Lambert, M.C.; Raulier, F.; Guo, X.J.; Bernier, P.Y. Biomass of trees sampled across Canada as part of the Energy from the Forest Biomass (ENFOR) Program 2017. Available online: https://doi.org/10.23687/fbad665e-8ac9-4635-9f84-e4fd53a6253c (accessed on 21 December 2018).
- Clarke, P. When can group level clustering be ignored? Multilevel models versus single-level models with sparse data. J. Epidemiol. Community Health 2008, 62, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, H.; Browne, W.; Rasbash, J. Partitioning Variation in Multilevel Models. Underst. Stat. 2002, 1, 223–231. [Google Scholar] [CrossRef]
- Kish, L. Survey Sampling; John Wiley & Sons: New York, NY, USA, 1965. [Google Scholar]
- Enquist, B.J.; Niklas, K.J. Invariant scaling relations across tree-dominated communities. Nature 2001, 410, 655–660. [Google Scholar] [CrossRef]
- Baskerville, G.L. Use of Logarithmic Regression in the Estimation of Plant Biomass. Can. J. For. Res. 1972, 2, 49–53. [Google Scholar] [CrossRef]
- Sprugel, D.G. Correcting for Bias in Log-Transformed Allometric Equations. Ecology 1983, 64, 209–210. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Murtagh, F.; Legendre, P. Ward′s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Hollister, J.; Shah, T. Elevatr: Access Elevation Data from Various APIs; US EPA Office of Research and Development: Washington, DC, USA, 2017.
- Pastore, M. Overlapping: A R package for Estimating Overlapping in Empirical Distributions. J. Open Source Softw. 2018, 3, 1–4. [Google Scholar] [CrossRef]
- Sarkar, D. Lattice: Multivariate Data Visualization with R, 1st ed.; Springer: Seattle, WA, USA, 2008. [Google Scholar]
- Galili, T. dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef] [PubMed]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling 2019. R package version 3.0-2. Available online: https://CRAN.R-project.org/package=raster (accessed on 10 September 2019).
- Iida, Y.; Poorter, L.; Sterck, F.J.; Kassim, A.R.; Kubo, T.; Potts, M.D.; Kohyama, T.S. Wood density explains architectural differentiation across 145 co-occurring tropical tree species. Funct. Ecol. 2012, 26, 274–282. [Google Scholar] [CrossRef]
- Henry, M.; Besnard, A.; Asante, W.A.; Eshun, J.; Adu-Bredu, S.; Valentini, R.; Bernoux, M.; Saint-André, L. Wood density, phytomass variations within and among trees, and allometric equations in a tropical rainforest of Africa. For. Ecol. Manag. 2010, 260, 1375–1388. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Swenson, N.G.; Enquist, B.J. Ecological and evolutionary determinants of a key plant functional trait: Wood density and its community-wide variation across latitude and elevation. Am. J. Bot. 2007, 94, 451–459. [Google Scholar] [CrossRef]
- Feldpausch, T.R.; Lloyd, J.; Lewis, S.L.; Brienen, R.J.W.; Gloor, M.; Monteagudo Mendoza, A.; Lopez-Gonzalez, G.; Banin, L.; Abu Salim, K.; Affum-Baffoe, K.; et al. Tree height integrated into pantropical forest biomass estimates. Biogeosciences 2012, 9, 3381–3403. [Google Scholar] [CrossRef] [Green Version]

| Characteristic | Dataset 1 | Dataset 2 |
|---|---|---|
| Region | Europe and Asia | Canada |
| Sample size | 4921 | 5199 |
| D range (cm) | 5.0–72.9 | 5.0–74.3 |
| H range (m) | 3.1–42.8 | 2.5–40.8 |
| AGB range (kg) | 2.2–4291.3 | 2.2–2951.4 |
| Number of tree species | 56 | 38 |
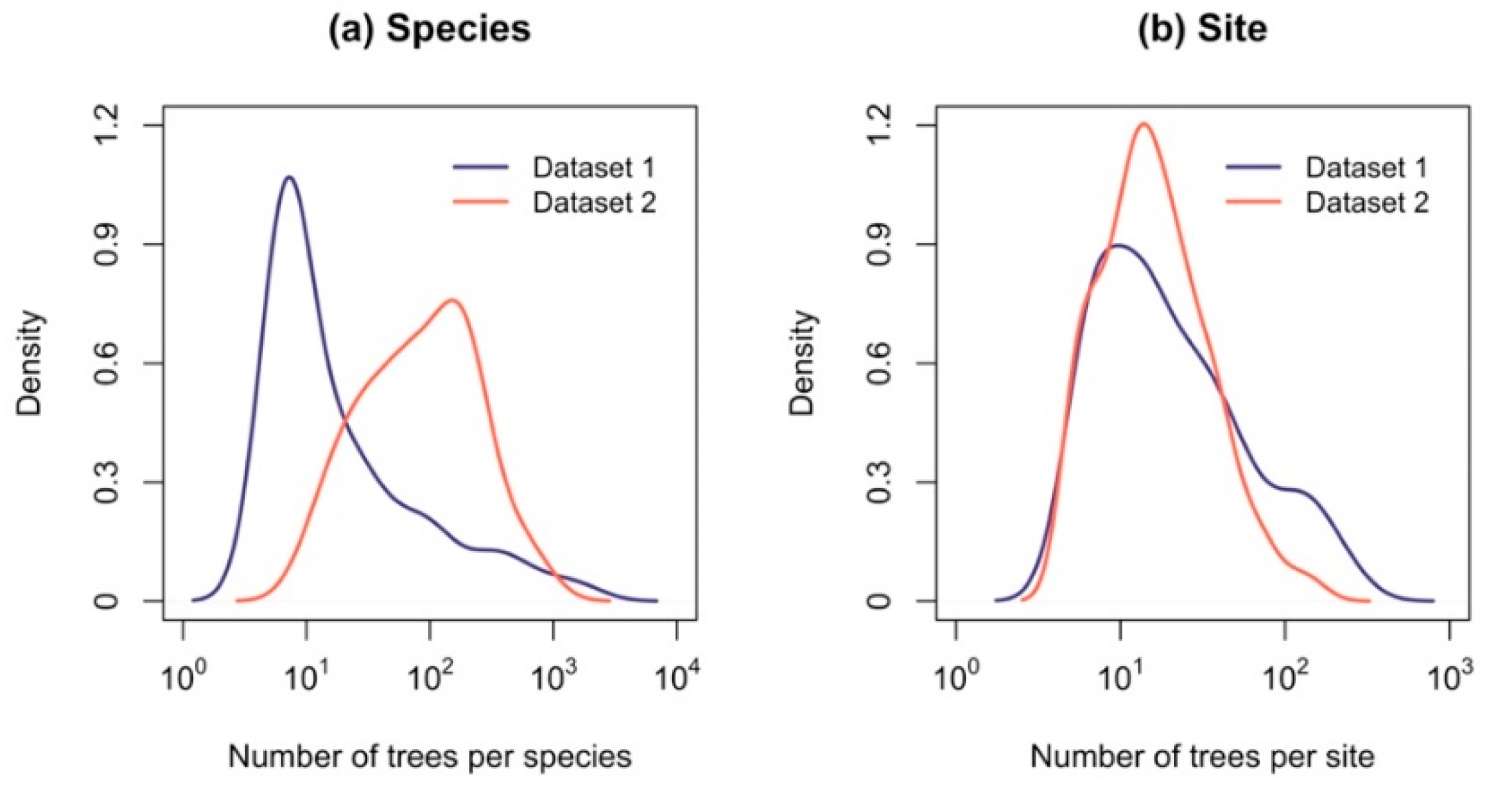
| Number of trees per species (range) | 5–1656 | 11–704 |
| Number of sites | 133 | 237 |
| Number of trees per site (range) | 5–278 | 5–164 |
| References | [34] | [35,36] |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dutcă, I. The Variation Driven by Differences between Species and between Sites in Allometric Biomass Models. Forests 2019, 10, 976. https://doi.org/10.3390/f10110976
Dutcă I. The Variation Driven by Differences between Species and between Sites in Allometric Biomass Models. Forests. 2019; 10(11):976. https://doi.org/10.3390/f10110976
Chicago/Turabian StyleDutcă, Ioan. 2019. "The Variation Driven by Differences between Species and between Sites in Allometric Biomass Models" Forests 10, no. 11: 976. https://doi.org/10.3390/f10110976






