Genome-Wide Identification of WRKY Genes and Their Response to Cold Stress in Coffea canephora
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Cold Treatment
2.2. WRKY Gene Family Identification
2.3. Chromosomal Location of CcWRKYs
2.4. Sequence Alignment and Phylogenetic Analysis
2.5. Exon-Intron Structure Analysis of CcWRKY Ggenes
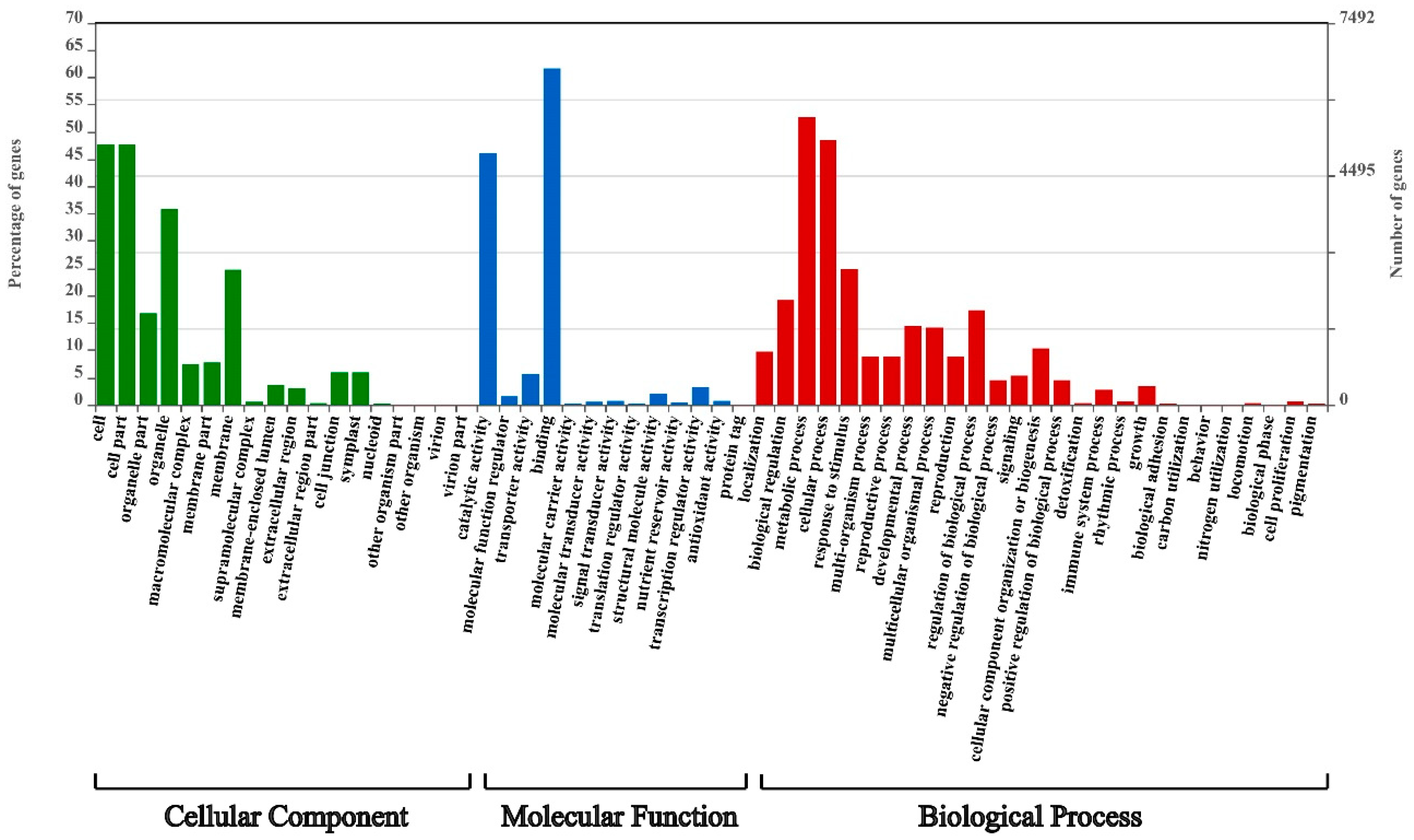
2.6. Conserved Motifs Search and GO Enrichment
2.7. Identification and Analysis of Potential CcWRKY Target Genes
2.8. qRT-PCR Analysis
3. Results
3.1. Isolation of the WRKY Genes in C. canephora
3.2. Phylogenetic Analysis and Motif Identification of the CcWRKY Family
3.3. Exon–Intron Organization of CcWRKY Genes
3.4. CcWRKYs Expression in Response to Cold Stress
3.5. CcWRKYs Response to Cold Stress by Mediating Multiple Biological Processes
3.6. Confirmations of the Expression Patterns of the Putative CcWRKY Target Genes under Cold Stress
4. Discussion
4.1. WRKY Genes in Coffee (Coffea spp.)
4.2. Phylogenetic Analysis, Conserved Motifs, and Structure of the CcWRKYs
4.3. Identification of CcWRKYs and Their Target’s Responses to Cold Stress in C. canephora
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ishiguro, S.; Nakamura, K. Characterization of a cdna encoding a novel DNA-binding protein, spf1, that recognizes sp8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Ulker, B.; Somssich, I.E. Wrky transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The wrky superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. Wrky transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Song, H.; Wang, P.; Lin, J.Y.; Zhao, C.; Bi, Y.; Wang, X. Genome-wide identification and characterization of wrky gene family in peanut. Front Plant Sci. 2016, 7, 534. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.; Liu, Y.; Shen, Q.J. The wrky gene family in rice (oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of wrky gene family in cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of wrky transcription factors in solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef]
- Guo, C.; Guo, R.; Xu, X.; Gao, M.; Li, X.; Song, J.; Zheng, Y.; Wang, X. Evolution and expression analysis of the grape (vitis vinifera l.) wrky gene family. J. Exp. Bot. 2014, 65, 1513–1528. [Google Scholar] [CrossRef]
- Pan, L.J.; Jiang, L. Identification and expression of the wrky transcription factors of carica papaya in response to abiotic and biotic stresses. Mol. Biol. Rep. 2014, 41, 1215–1225. [Google Scholar] [CrossRef]
- Kayum, M.A.; Jung, H.J.; Park, J.I.; Ahmed, N.U.; Saha, G.; Yang, T.J.; Nou, I.S. Identification and expression analysis of wrky family genes under biotic and abiotic stresses in brassica rapa. Mol. Genet. Genom. 2015, 290, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Tan, Q.; Sun, M.; Li, D.; Fu, X.; Chen, X.; Xiao, W.; Li, L.; Gao, D. Genome-wide identification of wrky family genes in peach and analysis of wrky expression during bud dormancy. Mol. Genet. Genom. 2016, 291, 1319–1332. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Hu, Y.; Han, Y.; Zhang, K.; Zhao, F.; Feng, J. The wrky transcription factors in the diploid woodland strawberry fragaria vesca: Identification and expression analysis under biotic and abiotic stresses. Plant Physiol. Biochem. 2016, 105, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, N.; Hu, R.; Xiang, F. Genome-wide identification of soybean wrky transcription factors in response to salt stress. SpringerPlus 2016, 5, 920. [Google Scholar] [CrossRef]
- He, C.; Teixeira da Silva, J.A.; Tan, J.; Zhang, J.; Pan, X.; Li, M.; Luo, J.; Duan, J. A genome-wide identification of the wrky family genes and a survey of potential wrky target genes in dendrobium officinale. Sci. Rep. 2017, 7, 9200. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Mao, M.; Wan, D.; Yang, Q.; Yang, F.; Mandlaa; Li, G.; Wang, R. Identification of the wrky gene family and functional analysis of two genes in caragana intermedia. BMC Plant Biol. 2018, 18, 31. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Jiang, W.; Yu, D. Male gametophyte-specific wrky34 transcription factor mediates cold sensitivity of mature pollen in arabidopsis. J. Exp. Bot. 2010, 61, 3901–3914. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, W.; Fang, L.; Sun, X.; Su, L.; Liang, Z.; Wang, N.; Londo, J.P.; Li, S.; Xin, H. Genome-wide identification of wrky family genes and their response to cold stress in vitis vinifera. BMC Plant Biol. 2014, 14, 103. [Google Scholar] [CrossRef]
- Li, P.; Song, A.; Gao, C.; Wang, L.; Wang, Y.; Sun, J.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum wrky gene cmwrky17 negatively regulates salt stress tolerance in transgenic chrysanthemum and arabidopsis plants. Plant Cell Rep. 2015, 34, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. Cswrky46, a wrky transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an aba-dependent manner. Plant Physiol. Biochem. 2016, 108, 478–487. [Google Scholar] [CrossRef]
- Kim, C.-Y.; Vo, K.T.X.; Nguyen, C.D.; Jeong, D.-H.; Lee, S.-K.; Kumar, M.; Kim, S.-R.; Park, S.-H.; Kim, J.-K.; Jeon, J.-S. Functional analysis of a cold-responsive rice wrky gene, oswrky71. Plant Biotechnol. Rep. 2016, 10, 13–23. [Google Scholar] [CrossRef]
- Yokotani, N.; Sato, Y.; Tanabe, S.; Chujo, T.; Shimizu, T.; Okada, K.; Yamane, H.; Shimono, M.; Sugano, S.; Takatsuji, H.; et al. Wrky76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013, 64, 5085–5097. [Google Scholar] [CrossRef]
- Tao, Z.; Liu, H.; Qiu, D.; Zhou, Y.; Li, X.; Xu, C.; Wang, S. A pair of allelic wrky genes play opposite roles in rice-bacteria interactions. Plant Physiol. 2009, 151, 936–948. [Google Scholar] [CrossRef]
- Marchive, C.; Mzid, R.; Deluc, L.; Barrieu, F.; Pirrello, J.; Gauthier, A.; Corio-Costet, M.F.; Regad, F.; Cailleteau, B.; Hamdi, S.; et al. Isolation and characterization of a vitis vinifera transcription factor, vvwrky1, and its effect on responses to fungal pathogens in transgenic tobacco plants. J. Exp. Bot. 2007, 58, 1999–2010. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. Transparent testa glabra2, a trichome and seed coat development gene of arabidopsis, encodes a wrky transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the wrky53 transcription factor and its role during leaf senescence in arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. Miniseed3 (mini3), a wrky family gene, and haiku2 (iku2), a leucine-rich repeat (lrr) kinase gene, are regulators of seed size in arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef]
- Miao, Y.; Zentgraf, U. A hect e3 ubiquitin ligase negatively regulates arabidopsis leaf senescence through degradation of the transcription factor wrky53. Plant J. 2010, 63, 179–188. [Google Scholar] [CrossRef]
- Grunewald, W.; De Smet, I.; Lewis, D.R.; Löfke, C.; Jansen, L.; Goeminne, G.; Vanden Bossche, R.; Karimi, M.; De Rybel, B.; Vanholme, B.; et al. Transcription factor wrky23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 1554–1559. [Google Scholar] [CrossRef]
- Luo, X.; Sun, X.; Liu, B.; Zhu, D.; Bai, X.; Cai, H.; Ji, W.; Cao, L.; Wu, J.; Wang, M.; et al. Ectopic expression of a wrky homolog from glycine soja alters flowering time in arabidopsis. PLoS ONE 2013, 8, e73295. [Google Scholar] [CrossRef]
- Li, W.; Wang, H.; Yu, D. Arabidopsis wrky transcription factors wrky12 and wrky13 oppositely regulate flowering under short-day conditions. Mol. Plant 2016, 9, 1492–1503. [Google Scholar] [CrossRef]
- Batista-Santos, P.; Lidon, F.C.; Fortunato, A.; Leitao, A.E.; Lopes, E.; Partelli, F.; Ribeiro, A.I.; Ramalho, J.C. The impact of cold on photosynthesis in genotypes of Coffea spp.—Photosystem sensitivity, photoprotective mechanisms and gene expression. J. Plant Physiol. 2011, 168, 792–806. [Google Scholar] [CrossRef]
- Ramalho, J.C.; DaMatta, F.M.; Rodrigues, A.P.; Scotti-Campos, P.; Pais, I.; Batista-Santos, P.; Partelli, F.L.; Ribeiro, A.; Lidon, F.C.; Leitão, A.E. Cold impact and acclimation response of coffea spp. Plants. Theor. Exp. Plant Physiol. 2014, 26, 5–18. [Google Scholar] [CrossRef]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef] [Green Version]
- Perrois, C.; Strickler, S.R.; Mathieu, G.; Lepelley, M.; Bedon, L.; Michaux, S.; Husson, J.; Mueller, L.; Privat, I. Differential regulation of caffeine metabolism in coffea arabica (arabica) and coffea canephora (robusta). Planta 2015, 241, 179–191. [Google Scholar] [CrossRef]
- Castro Caicedo, B.L.; Cortina Guerrero, H.A.; Roux, J.; Wingfield, M.J. New coffee (coffea arabica) genotypes derived from coffea canephora exhibiting high levels of resistance to leaf rust and ceratocystis canker. Trop. Plant Pathol. 2013, 38, 485–494. [Google Scholar] [CrossRef]
- Santos, T.B.; de Lima, R.B.; Nagashima, G.T.; Petkowicz, C.L.; Carpentieri-Pipolo, V.; Pereira, L.F.; Domingues, D.S.; Vieira, L.G. Galactinol synthase transcriptional profile in two genotypes of coffea canephora with contrasting tolerance to drought. Genet. Mol. Biol. 2015, 38, 182–190. [Google Scholar] [CrossRef]
- Fortunato, A.S.; Lidon, F.C.; Batista-Santos, P.; Eduardo Leitão, A.; Pais, I.P.; Ribeiro, A.I.; Cochicho Ramalho, J. Biochemical and molecular characterization of the antioxidative system of coffea sp. Under cold conditions in genotypes with contrasting tolerance. J. Plant Physiol. 2010, 167, 333–342. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. Hmmer web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal w and clustal x version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. Gsds 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. Agrigo v2.0: A go analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. Wego: A web tool for plotting go annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Van Bel, M.; Diels, T.; Vancaester, E.; Kreft, L.; Botzki, A.; Van de Peer, Y.; Coppens, F.; Vandepoele, K. Plaza 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018, 46, D1190–D1196. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−δδct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. Tm4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide survey and characterization of the wrky gene family in populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [Green Version]
- Ramiro, D.; Jalloul, A.; Petitot, A.; De Sa, M.F.G.; Maluf, M.P.; Fernandez, D. Identification of coffee wrky transcription factor genes and expression profiling in resistance responses to pathogens. Tree Genet. Genomes 2010, 6, 767–781. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, L.; Zhu, Y.; Li, Y.; Yan, H.; Xiang, Y. Comparative genomic analysis of the wrky iii gene family in populus, grape, arabidopsis and rice. Biol. Direct. 2015, 10, 48. [Google Scholar] [CrossRef]
- Karanja, B.K.; Fan, L.; Xu, L.; Wang, Y.; Zhu, X.; Tang, M.; Wang, R.; Zhang, F.; Muleke, E.M.; Liu, L. Genome-wide characterization of the wrky gene family in radish (raphanus sativus l.) reveals its critical functions under different abiotic stresses. Plant Cell Rep. 2017, 36, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]




| Motif | E Value | Sites | Width | Best Possible Match | Groups |
|---|---|---|---|---|---|
| 1 | 9.2 × 10−1082 | 49 | 31 | EVDILDDGYRWRKYGQKVVKGNPNPRSYYKC | I, II a, II b, II c, II d, II e, III |
| 2 | 1.2 × 10−850 | 49 | 29 | GCPVRKQVQRCLEDMSILITTYEGTHNHP | I, II a, II b, II c, II d, II e, III |
| 3 | 8.2 × 10−242 | 10 | 40 | AEDGYNWRKYGQKQVKGSEYPRSYYKCTHPNCPVKKKVER | I |
| 4 | 1.1 × 10−74 | 10 | 24 | LDGQITEIVYKGNHNHPKPQSTRR | I |
| 5 | 5.8 × 10−65 | 37 | 15 | QKKTRKPRFAFQTRS | I, II a, II b, II c, II d, III |
| 6 | 3.3 × 10−59 | 8 | 39 | MGEVMEENQKLRMHLDRVMKEYRALQMQFHDMVQQEPNK | II a, II b |
| 7 | 2.4 × 10−53 | 9 | 32 | SFADTLSAATAAITADPNFTAALAAAISSIIG | II a, II b |
| 8 | 1.7 × 10−31 | 6 | 29 | VAATAMASTTSAAASMLMSGSTTSTSGLL | II b |
| 9 | 5.3 × 10−21 | 6 | 33 | LTIPPGLSPTSFLESPVLLSNIKAEPSPTTGTF | I |
| 10 | 2.7 × 10−11 | 7 | 19 | ISASAPFPTVTLDLTQNPN | II a, II b |
| GO Term | Ontology | Description | No. 1 | FDR 2 |
|---|---|---|---|---|
| GO:0019320 | P | Hexose catabolic process | 9 | 5.9 × 10−5 |
| GO:0044262 | P | Cellular carbohydrate metabolic process | 17 | 5.9 × 10−5 |
| GO:0006006 | P | Glucose metabolic process | 10 | 5.9 × 10−5 |
| GO:0006007 | P | Glucose catabolic process | 9 | 5.9 × 10−5 |
| GO:0046365 | P | Monosaccharide catabolic process | 9 | 5.9 × 10−5 |
| GO:0006091 | P | Generation of precursor metabolites and energy | 11 | 5.9 × 10−5 |
| GO:0006096 | P | Glycolysis | 8 | 5.9 × 10−5 |
| GO:0005975 | P | Carbohydrate metabolic process | 28 | 5.9 × 10−5 |
| GO:0005544 | F | Calcium-dependent phospholipid binding | 6 | 6.0 × 10−5 |
| GO:0044275 | P | Cellular carbohydrate catabolic process | 9 | 1.1 × 10−4 |
| GO:0046164 | P | Alcohol catabolic process | 9 | 1.1 × 10−4 |
| GO:0016052 | P | Carbohydrate catabolic process | 10 | 1.5 × 10−4 |
| GO:0019318 | P | Hexose metabolic process | 10 | 1.5 × 10−4 |
| GO:0005996 | P | Monosaccharide metabolic process | 10 | 2.3 × 10−4 |
| GO:0006066 | P | Alcohol metabolic process | 11 | 4.0 × 10−4 |
| GO:0005509 | F | Calcium ion binding | 12 | 8.3 × 10−4 |
| GO:0005543 | F | Phospholipid binding | 6 | 9.9 × 10−4 |
| GO:0044282 | P | Small-molecule catabolic process | 9 | 1.8 × 10−3 |
| GO:0006414 | P | Translational elongation | 5 | 2.9 × 10−3 |
| GO:0044265 | P | Cellular macromolecule catabolic process | 10 | 3.2 × 10−3 |
| GO:0009057 | P | Macromolecule catabolic process | 11 | 5.2 × 10−3 |
| GO:0044248 | P | Cellular catabolic process | 11 | 9.3 × 10−3 |
| GO:0044281 | P | Small-molecule metabolic process | 23 | 1.6 × 10−2 |
| GO:0009058 | P | Biosynthetic process | 43 | 1.6 × 10−2 |
| GO:0035251 | F | UDP-glucosyltransferase activity | 5 | 1.8 × 10−2 |
| GO:0008289 | F | Lipid binding | 6 | 1.8 × 10−2 |
| GO:0009056 | P | Catabolic process | 12 | 2.2 × 10−2 |
| GO:0046527 | F | Glucosyltransferase activity | 5 | 2.5 × 10−2 |
| GO:0044237 | P | Cellular metabolic process | 77 | 2.9 × 10−2 |
| GO:0006633 | P | Fatty acid biosynthetic process | 5 | 3.6 × 10−2 |
| GO:0050794 | P | Regulation of cellular process | 23 | 4.3 × 10−2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Yang, Y.; Zhang, Z.; Xiao, Z.; Bai, X.; Gao, J.; Hur, Y.; Hao, S.; He, F. Genome-Wide Identification of WRKY Genes and Their Response to Cold Stress in Coffea canephora. Forests 2019, 10, 335. https://doi.org/10.3390/f10040335
Dong X, Yang Y, Zhang Z, Xiao Z, Bai X, Gao J, Hur Y, Hao S, He F. Genome-Wide Identification of WRKY Genes and Their Response to Cold Stress in Coffea canephora. Forests. 2019; 10(4):335. https://doi.org/10.3390/f10040335
Chicago/Turabian StyleDong, Xiangshu, Yanan Yang, Ziying Zhang, Ziwei Xiao, Xuehui Bai, Jing Gao, Yoonkang Hur, Shumei Hao, and Feifei He. 2019. "Genome-Wide Identification of WRKY Genes and Their Response to Cold Stress in Coffea canephora" Forests 10, no. 4: 335. https://doi.org/10.3390/f10040335





