Soil Organic Carbon Chemical Functional Groups under Different Revegetation Types Are Coupled with Changes in the Microbial Community Composition and the Functional Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experiment Site
2.2. Soil Samples Collection
2.3. Determination of Soil Characteristics
2.4. Cross Polarization Magic Angle Spinning (CPMAS)-13C NMR Analysis
2.5. Metagenome DNA Extraction and Shotgun Sequencing
2.6. Metagenome Sequence Analysis, Assembly, and Functional Annotation
2.7. Statistical Analysis
3. Results
3.1. Soil Chemical Characteristics under Different Revegetation Types
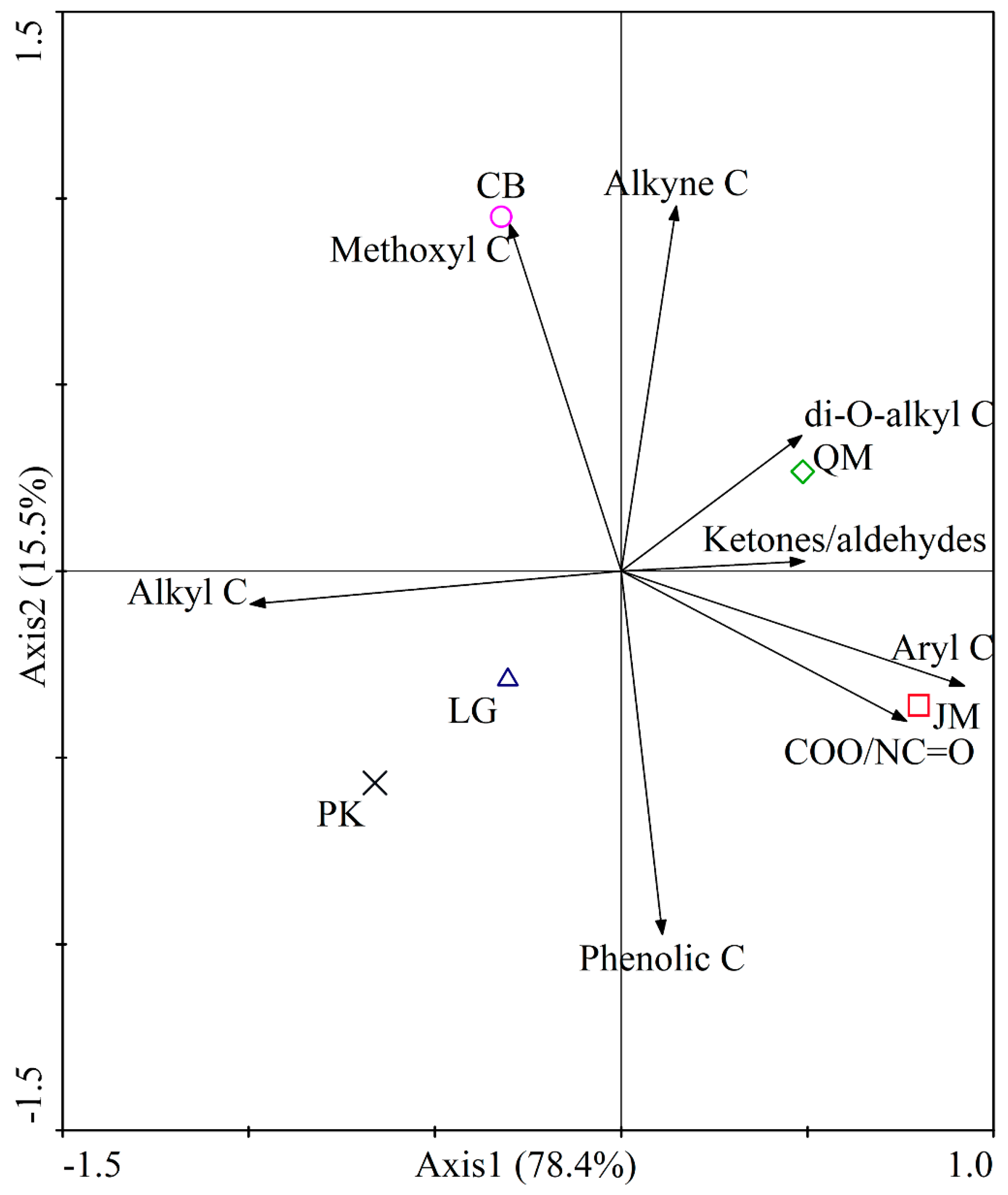
3.2. SOC Chemical Composition under Different Revegetation Types
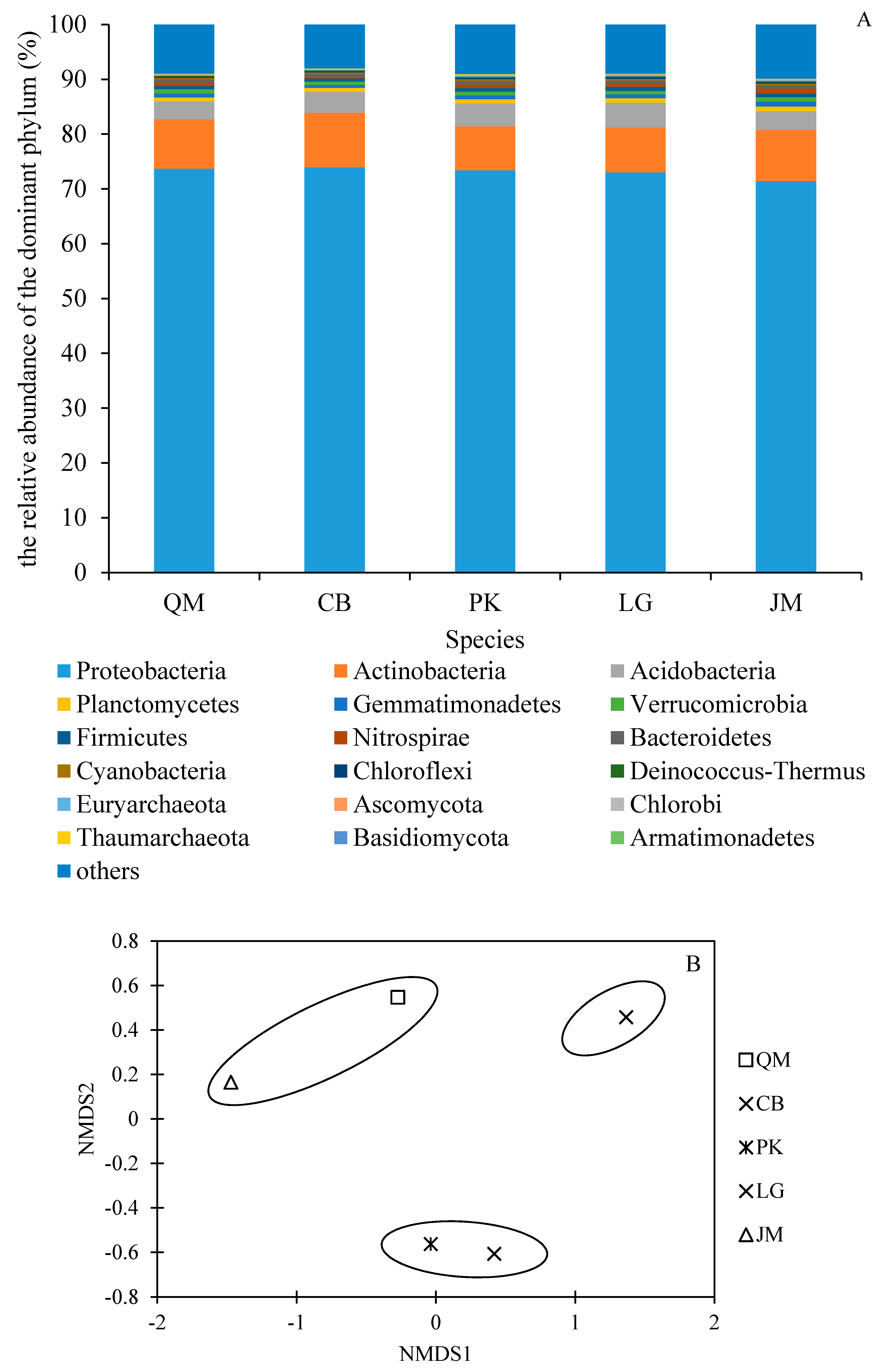
3.3. Soil Microbial Community Characteristics under Different Revegetation Types
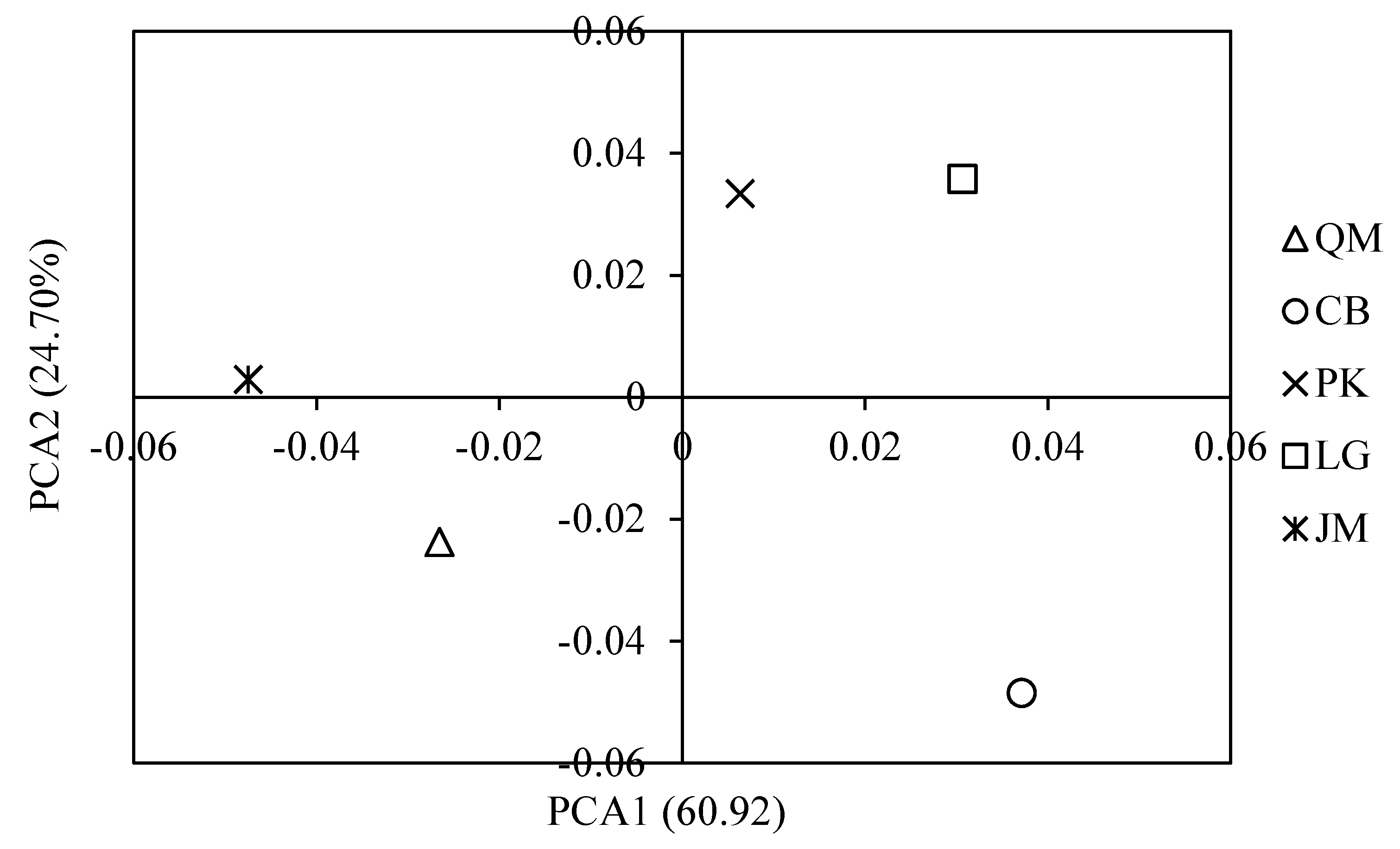
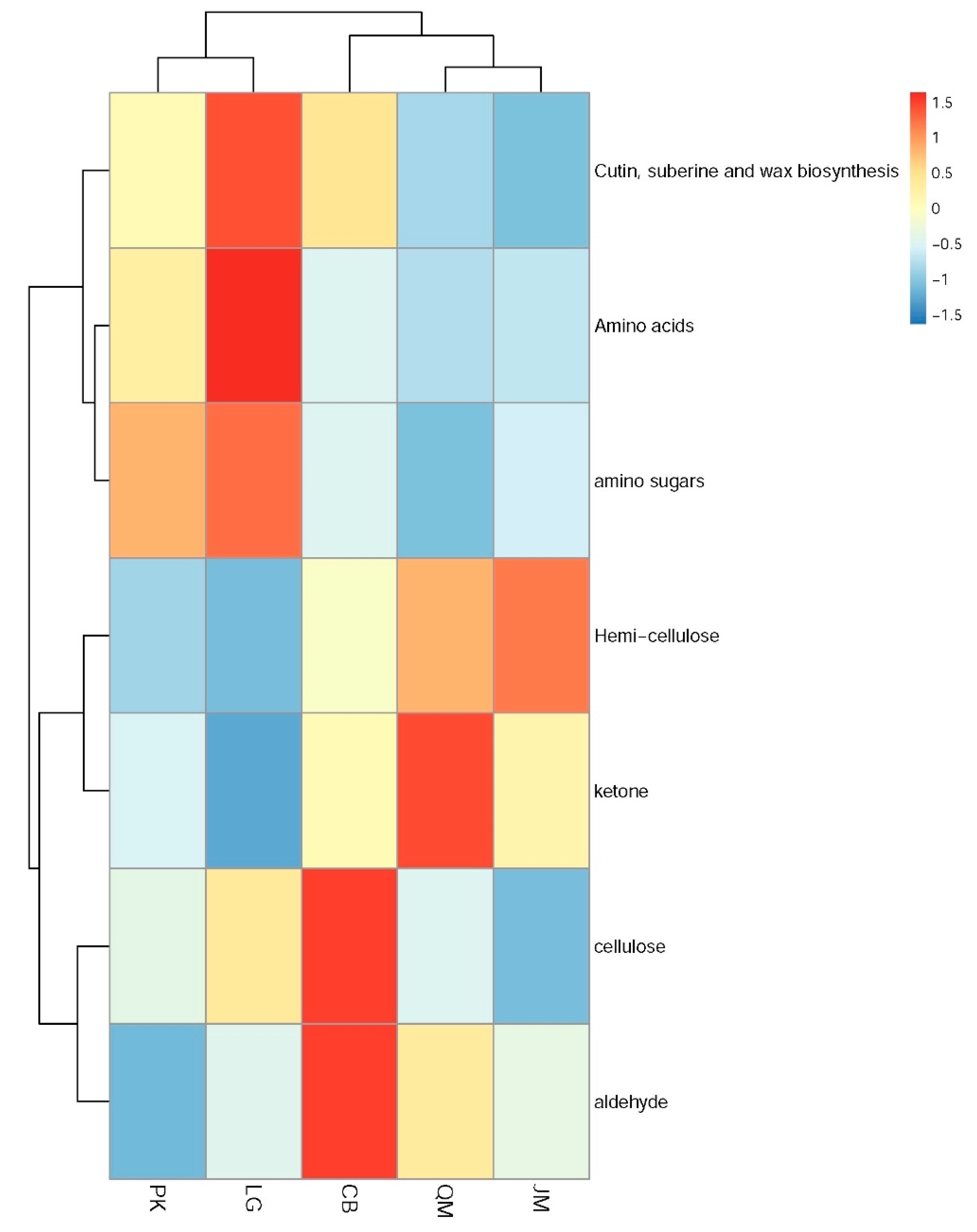
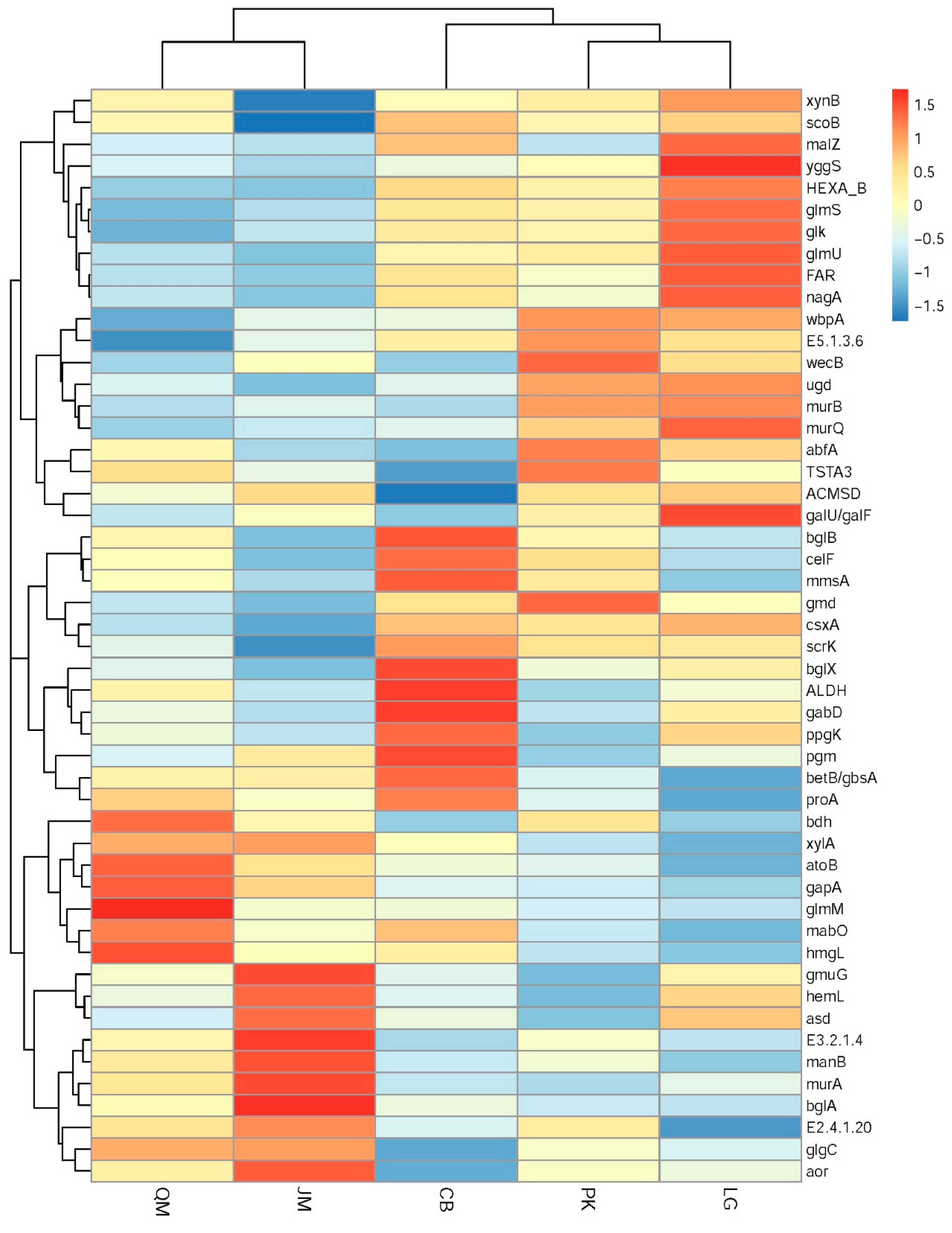
3.4. Functional Characteristics of Soil Microbial Communities
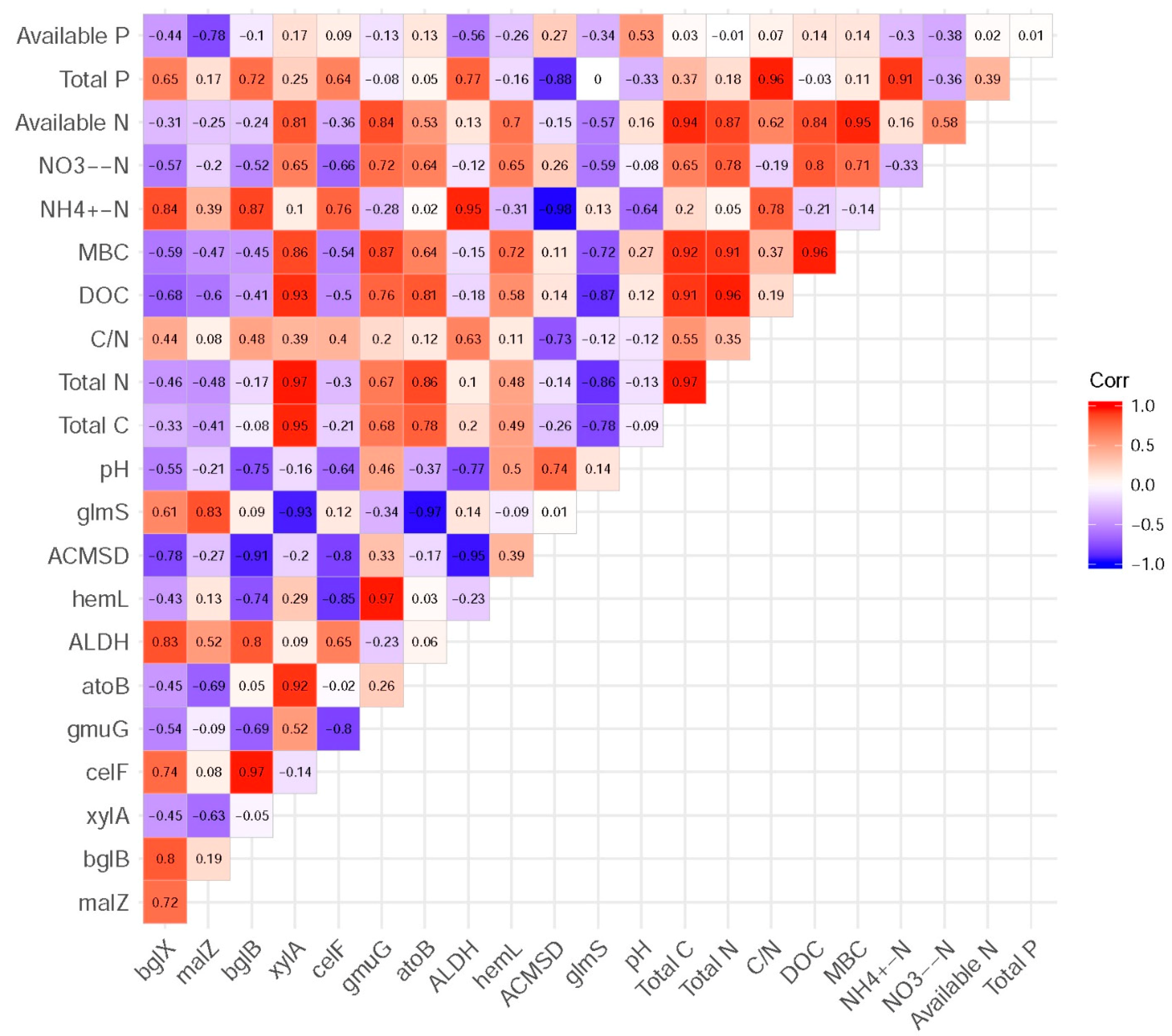
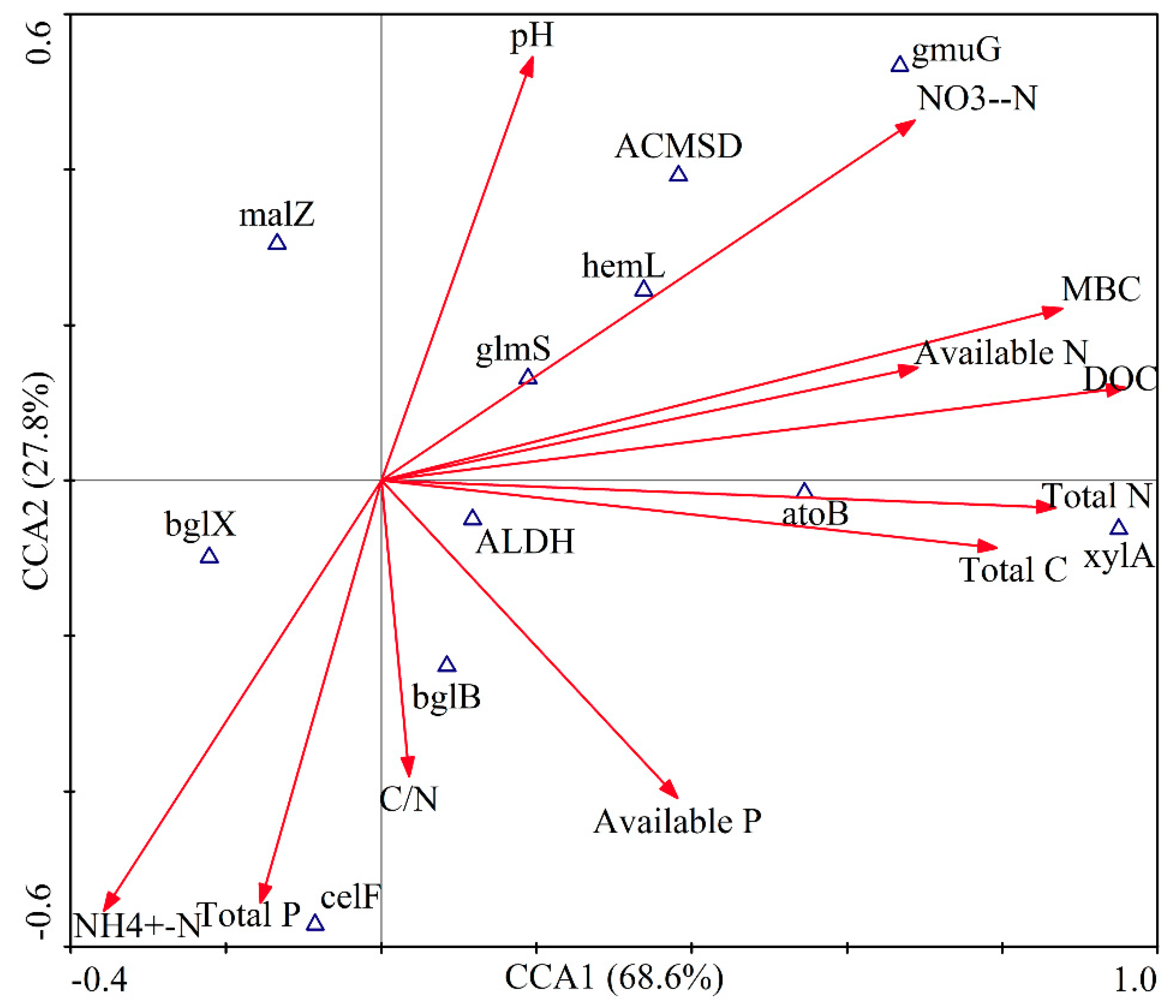
3.5. Functional Genes and Their Relationship with Measured Soil Characteristics
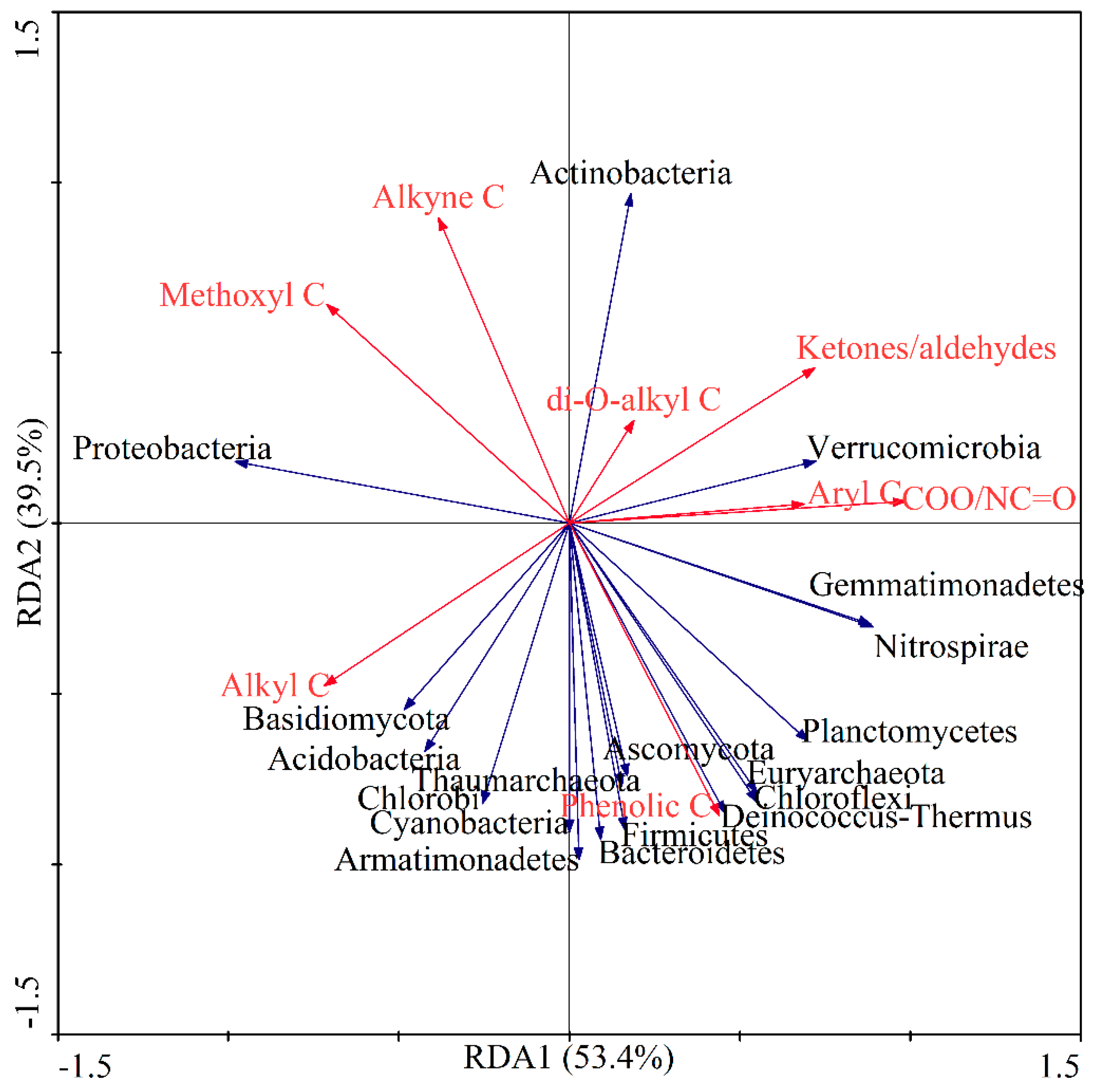
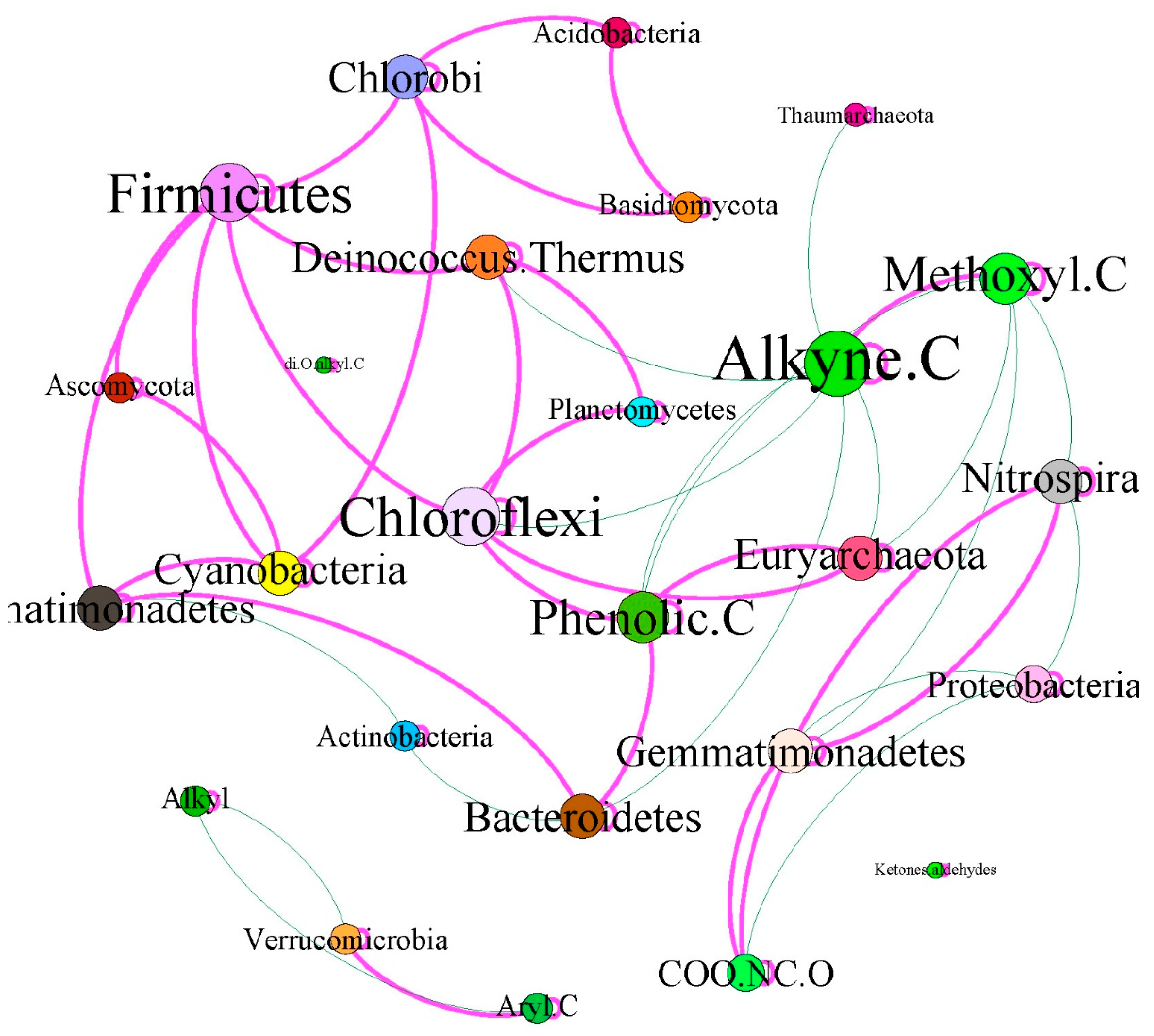
3.6. Correlations between Microbial Community and SOC Chemical Functional Groups
4. Discussion
4.1. Soil Characteristics Response to Different Revegetation Types
4.2. The Effect of Different Revegetation Types on SOC Chemical Composition
4.3. Responses of Soil Microbial Functional Genes Involved in Carbon Cycling under Different Revegetation Types
4.4. Response of Soil C Fractions to Soil Microbial Composition under Different Revegetation Types
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, X.; Herbert, S.; Hashemi, A.M.; Zhang, X.; Ding, G.W. Effects of agricultural management on soil organic matter and carbon transformation—A review. Plant Soil Environ. 2006, 52, 531–543. [Google Scholar] [CrossRef]
- Solomon, D.; Lehmann, J.; Thies, J.; Schäfer, T.; Liang, B.; James, K.; Neves, E.G.; Petersen, J.; Luizao, F.; Skjemstad, J. Molecular signature and sources of biochemical recalcitrance of organic C in Amazonian Dark Earths. Geochim. Cosmochim. Acta 2007, 71, 2290–2298. [Google Scholar] [CrossRef]
- Jiménez-González, M.A.; Álvarez, A.M.; Hernández, Z.; Almendros, G. Soil carbon storage predicted from the diversity of pyrolytic alkanes. Biol. Fert. Soils 2018, 54, 617–629. [Google Scholar] [CrossRef]
- Banning, N.C.; Grant, C.D.; Jones, D.L.; Murphy, D.V. Recovery of soil organic matter, organic matter turnover and nitrogen cycling in a post-mining forest rehabilitation chronosequence. Soil Biol. Biochem. 2008, 40, 2021–2031. [Google Scholar] [CrossRef]
- Carvalhais, N.; Forkel, M.; Khomik, M.; Bellarby, J.; Jung, M.; Migliavacca, M.; Mu, M.; Saatchi, S.; Santoro, M.; Thurner, M.; et al. Global covariation of carbon turnover times with climate in terrestrial ecosystems. Nature 2014, 514, 213–217. [Google Scholar] [CrossRef]
- Paquette, A.; Messier, C. The role of plantations in managing the world’s forests in the Anthropocene. Front. Ecol. Environ. 2010, 8, 27–34. [Google Scholar] [CrossRef]
- Liu, B.J.; Lu, F.; Wang, X.K.; Liu, W.W.; Wang, L.Y.; Rao, E.; Zhang, L.; Zheng, H. Greenhouse gas emissions and net carbon sequestration of the natural forest protection program in China. Acta Ecol. Sin. 2016, 36, 4266–4278. [Google Scholar]
- Lal, R. Soil carbon sequestration impacts on global climate change and food security. Science 2004, 304, 1623–1627. [Google Scholar] [CrossRef]
- Shrestha, B.M.; Certini, G.; Forte, C.; Singh, B.R. Soil organic matter quality under different land uses in a mountain watershed of Nepal. Soil Sci. Soc. Am. J. 2008, 72, 1563–1569. [Google Scholar] [CrossRef]
- Tuo, D.; Gao, G.; Chang, R.; Li, Z.; Ma, Y.; Wang, S.; Wang, C.; Fu, B. Effects of revegetation and precipitation gradient on soil carbon and nitrogen variations in deep profiles on the Loess Plateau of China. Sci. Total Environ. 2018, 626, 399–411. [Google Scholar] [CrossRef]
- Schulp, C.J.E.; Nabuurs, G.J.; Verburg, P.H.; de Waal, R.W. Effect of tree species on carbon stocks in forest floor and mineral soil and implications for soil carbon inventories. For. Ecol. Manag. 2008, 256, 482–490. [Google Scholar] [CrossRef]
- Fekete, I.; Lajtha, K.; Kotroczó, Z.; Várbíró, G.; Varga, C.; Tóth, J.A.; Demeter, I.; Veperdi, G.; Berki, I. Long-term effects of climate change on carbon storage and tree species composition in a dry deciduous forest. Glob. Chang. Biol. 2017, 23, 3154–3168. [Google Scholar] [CrossRef]
- Lajtha, K.; Bowden, R.D.; Crow, S.; Fekete, I.; Kotroczó, Z.; Plante, A.F.; Simpson, M.; Nadelhoffer, K.J. The Detrital Input and Removal Treatment (DIRT) Network. Sci. Total Environ. 2017, 640–641, 1112–1120. [Google Scholar]
- Liu, S.; Bondlamberty, B.; Boysen, L.R.; Ford, J.D.; Fox, A.; Gallo, K.; Hatfield, J.; Henebry, G.M.; Huntington, T.G.; Liu, Z.; et al. Grand challenges in understanding the interplay of climate and land changes. Earth Interact. 2017, 21, 1–43. [Google Scholar] [CrossRef]
- Huang, Z.; Xu, Z.; Chen, C.; Boyd, S. Changes in soil carbon during the establishment of a hardwood plantation in subtropical Australia. For. Ecol. Manag. 2008, 254, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Song, B.; Luo, M.; Li, H.; He, S. Chemical composition and structural features of soil organic carbon (SOC) in plantation. J. Southwest Minzu Univ. 2017, 43, 474–479. [Google Scholar]
- Fan, F.; Li, Z.; Wakelin, S.A.; Steven, W.; Yu, W.; Liang, Y.C. Mineral fertilizer alters cellulolytic community structure and suppresses soil cellobiohydrolase activity in a long-term fertilization experiment. Soil Biol. Biochem. 2012, 55, 70–77. [Google Scholar] [CrossRef]
- Schmidt, M.W.; Torn, M.S.; Abiven, S.; Dittmar, T.; Guggenberger, G.; Janssens, I.A.; Kleber, M.; Kögel-Knabner, I.; Lehmann, J.; Manning, D.A.; et al. Persistence of soil organic matter as an ecosystem property. Nature 2011, 478, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Gavazov, K.S. Dynamics of alpine plant litter decomposition in a changing climate. Plant Soil 2010, 337, 19–32. [Google Scholar] [CrossRef]
- Djukic, I.; Kepfer-Rojas, S.; Schmidt, I.K.; Larsen, K.S.; Beier, C.; Berg, B.; Verheyen, K. Early stage litter decomposition across biomes. Sci. Total Environ. 2018, 628–629, 1369–1394. [Google Scholar] [CrossRef]
- Schimel, J.P.; Schaeffer, S.M. Microbial control over carbon cycling in soil. Front. Microbiol. 2012, 3, 348. [Google Scholar] [CrossRef]
- Heimann, M.; Reichstein, M. Terrestrial ecosystem carbon dynamics and climate feedbacks. Nature 2008, 451, 289–292. [Google Scholar] [CrossRef] [Green Version]
- Harris, J. Soil microbial communities and restoration ecology: Facilitators or Followers? Science 2009, 325, 573–574. [Google Scholar] [CrossRef]
- Palomo, A.; Jane Fowler, S.; Gülay, A.; Rasmussen, S.; Sicheritz-Ponten, T.; Smets, B.F. Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. ISME J. 2016, 10, 2569–2581. [Google Scholar] [CrossRef]
- Kuramae, E.E.; Gamper, H.A.; Yergeau, E.; Piceno, Y.M.; Brodie, E.L.; Desantis, T.Z.; Andersen, G.L.; van Veen, J.A.; Kowalchuk, G.A. Microbial secondary succession in a chronosequence of chalk grasslands. ISME J. 2010, 4, 711–715. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.; OLK, D.C.; Fang, X.W.; He, Z.Q.; Schmidt-Rohr, K. Influence of animal manure application on the chemical structures of soil organic matter as investigated by advanced solid-state NMR and FT-IR spectroscopy. Geoderma 2008, 146, 353–362. [Google Scholar] [CrossRef]
- Stursová, M.; Zifčáková, L.; Leigh, M.B.; Burgess, R.; Baldrian, P. Cellulose utilization in forest litter and soil: Identification of bacterial and fungal decomposers. FEMS Microbiol. Ecol. 2012, 80, 735–746. [Google Scholar] [CrossRef]
- Prescott, C.E.; Grayston, S.J. Tree species influence on microbial communities in litter and soil: Current knowledge and research needs. For. Ecol. Manag. 2013, 309, 19–27. [Google Scholar] [CrossRef]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Ding, J.; Zhang, Y.; Wang, M.; Sun, X.; Cong, J.; Deng, Y.; Lu, H.; Yuan, T.; van Nostrand, J.D.; Li, D.; et al. Soil organic matter quantity and quality shape microbial community compositions of subtropical broadleaved forests. Mol. Ecol. 2015, 24, 5175–5185. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Mccormack, M.L.; Wang, J.Y.; Guo, D.; Wang, Q.F.; Zhang, X.Y.; Yu, G.; Blagodatskaya, E.; Kuzyakov, Y. Linkages between the soil organic matter fractions and the microbial metabolic functional diversity within a broad-leaved Koreanpine forest. Eur. J. Soil Biol. 2015, 66, 57–64. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Chang, S.X.; Liang, X.; Qin, H.; Chen, J.H.; Xu, Q.F. Linking soil fungal community structure and function to soil organic carbon chemical composition in intensively managed subtropical bamboo forests. Soil Biol. Biochem. 2017, 107, 19–31. [Google Scholar] [CrossRef]
- Zhao, F.Z.; Ren, C.J.; Zhang, L.; Han, X.H.; Yang, G.H.; Wang, J. Changes in soil microbial community are linked to soil carbon fractions after afforestation: Soil microbial community affects carbon fractions. Eur. J. Soil Biol. 2018, 69, 370–379. [Google Scholar] [CrossRef]
- Qi, J.H. Content of soil organic carbon and its relations with physicochemical properties of secondary natural Oak forests in eastern mountain Area of Liaoning Province. J. Soil Water Conserv. 2017, 31, 135–140. [Google Scholar]
- Yin, Y.; Liu, Y.Y.; Jing, Y.L.; Wei, Y.W.; An, Y.Q.; Yan, P.C.; Zhou, Y.B. Contents of soil organic carbon and components in three types of forests in the mountain area of Eastern Liaoning. Chin. J. Ecol. 2018, 37, 2100–2106. [Google Scholar]
- Deng, J.; Yin, Y.; Zhu, W.; Zhou, Y. Variations in soil bacterial community diversity and structures among different revegetation types in the Baishilazi Nature Reserve. Front. Microbiol. 2018, 9, 2874. [Google Scholar] [CrossRef]
- Deng, J.; Yin, Y.; Luo, J.; Zhu, W.; Zhou, Y. Different revegetation types alter soil physical-chemical characteristics and fungal community in the Baishilazi Nature Reserve. PeerJ 2019, 6, e6251. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, L.; Yi, L.T. Plant floristic diversity in the Baishilazi National Nature Reserve of Liaoning, China. J. Zhejiang A F Univ. 2014, 31, 210–216. [Google Scholar]
- Zhang, H.; You, W.; Wei, W.; Zhou, M. Soil physical and chemical properties and correlation with organic carbon in original Korean pine forest in Eastern Liaoning mountainous area. J. Northwest A F Univ. 2017, 45, 76–82. [Google Scholar]
- Ren, C.; Zhao, F.; Kang, D.; Yang, G.; Han, X.H.; Tong, X.; Feng, Y.; Ren, G. Linkages of C:N:P stoichiometry and bacterial community in soil following afforestation of former farmland. Forest Ecol. Manag. 2016, 376, 59–66. [Google Scholar] [CrossRef]
- Schrumpf, M.; Schulze, E.D.; Kaiser, K.; Schumacher, J. How accurately can soil organic carbon stocks and stock changes be quantified by soil inventories? Biogeosciences 2011, 8, 1193–1212. [Google Scholar] [CrossRef]
- Jackson, M.L. Soil chemical analysis. Verlag: Prentice Hall, Inc. Englewood Cliffs, NJ. 498 S. DM 39.40. J. Soil Sci. Plant Nutr. 1958, 85, 251–252. [Google Scholar]
- Qing, W.; Cong, W.; Yu, W.; Turak, A.; Chen, D.; Huang, Y.; Ao, J.; Jiang, Y.; Huang, Z. Effects of nitrogen and phosphorus inputs on soil bacterial abundance, diversity, and community composition in Chinese Fir plantations. Front. Microbiol. 2018, 9, 1543. [Google Scholar]
- Cai, Z.Q.; Zhang, Y.H.; Yang, C.; Wang, S. Land-use type strongly shapes community composition, but not always diversity of soil microbes in tropical China. Catena 2018, 165, 369–380. [Google Scholar] [CrossRef]
- Gong, W.; Yan, X.Y.; Wang, J.Y.; Hu, T.X.; Gong, Y.B. Long-term manuring and fertilization effects on soil organic carbon pools under a wheat–maize cropping system in North China Plain. Plant Soil 2009, 314, 67–76. [Google Scholar] [CrossRef]
- Bao, S.D. Soil and Agricultural Chemistry Analysis; China Agriculture Press: Beijing, China, 2000. (In Chinese) [Google Scholar]
- Brookes, P.C. Chloroform fumigation and the release of soil nitrogen: A rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 1985, 17, 837–842. [Google Scholar] [CrossRef]
- Schmidt, M.W.I. Improvement of 13C and 15N CPMAS NMR spectra of bulk soils, particle size fractions and organic material by treatment with 10% hydrofluoric acid. Eur. J. Soil Biol. 2010, 48, 319–328. [Google Scholar] [CrossRef]
- MAO, J.D.; Hu, W.G.; Schmidt-Rohr, K.; Davies, G.; Ghabbour, E.A.; Xing, B.S. Quantitative characterization of humic substances by solid-state carbon-13 nuclear magnetic resonance. Soil Sci. Soc. Am. J. 2000, 64, 873–884. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Leung, H.C.; Yiu, S.M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010, 38, e132. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Banerjee, S. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Gao, Q.; Yang, Z.L. Diversity and distribution patterns of root-associated fungi on herbaceous plants in alpine meadows of southwestern China. Mycologia 2016, 108, 281–291. [Google Scholar] [CrossRef]
- Rahimabady, M.S.; Akbarinia, M.; Kooch, Y. The effect of land covers on soil quality properties in the Hyrcanian regions of Iran. J. Biosci. Bioeng. 2015, 4, 73–79. [Google Scholar]
- Jahed, R.R.; Hosseini, S.M. The effect of natural and planted forest stands on soil fertility in the Hyrcanian region, Iran. Biodiver. J. Biol. Diver. 2014, 15, 206–214. [Google Scholar] [CrossRef]
- Grayston, S.J.; Prescott, C.E. Microbial communities in forest floors under four tree species in coastal British Columbia. Soil Biol. Biochem. 2005, 37, 1157–1167. [Google Scholar] [CrossRef]
- Yang, X.Q.; Han, Y.Z. Spatial variations of soil organic carbon and nitrogen of forest land in Guandi Mountain. For. Res. 2001, 24, 223–229. [Google Scholar]
- Pérez, M.G.; Martin-Neto, L.; Saab, S.C.; Novotny, E.; Pereira Milori, D.M.B.; Bagnato, V.S.; Colnago, L.A.; Melo, W.J.; Knicker, H. Characterization of humic acids from a Brazilian Oxisol under different tillage systems by EPR, 13C NMR, FTIR and fluorescence spectroscopy. Geoderma 2004, 118, 181–190. [Google Scholar] [CrossRef]
- Zhao, H.; Lv, Y.; Wang, X.; Zhang, H.; Yang, X. Tillage impacts on the fractions and compositions of soil organic carbon. Geoderma 2012, 189–190, 397–403. [Google Scholar] [CrossRef]
- Rumpel, C.; KögelKnabner, I.; Bruhn, F. Vertical distribution, age, and chemical composition of organic carbon in two forest soils of different pedogenesis. Org. Geochem. 2002, 33, 1131–1142. [Google Scholar] [CrossRef]
- Li, Y.F.; Zhang, J.; Chang, S.X.; Jiang, P.K.; Zhou, G.; Shen, Z.; Wu, J.; Lin, L.; Wang, Z.; Shen, M.C. Converting native shrub forests to Chinese chestnut plantations and subsequent intensive management affected soil C and N pools. For. Ecol. Manag. 2014, 312, 161–169. [Google Scholar] [CrossRef]
- Chung, T.L.; Chen, J.S.; Chiu, C.Y.; Tian, G.L. 13C-NMR spectroscopy studies of humic substances in subtropical perhumid montane forest soil. J. For. Res. 2012, 17, 458–467. [Google Scholar] [CrossRef]
- Fang, X.; Zhang, J.; Meng, M.; Guo, X.; Wu, Y.; Liu, X.; Zhao, K.; Ding, L.; Shao, Y.; Fu, W. Forest-type shift and subsequent intensive management affected soil organic carbon and microbial community in southeastern China. Eur. J. For. Res. 2017, 136, 689–697. [Google Scholar] [CrossRef]
- Ussiri, D.A.N.; Johnson, C.E. Organic matter composition and dynamics in a northern hardwood forest ecosystem 15 years after clear-cutting. For. Ecol. Manag. 2007, 240, 131–142. [Google Scholar] [CrossRef]
- Solomon, D.; Lehmann, J.; Kinyangi, J.; Amelung, W.; Lone, I.; Pell, A.N.; Riha, S.; Ngoze, S.; Verchot, L.; Mbugua, D.; et al. Long-term impacts of anthropogenic perturbations on dynamics and speciation of organic carbon in tropical forest and subtropical grassland ecosystems. Glob. Chang. Biol. 2010, 13, 511–530. [Google Scholar] [CrossRef]
- Ussiri, D.A.N.; Johnson, C.E. Characterization of organic matter in a northern hardwood forest soil by 13 C NMR spectroscopy and chemical methods. Geoderma 2003, 111, 123–149. [Google Scholar] [CrossRef]
- Bonanomi, G.; Incerti, G.; Giannino, F.; Mingo, A.; Lanzotti, V.; Mazzoleni, S. Litter quality assessed by solid state 13C NMR spectroscopy predicts decay rate better than C/N and Lignin/N ratios. Soil Biol. Biochem. 2013, 56, 40–48. [Google Scholar] [CrossRef]
- Lorenz, K.; Lal, R.; Jiménez, J.J. Soil organic carbon stabilization in dry tropical forests of Costa Rica. Geoderma 2009, 152, 95–103. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, B.; Wang, Q.; Cao, X.; Zhang, J. Differences in chemical composition of soil organic carbon resulting from long-term fertilization strategies. PLoS ONE 2015, 10, e0124359. [Google Scholar] [CrossRef]
- Helfrich, M.; Ludwig, B.; Buurman, P.; Flessa, H. Effect of land use on the composition of soil organic matter in density and aggregate fractions as revealed by solid-state 13C NMR spectroscopy. Geoderma 2006, 136, 331–341. [Google Scholar] [CrossRef]
- Dieckow, J.; Bayer, C.; Conceição, P.C.; Zanatta, J.A.; Martin-Neto, L.; Milori, D.B.M.; Salton, J.C.; Macedo, M.M.; Mielniczuk, J.; Hernani, L.C. Land use, tillage, texture and organic matter stock and composition in tropical and subtropical Brazilian soils. Eur. J. Soil Biol. 2010, 60, 240–249. [Google Scholar] [CrossRef]
- Webster, E.A.; Hopkins, D.W.; Chudek, J.A.; Haslam, S.F.; Simek, M.; Pîcek, T. The relationship between microbial carbon and the resource quality of soil carbon. J. Environ. Qual. 2001, 30, 147–150. [Google Scholar] [CrossRef]
- Yan, H.; Chen, C.; Xu, Z.; Williams, D.; Xu, J.M. Assessing management impacts on soil organic matter quality in subtropical Australian forests using physical and chemical fractionation as well as 13C NMR spectroscopy. Soil Biol. Biochem. 2009, 41, 640–650. [Google Scholar]
- Dou, S.; Zhang, J.J.; Li, K. Effect of organic matter applications on 13C-NMR spectra of humic acids of soil. Eur. J. Soil Biol. 2010, 59, 532–539. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.R.; Mo, J.M.; Wang, J.X.; Makeschin, F.; Wolff, M. Soil organic carbon stock and chemical composition in four plantations of indigenous tree species in subtropical China. Ecol. Res. 2010, 25, 1071–1079. [Google Scholar] [CrossRef]
- Liang, C.; Balser, T.C. Warming and nitrogen deposition lessen microbial residue contribution to soil carbon pool. Nat. Commun. 2012, 3, 1222. [Google Scholar] [CrossRef] [Green Version]
- Manzoni, S.; Porporato, A. Soil carbon and nitrogen mineralization: Theory and models across scales. Soil Biol. Biochem. 2009, 41, 1355–1379. [Google Scholar] [CrossRef]
- Strickland, M.S.; Lauber, C.; Fierer, N.; Bradford, M.A. Testing the functional significance of microbial community composition. Ecology 2009, 90, 441–451. [Google Scholar] [CrossRef]
- Sergey, B.; Evgenia, B.; Tatyana, Y.; Yakov, K. Model of apparent and real priming effects: Linking microbial activity with soil organic matter decomposition. Soil Biol. Biochem. 2010, 42, 1275–1283. [Google Scholar]
- Carney, K.M.; Hungate, B.A.; Drake, B.G.; Megonigal, J.P. Altered soil microbial community at elevated CO(2) leads to loss of soil carbon. Proc. Natl. Acad. Sci. USA 2007, 104, 4990–4995. [Google Scholar] [CrossRef]
- Billings, S.A.; Ziegler, S.E. Altered patterns of soil carbon substrate usage and heterotrophic respiration in a pine forest with elevated CO2 and N fertilization. Glob. Chang. Biol. 2008, 14, 1025–1036. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Salles, J.F.; Poly, F.; Schmid, B.; Le Roux, X. Community niche predicts the functioning of denitrifying bacterial assemblages. Ecology 2009, 90, 3324–3332. [Google Scholar] [CrossRef] [Green Version]
- Rui, J.; Li, J.; Wang, S.; An, J.; Liu, W.T.; Lin, Q.; Yang, Y.; He, Z.; Li, X. Responses of bacterial communities to simulated climate changes in alpine meadow soil of the Qinghai-Tibet plateau. Appl. Environ. Microbiol. 2015, 81, 6070–6077. [Google Scholar] [CrossRef]
- Trivedi, P. Microbial regulation of the soil carbon cycle: Evidence from gene|[ndash]|enzyme relationships. ISME J. 2016, 10, 2593–2604. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef]
- Zhong, Y.; Yan, W.; Wang, R.; Wang, W.; Shangguan, Z.P. Decreased occurrence of carbon cycle functions in microbial communities along with long-term secondary succession. Soil Biol. Biochem. 2018, 123, 207–217. [Google Scholar] [CrossRef]
- Degens, B.P.; Schipper, L.A.; Sparling, G.P.; Vojvodic-Vukovic, M. Decreases in organic C reserves in soils can reduce the catabolic diversity of soil microbial communities. Soil Biol. Biochem. 2000, 32, 189–196. [Google Scholar] [CrossRef]
- Tardy, V.; Spor, E.; Mathieu, O.; Eque, J.L.E.; Terrat, S.; Plassart, P.; Regnier, T.; Bardgett, P.; van Der Putten, W.H.; Roggero, P.P.; et al. Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol. Biochem. 2015, 90, 204–213. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Reed, S.C.; Keller, A.B.; Nemergut, D.R.; O’Neill, S.P.; Ostertag, R.; Vitousek, P.M. Litter quality versus soil microbial community controls over decomposition: A quantitative analysis. Oecologia 2014, 174, 283–294. [Google Scholar] [CrossRef]
- Xiao, Y.; Huang, Z.G.; Lu, X.G. Changes of soil labile organic carbon fractions and their relation to soil microbial characteristics in four typical wetlands of Sanjiang Plain, Northeast China. Ecol. Eng. 2015, 82, 381–389. [Google Scholar] [CrossRef]
- Ren, C.; Sun, P.; Kang, D.; Zhao, F.; Feng, Y.; Ren, G.; Han, X.; Yang, G. Responsiveness of soil nitrogen fractions and bacterial communities to afforestation in the Loess Hilly Region (LHR) of China. Sci. Rep. 2016, 6, 28469. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Wang, H.; Liu, X.; Zhao, X.; Lu, D.; Zhou, J.; Li, C. Changes in soil microbial community and organic carbon fractions under short-term straw return in a rice-wheat cropping system. Soil Till. Res. 2017, 165, 121–127. [Google Scholar] [CrossRef]
- Cusack, D.F.; Silver, W.L.; Torn, M.S.; Burton, S.D.; Firestone, M.K. Changes in microbial community characteristics and soil organic matter with nitrogen additions in two tropical forests. Ecology 2011, 92, 621–632. [Google Scholar] [CrossRef]
- Ng, E.L.; Patti, A.F.; Rose, M.T.; Schefe, C.; Wilkinson, K.; Smernik, R.J.; Smernik, T.R. Does the chemical nature of soil carbon drive the structure and functioning of soil microbial communities? Soil Biol. Biochem. 2014, 70, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Liu, S.; Scott, C.; Wang, J.; Shi, Z.; Huang, X.; Wen, Y.; Lu, L.; Cai, D.X. Soil microbial community composition rather than litter quality is linked with soil organic carbon chemical composition in plantations in subtropical China. J. Soil Sediment 2015, 15, 1094–1103. [Google Scholar] [CrossRef]




| Types | JM | QM | CB | LG | PK | F | p Value |
|---|---|---|---|---|---|---|---|
| pH | 5.70a | 4.89c | 4.99c | 5.40b | 5.48b | 111.797 | 0.0001 |
| Total C (g·kg−1) | 100.53a | 84.62b | 75.49c | 43.79d | 41.70d | 112.275 | 0.0001 |
| Total N (g·kg−1) | 7.80a | 7.38a | 5.47b | 3.85c | 3.58c | 29.718 | 0.0001 |
| C/N | 12.89ab | 11.64bc | 13.81a | 11.36c | 11.65bc | 6.489 | 0.0077 |
| Dissolved organic carbon (DOC) (mg·kg−1) | 171.80a | 158.71ab | 130.31b | 123.82b | 123.45b | 3.648 | 0.0441 |
| Microbial biomass carbon (MBC) (mg·kg−1) | 53.35a | 33.11b | 23.96c | 17.91d | 14.26e | 462.166 | 0.0001 |
| NH4+-N (mg·kg−1) | 4.63b | 11.84b | 42.74a | 5.12b | 4.43b | 14.955 | 0.0003 |
| NO3−-N (mg·kg−1) | 13.17ab | 14.35a | 7.78c | 10.94b | 6.24c | 15.595 | 0.0003 |
| Available N (mg·kg−1) | 57.15a | 41.25b | 43.60b | 33.35c | 28.04c | 14.99 | p < 0.001 |
| Total P (g·kg−1) | 0.93b | 0.74bc | 1.46a | 0.62c | 0.77bc | 26.59 | p < 0.001 |
| Available P (mg·kg−1) | 4.42ab | 2.39ab | 2.53ab | 1.21b | 5.65a | 1.99 | p = 0.172 |
| Alkyl C | O-alkyl C | Aromatic C | Carbonyl C | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0–45 Alkyl C | 45–60 Methoxyl C | 60–93 Alkyne C | 93–110 di-O-alkyl C | Total | 110–142 Aryl C | 142–160 Phenolic C | Total | 160–190 COO/NC=O | 190–220 Ketones/Aldehydes | Total | |
| JM | 17.14 ± 0.69cC | 9.26 ± 0.41aA | 28.53 ± 0.86bBC | 8.55 ± 0.58abA | 46.34 ± 1.72abAB | 14.45 ± 1.00aA | 6.24 ± 0.14aAB | 20.69 ± 1.13aA | 13.60 ± 0.99aA | 2.22 ± 1.22aA | 15.82 ± 2.19aA |
| QM | 18.15 ± 1.49bcBC | 9.41 ± 0.21aA | 29.59 ± 0.23aAB | 8.62 ± 0.22abA | 47.63 ± 0.43aAB | 14.26 ± 0.99abA | 5.99 ± 0.45abAB | 20.25 ± 1.43abA | 12.74 ± 0.59aA | 1.23 ± 0.13abA | 13.97 ± 0.71aA |
| LG | 20.62 ± 1.04aAB | 9.36 ± 0.31aA | 28.49 ± 0.16bBC | 8.77 ± 0.05aA | 46.62 ± 0.38abAB | 12.86 ± 0.08bcAB | 6.28 ± 0.08aA | 19.14 ± 0.06abcAB | 12.64 ± 0.80aA | 0.97 ± 0.59bA | 13.61 ± 1.39aA |
| PK | 21.84 ± 1.26aA | 9.31 ± 0.19aA | 28.12 ± 0.76bC | 7.93 ± 0.51bA | 45.36 ± 1.43bB | 12.38 ± 0.31cAB | 6.27 ± 0.22aAB | 18.65 ± 0.52bcAB | 12.70 ± 0.27aA | 1.45 ± 0.43abA | 14.15 ± 0.69aA |
| CB | 20.29 ± 1.62abABC | 9.51 ± 0.05aA | 30.08 ± 0.22aA | 8.53 ± 0.45abA | 48.12 ± 0.45aA | 11.87 ± 1.07cB | 5.47 ± 0.46bB | 17.34 ± 1.53cB | 12.57 ± 0.32aA | 1.69 ± 0.18abA | 14.26 ± 0.48aA |
| Samples | Alkyl C/O-alkyl C | Aromaticity | Aliphatic/Aromatic | Aromatic C /O-alkyl C | Aliphaticity |
|---|---|---|---|---|---|
| JM | 0.37 ± 0.02cB | 0.25 ± 0.01aA | 3.07 ± 0.18cB | 0.45 ± 0.02aA | 0.75 ± 0.01cB |
| QM | 0.38 ± 0.03cB | 0.24 ± 0.02abA | 3.26 ± 0.27bcB | 0.43 ± 0.03aA | 0.76 ± 0.02bcB |
| LG | 0.44 ± 0.02abAB | 0.22 ± 0.00bcAB | 3.51 ± 0.08bcAB | 0.41 ± 0.00aAB | 0.78 ± 0.00abAB |
| PK | 0.48 ± 0.04aA | 0.22 ± 0.01bcAB | 3.61 ± 0.12abAB | 0.41 ± 0.01aAB | 0.78 ± 0.01abAB |
| CB | 0.42 ± 0.04bcAB | 0.20 ± 0.02cB | 3.97 ± 0.43aA | 0.36 ± 0.03bB | 0.80 ± 0.02aA |
| Types | Cutin, Suberine and Wax Biosynthesis | Cellulose | Hemi-Cellulose | Amino Acids | Amino Sugars | Ketone | Aldehyde |
|---|---|---|---|---|---|---|---|
| QM | 0.03% | 23.49% | 3.04% | 0.03% | 1.15% | 0.33% | 2.88% |
| CB | 0.07% | 26.63% | 2.73% | 0.03% | 1.18% | 0.31% | 3.03% |
| PK | 0.06% | 23.65% | 2.45% | 0.04% | 1.24% | 0.30% | 2.70% |
| LG | 0.09% | 24.81% | 2.37% | 0.04% | 1.27% | 0.29% | 2.78% |
| JM | 0.03% | 22.51% | 3.17% | 0.03% | 1.18% | 0.31% | 2.79% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, J.; Zhu, W.; Zhou, Y.; Yin, Y. Soil Organic Carbon Chemical Functional Groups under Different Revegetation Types Are Coupled with Changes in the Microbial Community Composition and the Functional Genes. Forests 2019, 10, 240. https://doi.org/10.3390/f10030240
Deng J, Zhu W, Zhou Y, Yin Y. Soil Organic Carbon Chemical Functional Groups under Different Revegetation Types Are Coupled with Changes in the Microbial Community Composition and the Functional Genes. Forests. 2019; 10(3):240. https://doi.org/10.3390/f10030240
Chicago/Turabian StyleDeng, Jiaojiao, Wenxu Zhu, Yongbin Zhou, and You Yin. 2019. "Soil Organic Carbon Chemical Functional Groups under Different Revegetation Types Are Coupled with Changes in the Microbial Community Composition and the Functional Genes" Forests 10, no. 3: 240. https://doi.org/10.3390/f10030240
APA StyleDeng, J., Zhu, W., Zhou, Y., & Yin, Y. (2019). Soil Organic Carbon Chemical Functional Groups under Different Revegetation Types Are Coupled with Changes in the Microbial Community Composition and the Functional Genes. Forests, 10(3), 240. https://doi.org/10.3390/f10030240





