Soil Depth Determines the Composition and Diversity of Bacterial and Archaeal Communities in a Poplar Plantation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Sampling
2.3. Soil Physicochemical Characteristics
2.4. DNA Extraction, Polymerase Chain Reaction (PCR) Amplification, and Illumina MiSeq Sequencing
2.5. Data Analysis
3. Results
3.1. Soil Physicochemical Characteristics with Depth
3.2. Alpha and Beta Diversity Patterns
3.3. Change in Bacterial and Archaeal Community Compositions with Depth
3.4. Relationship between Bacterial and Archaeal Community Compositions and Edaphic Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bardgett, R.D.; Freeman, C.; Ostle, N.J. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008, 2, 805–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive Earth‘s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Seuradge, B.J.; Oelbermann, M.; Neufeld, J.D. Depth-dependent influence of different land-use systems on bacterial biogeography. FEMS Microbiol. Ecol. 2017, 93, 239. [Google Scholar] [CrossRef] [PubMed]
- Eilers, K.G.; Debenport, S.; Anderson, S.; Fierer, N. Digging deeper to find unique microbial communities: The strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biol. Biochem. 2012, 50, 58–65. [Google Scholar] [CrossRef]
- Tang, Y.; Yu, G.; Zhang, X.; Wang, Q.; Ge, J.; Liu, S. Changes in nitrogen-cycling microbial communities with depth in temperate and subtropical forest soils. Appl. Soil Ecol. 2018, 124, 218–228. [Google Scholar] [CrossRef]
- FAOSTAT. Statistical Division. FAO. Available online: http://www.fao.org/faostat/en/#data/FO (accessed on 20 August 2018).
- Quesada, C.A.; Lloyd, J.; Schwarz, M.; Patino, S.; Baker, T.R.; Czimczik, C.; Fyllas, N.M.; Martinelli, L.; Nardoto, G.B.; Schmerler, J.; et al. Variations in chemical and physical properties of Amazon forest soils in relation to their genesis. Biogeosciences 2010, 7, 1515–1541. [Google Scholar] [CrossRef] [Green Version]
- Hansel, C.M.; Fendorf, S.; Jardine, P.M.; Francis, C.A. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 2008, 74, 1620–1633. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Breznak, J.A.; Schmidt, T.M. Isolation and characterization of soil bacteria that define Teniglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 2007, 73, 2708–2717. [Google Scholar] [CrossRef]
- Zhou, J.Z.; Xia, B.C.; Treves, D.S.; Wu, L.Y.; Marsh, T.L.; O‘Neill, R.V.; Palumbo, A.V.; Tiedje, J.M. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 2002, 68, 326–334. [Google Scholar] [CrossRef]
- Liu, D.; Huang, Y.; An, S.; Sun, H.; Bhople, P.; Chen, Z. Soil physicochemical and microbial characteristics of contrasting land-use types along soil depth gradients. Catena 2018, 162, 345–353. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P.; Holden, P.A. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef]
- Fritze, H.; Pietikäinen, J.; Pennanen, T. Distribution of microbial biomass and phospholipid fatty acids in Podzol profiles under coniferous forest. Eur. J. Soil Sci. 2010, 51, 565–573. [Google Scholar] [CrossRef]
- Tripathi, B.M.; Kim, M.; Kim, Y.; Byun, E.; Yang, J.-W.; Ahn, J.; Lee, Y.K. Variations in bacterial and archaeal communities along depth profiles of Alaskan soil cores. Sci. Rep. 2018, 8, 504. [Google Scholar] [CrossRef] [PubMed]
- Cervera, M.T.; Storme, V.; Ivens, B.; Gusmao, J.; Liu, B.H.; Hostyn, V.; Van Slycken, J.; Van Montagu, M.; Boerjan, W. Dense genetic linkage maps of three Populus species (Populus deltoides, P-nigra and P-trichocarpa) based on AFLP and microsatellite markers. Genetics 2001, 158, 787–809. [Google Scholar] [PubMed]
- Liang, W.-J.; Hu, H.-Q.; Liu, F.-J.; Zhang, D.-M. Research advance of biomass and carbon storage of poplar in China. J. For. Res. 2006, 17, 75–79. [Google Scholar] [CrossRef]
- IUSS Working Group, WRB. World Reference Base for Soil Resources 2014, Update 2015 International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; World Soil Resources Reports No. 106; FAO: Rome, Italy, 2015. [Google Scholar]
- Rayment, G.E.; Lyons, D.J. Soil Chemical Methods-Australasia; CSIRO Publishing: Clayton, Australia, 2010. [Google Scholar]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. Msystems 2016, 1, e00009-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, S.; Li, L.; Li, L.; Liu, Y.; Ren, Q.; Shi, M.; Liu, J.; Jiang, J.; Ma, H.; Huang, Z.; et al. Understanding the gut–kidney axis among biopsy-proven diabetic nephropathy, type 2 diabetes mellitus and healthy controls: An analysis of the gut microbiota composition. Acta Diabetol. 2019, 56, 581–592. [Google Scholar] [CrossRef]
- Wu, B.; Hou, S.; Peng, D.; Wang, Y.; Wang, C.; Xu, F.; Xu, H. Response of soil micro-ecology to different levels of cadmium in alkaline soil. Ecotoxicol. Environ. Saf. 2018, 166, 116–122. [Google Scholar] [CrossRef]
- Li, X.; Meng, D.; Li, J.; Yin, H.; Liu, H.; Liu, X.; Liu, X.; Cheng, C.; Xiao, Y.; Liu, Z.; et al. Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ. Pollut. 2017, 231, 908–917. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Good, I.J. The population frequencies of species and the estimation of population parameters. Biometrika 1953, 40, 237–264. [Google Scholar] [CrossRef]
- Bai, R.; Xi, D.; He, J.-Z.; Hu, H.-W.; Fang, Y.-T.; Zhang, L.-M. Activity, abundance and community structure of anammox bacteria along depth profiles in three different paddy soils. Soil Biol. Biochem. 2015, 91, 212–221. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, R.I.; Whiteley, A.S.; O‘Donnell, A.G.; Bailey, M.J. Influence of depth and sampling time on bacterial community structure in an upland grassland soil. FEMS Microbiol. Ecol. 2003, 43, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Levy-Booth, D.J.; Prescott, C.E.; Christiansen, J.R.; Grayston, S.J. Site preparation and fertilization of wet forests alter soil bacterial and fungal abundance, community profiles and CO2 fluxes. For. Ecol. Manag. 2016, 375, 159–171. [Google Scholar] [CrossRef]
- Deng, J.; Gu, Y.; Zhang, J.; Xue, K.; Qin, Y.; Yuan, M.; Yin, H.; He, Z.; Wu, L.; Schuur, E.A.; et al. Shifts of tundra bacterial and archaeal communities along a permafrost thaw gradient in Alaska. Mol. Ecol. 2015, 24, 222–234. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Allen, A.S.; Schimel, J.P.; Holden, P.A. Controls on microbial CO2 production: a comparison of surface and subsurface soil horizons. Glob. Chang. Biol. 2003, 9, 1322–1332. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Will, C.; Thuermer, A.; Wollherr, A.; Nacke, H.; Herold, N.; Schrumpf, M.; Gutknecht, J.; Wubet, T.; Buscot, F.; Daniel, R. Horizon-Specific Bacterial Community Composition of German Grassland Soils, as Revealed by Pyrosequencing-Based Analysis of 16S rRNA Genes. Appl. Environ. Microbiol. 2010, 76, 6751–6759. [Google Scholar] [CrossRef] [Green Version]
- Daims, H.; Nielsen, J.L.; Nielsen, P.H.; Schleifer, K.H.; Wagner, M. In situ characterization of Nitrospira-like nitrite oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 2001, 67, 5273–5284. [Google Scholar] [CrossRef] [PubMed]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.-F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, M.C.; Mori, T.; Rueckert, C.; Uria, A.R.; Helf, M.J.; Takada, K.; Gernert, C.; Steffens, U.A.; Heycke, N.; Schmitt, S.; et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014, 506, 58–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Badger, M.R.; Bek, E.J. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 2008, 59, 1525–1541. [Google Scholar] [CrossRef]
- Velázquez, E.; García-Fraile, P.; Ramírez-Bahena, M.; Peix, A.; Rivas, R. Proteobacteria Forming Nitrogen Fixing Symbiosis with Higher Plants; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2010; pp. 37–56. [Google Scholar]
- Imhoff, J.F.; Wiese, J. The Order Kiloniellales. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 301–306. [Google Scholar]
- Knief, C.; Lipski, A.; Dunfield, F.P. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 2003, 69, 6703–6714. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.; Wang, J.-T.; Deng, Y.; He, J.-Z.; Feng, K.; Zhang, L.-M. Microbial Community and Functional Structure Significantly Varied among Distinct Types of Paddy Soils but Responded Differently along Gradients of Soil Depth Layers. Front. Microbiol. 2017, 8, 945. [Google Scholar] [CrossRef]
- Hartmann, M.; Lee, S.; Hallam, S.J.; Mohn, W.W. Bacterial, archaeal and eukaryal community structures throughout soil horizons of harvested and naturally disturbed forest stands. Environ. Microbiol. 2009, 11, 3045–3062. [Google Scholar] [CrossRef]
- Li, C.; Yan, K.; Tang, L.; Jia, Z.; Li, Y. Change in deep soil microbial communities due to long-term fertilization. Soil Biol. Biochem. 2014, 75, 264–272. [Google Scholar] [CrossRef]
- Jetten, M.S.; Niftrik, L.; Strous, M.; Kartal, B.; Keltjens, J.T.; Op den Camp, H.J. Biochemistry and molecular biology of anammox bacteria. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 65–84. [Google Scholar] [CrossRef]
- Islam, T.; Jensen, S.; Reigstad, L.J.; Larsen, O.; Birkeland, N.-K. Methane oxidation at 55 degrees C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. USA 2008, 105, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Pol, A.; Heijmans, K.; Harhangi, H.R.; Tedesco, D.; Jetten, M.S.M.; den Camp, H.J.M.O. Methanotrophy below pH1 by a new Verrucomicrobia species. Nature 2007, 450, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, G.T.; Bates, S.T.; Eilers, K.G.; Lauber, C.L.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 2011, 43, 1450–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikkonen, A.; Santalahti, M.; Lappi, K.; Pulkkinen, A.-M.; Montonen, L.; Suominen, L. Bacterial and archaeal communities in long-term contaminated surface and subsurface soil evaluated through coextracted RNA and DNA. FEMS Microbiol. Ecol. 2014, 90, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Konneke, M.; Bernhard, A.E.; de la Torre, J.R.; Walker, C.B.; Waterbury, J.B.; Stahl, D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 2005, 437, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Treusch, A.H.; Leininger, S.; Kletzin, A.; Schuster, S.C.; Klenk, H.P.; Schleper, C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 2005, 7, 1985–1995. [Google Scholar] [CrossRef] [PubMed]
- Koenneke, M.; Schubert, D.M.; Brown, P.C.; Huegler, M.; Standfest, S.; Schwander, T.; Schada von Borzyskowski, L.; Erb, T.J.; Stahl, D.A.; Berg, I.A. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc. Natl. Acad. Sci. USA 2014, 111, 8239–8244. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.B.; de la Torre, J.R.; Klotz, M.G.; Urakawa, H.; Pinel, N.; Arp, D.J.; Brochier-Armanet, C.; Chain, P.S.; Chan, P.P.; Gollabgir, A. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. USA 2010, 107, 8818–8823. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Whitman, W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. In Incredible Anaerobes: From Physiology to Genomics to Fuels; Wiegel, J., Maier, R.J., Adams, M.W.W., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2008; pp. 171–189. [Google Scholar]
- Cabello, P.; Roldan, M.D.; Moreno-Vivian, C. Nitrate reduction and the nitrogen cycle in archaea. Microbiol. Sgm. 2004, 150, 3527–3546. [Google Scholar] [CrossRef]
- Jason, R.; Janet, L.S.; Christopher, R.S.; Robert, E.B. The natural history of nitrogen fixation. Mol. Biol. Evol. 2004, 21, 541–554. [Google Scholar]
- Michaelis, W.; Seifert, R.; Nauhaus, K.; Treude, T.; Thiel, V.; Blumenberg, M.; Knittel, K.; Gieseke, A.; Peterknecht, K.; Pape, T. Microbial reefs in the black sea fueled by anaerobic oxidation of methane. Science 2002, 297, 1013–1015. [Google Scholar] [CrossRef] [PubMed]
- Angel, R.; Claus, P.; Conrad, R. Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J. 2012, 6, 847–862. [Google Scholar] [CrossRef] [PubMed]
- Pesaro, M.; Widmer, F. Identification of novel Crenarchaeota and Euryarchaeota clusters associated with different depth layers of a forest soil. FEMS Microbiol. Ecol. 2002, 42, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Poplawski, A.B.; Martensson, L.; Wartiainen, I.; Rasmussen, U. Archaeal diversity and community structure in a Swedish barley field: Specificity of the EK510R/(EURY498) 16S rDNA primer. J. Microbiol. Methods 2007, 69, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.-W.; Zhang, L.-M.; Yuan, C.-L.; He, J.-Z. Contrasting Euryarchaeota communities between upland and paddy soils exhibited similar pH-impacted biogeographic patterns. Soil Biol. Biochem. 2013, 64, 18–27. [Google Scholar] [CrossRef]
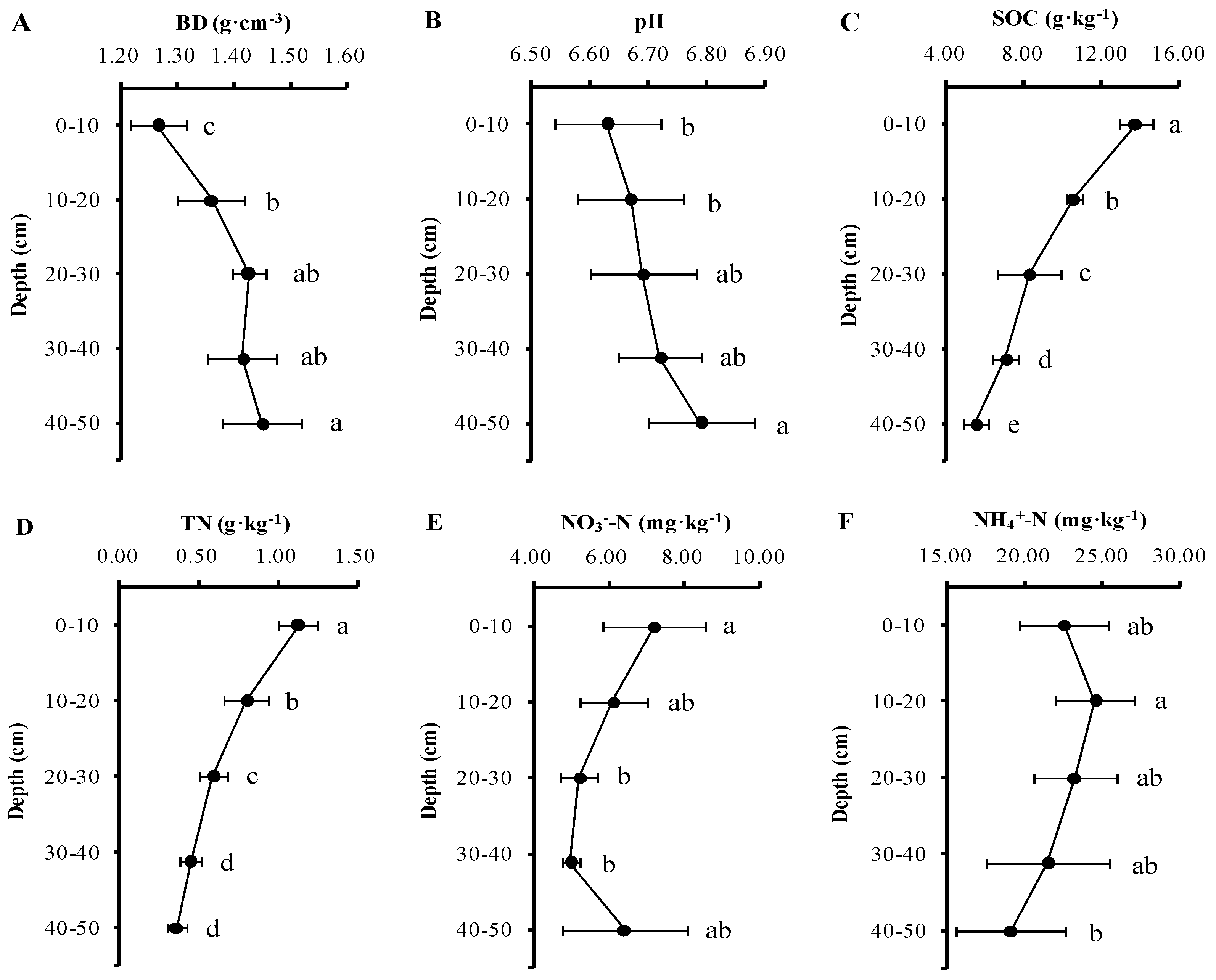




| Depth cm | Sequences | Shannon | Chao1 | Coverage % |
|---|---|---|---|---|
| D1 | 52417 | 6.58 | 2811.80 | 99.2 |
| D2 | 49206 | 6.44 | 2969.55 | 98.7 |
| D3 | 45874 | 6.23 | 2957.74 | 98.7 |
| D4 | 57609 | 6.01 | 2861.64 | 99.1 |
| D5 | 53086 | 5.89 | 2877.74 | 98.8 |
| BD | pH | SOC | TN | NO3−−N | NH4+−N | |
|---|---|---|---|---|---|---|
| Nitrospirae | 0.608 * | 0.646 ** | −0.736 ** | −0.741 ** | −0.558 * | −0.516 * |
| GAL15 | 0.605 * | 0.804 *** | −0.693 ** | −0.668 ** | −0.404 | −0.613 * |
| Euryarchaeota | 0.474 | 0.55 * | −0.636 * | −0.612 * | −0.622 * | −0.036 |
| unclassified_k__ norank_d__Bacteria | 0.554 * | 0.497 | −0.526 * | −0.478 | −0.765 ** | 0.119 |
| Latescibacteria | 0.542 * | 0.489 | −0.564 * | −0.497 | −0.676 ** | 0.086 |
| Chloroflexi | 0.476 | 0.534 * | −0.43 | −0.305 | −0.458 | −0.037 |
| SBR1093 | 0.328 | 0.358 | −0.458 | −0.405 | −0.472 | −0.001 |
| Saccharibacteria | −0.116 | −0.13 | 0.349 | 0.34 | 0.356 | 0.128 |
| Gemmatimonadetes | −0.59 * | −0.109 | 0.483 | 0.511 | 0.468 | 0.13 |
| Tectomicrobia | −0.533 * | −0.041 | 0.442 | 0.538 * | 0.23 | 0.29 |
| Thaumarchaeota | −0.574 * | −0.437 | 0.65 ** | 0.704 ** | 0.319 | 0.582 * |
| Proteobacteria | −0.765 ** | −0.253 | 0.619 * | 0.626 * | 0.584 * | 0.13 |
| Bacteroidetes | −0.847 *** | −0.374 | 0.733 ** | 0.713 ** | 0.696 ** | 0.037 |
| Planctomycetes | −0.131 | −0.121 | 0.148 | 0.186 | −0.202 | 0.596 * |
| Firmicutes | −0.202 | −0.211 | 0.043 | 0.107 | −0.18 | 0.742 ** |
| Verrucomicrobia | −0.361 | −0.265 | 0.323 | 0.313 | −0.046 | 0.567 * |
| Cyanobacteria | −0.233 | −0.366 | 0.214 | 0.196 | −0.036 | 0.614 * |
| Actinobacteria | −0.074 | 0.411 | 0.12 | 0.212 | 0.091 | −0.277 |
| Acidobacteria | −0.043 | 0.181 | −0.008 | 0.046 | −0.232 | 0.164 |
| Armatimonadetes | −0.069 | 0.15 | −0.016 | 0.049 | −0.204 | 0.101 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, H.; Guo, J.; Wang, W.; Song, X.; Yu, S. Soil Depth Determines the Composition and Diversity of Bacterial and Archaeal Communities in a Poplar Plantation. Forests 2019, 10, 550. https://doi.org/10.3390/f10070550
Feng H, Guo J, Wang W, Song X, Yu S. Soil Depth Determines the Composition and Diversity of Bacterial and Archaeal Communities in a Poplar Plantation. Forests. 2019; 10(7):550. https://doi.org/10.3390/f10070550
Chicago/Turabian StyleFeng, Huili, Jiahuan Guo, Weifeng Wang, Xinzhang Song, and Shuiqiang Yu. 2019. "Soil Depth Determines the Composition and Diversity of Bacterial and Archaeal Communities in a Poplar Plantation" Forests 10, no. 7: 550. https://doi.org/10.3390/f10070550
APA StyleFeng, H., Guo, J., Wang, W., Song, X., & Yu, S. (2019). Soil Depth Determines the Composition and Diversity of Bacterial and Archaeal Communities in a Poplar Plantation. Forests, 10(7), 550. https://doi.org/10.3390/f10070550





