Abstract
Erwinia billingiae S31R1 and Bacillus simplex S11R41, isolated from the rhizosphere of a healthy tree located in a Pinus radiata D. Don plantation with high presence of fungal pathogens, are antagonists of pine root rot fungi Heterobasidion annosum and Armillaria mellea in vitro and in young trees. For effective biocontrol of these pathogens, the bacteria must stably colonize P. radiata roots following their application. To determine root colonization patterns, the bacteria were transformed with stable plasmids encoding green fluorescent protein (GFP). Transformed E. billingiae was visualized on roots 24 days after soil inoculation by confocal and epifluorescence microscopy, and GFP was detected by ELISA 31 days after inoculation. The presence of E. billingiae microcolonies, in some cases in root intercellular spaces, suggests that bacterial growth was active and localized. Fluorescence of B. simplex S11R41 was visualized on P. radiata roots 31 days after inoculation and its colonization pattern changed from scattered cells to localized microcolonies. Although the populations decreased over time, microcolony formation and localization in specific regions of roots indicated that E. billingiae, normally considered to be an epiphyte, and B. simplex can stably colonize roots of P. radiata.
1. Introduction
Pinus radiata D. Don plantations are abundant in New Zealand, Chile, Australia, Spain and South Africa, covering over four million hectares [1]. In the Basque country (northern Spain), P. radiata supplies 46% (28.6 million m3) of the total wood stock [2] and occupies 33% of the total forest area [3], being the most extensively planted and highly productive conifer. It is however susceptible to several fungal pathogens, including root rot fungi Heterobasidion annosum (Fr.) Bref. sensu stricto and Armillaria (Fr.) Staude [4,5,6,7,8]. These fungi cause considerable economic losses in plantations due to tree mortality and/or volume losses [4,5,9] and yet there are few effective control measures available due to prohibitive legislation, high cost, environmental risks, high infection levels, and confounding environmental factors [10,11,12,13,14,15].
Soil bacteria that inhibit pathogenic fungi show promise as a component of integrated management strategies to protect forest trees against root rot diseases. Pine rhizosphere isolates Erwinia billingiae S31R1 and Bacillus simplex S11R41 are antagonists of H. annosum s.s. and A. mellea (Vahl) P. Kumm [16]. These strains inhibited the growth of H. annosum and A. mellea in vitro by 68%–99% [16]. Following infection with A. mellea, 36.5% of untreated one-year-old P. radiata died in comparison to 7.1% and 11.6% of trees treated with B. simplex S11R41 and E. billingiae S31R1, respectively [16]. While H. annosum was detected in 90% of young pine trees that were not inoculated with the bacteria, detection was reduced to 55% of trees treated with B. simplex S11R41 [16]. Thus, these strains may be applied as a nursery treatment to mitigate the damage caused by the fungal pathogens.
The success of bacteria as biological control agents depends on their ability to colonize host plants. Root colonization is a result of the attraction of the bacteria to plant roots by chemotaxis, attachment to root cells, and distribution along the roots. At root sites where nutrient exudation is abundant, bacterial populations increase and establish microcolonies [17]. Bacterial population size, root coverage, and colonization sites influence antagonism [18,19]. Very little is known about bacterial colonization of P. radiata roots or about root colonization by B. simplex and especially E. billingiae, which is mainly considered as an epiphyte [20]. B. simplex S11R41 and E. billingiae S31R1 were isolated from the rhizosphere of a healthy P. radiata tree located in a plantation with high presence of fungal pathogens [16], and are therefore expected to be well adapted to their host and environmental conditions, and to compete effectively with other indigenous microbes [21,22]. The objective of the research described herein was to assess the ability of these biocontrol bacterial strains to stably colonize P. radiata seedlings roots by labeling them with green fluorescent protein (GFP) and determining their location and survival following inoculation.
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
The bacterial strains E. billingiae S31R1 and B. simplex S11R41 (Table 1) were isolated from the roots of a healthy tree in a P. radiata plantation in Northern Spain as described by Mesanza et al. [16]. These strains were selected for further study based on their biocontrol potential which was previously characterized [16]. E. billingiae and B. simplex were routinely cultured on Luria Bertani (LB) medium at 30 °C. Cloning host Escherichia coli JM109 was grown on the same medium at 37 °C [23].

Table 1.
Bacterial strains and plasmids used in this study.
2.2. Plasmid Construction and Transformation of Biocontrol Bacterial Strains
Plasmid pNmI was constructed from the broad host range expression vector pRK415 [24], which carries the lac promoter to drive expression of gfp. To confer plasmid stability in the absence of selective pressure, the partitioning (par) locus of pTR102 [25] was inserted into the SmaI site located in the multiple cloning site of pRK415 to generate pRK415par. The gfp S65T gene was amplified from pVIK165 [26] using 300 nM of primer F: 5′AATTGCCTGCAGTGAGGAGGAAACAAAGATGAGTAAAGGAG, which included the restriction site for PstI (underlined), and 300 nM of primer R: 5′AATTGCTCTAGAGGTATCGATAAGCTTGATAGGCCCGGGAATTG, which included the restriction site for XbaI (underlined), 2.5 µL of 10× standard Taq buffer (NEB, Ipswich, MA, USA), 200 nM dNTPs (NEB), 1.25 U of Taq DNA polymerase (NEB) and 1 µL of template DNA in a final volume of 25 µL. The PCR conditions were as follows: 2 min at 94 °C, 30 cycles of 15 s at 94 °C, 15 s at 61 °C, and 30 s at 72 °C, and a final 1 min at 72 °C. Plasmid pRK415par and gfp amplicons were digested with PstI (Fermentas, Waltham, MA) and XbaI (NEB), and then purified using QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany). Ligation was carried out overnight at 4 °C in 20 µL reactions containing 17 ng of purified PCR products, 100 ng of purified pRK415par, 2 µL 10× buffer (NEB) and 1 µL T4 DNA ligase (NEB). Competent E. coli JM109 (Promega, Madison, WI, USA) cells were transformed with the ligation products and transformants selected on LB agar containing 5 µg/mL tetracycline. The presence of gfp in pNmI was confirmed by PCR using 300 nM of flanking primers F: 5′GTCAGTGGAGAGGGTGAAGG and R: 5′CCATGTGTAATCCCAGCAGC, 2.5 µL of 10× buffer (NEB), 200 nM dNTPs (NEB), 1.25 U of Taq DNA polymerase (NEB) and 1 µL of extracted plasmid DNA in a final volume of 25 µL. The PCR conditions were as follows: 2 min at 94 °C, 35 cycles of 15 s at 94 °C, 15 s at 58 °C, and 2 min at 72 °C, and a final 1 min at 72 °C.
E. billingiae S31R1 was transformed with pNmI by electroporation as follows. One ml of culture, grown overnight at 30 °C, was transferred into 50 mL LB and incubated with shaking until OD600nm reached a value of 0.5–0.6. After chilling on ice for 10 min in an ice-cold centrifuge tube, it was centrifuged at 4 °C (3000 rpm, 10 min) to pellet the cells; the pellet was finally resuspended in 0.5 mL ice cold 10% glycerol. On ice, 100 µL electrocompetent cells were mixed with 100 ng plasmid DNA, and electroporated (1.8 Kv). Immediately, 1 mL LB was added, and the cells were allowed to recover at 30 °C for 60 min. The presence of pNmI was selected on LB agar containing 4 µg/mL tetracycline. B. simplex S11R41 was transformed with pNF8, kindly provided by Dr. Rafael Garduno (Canadian Food Inspection Agency, NS), by protoplast electroporation method [28]. Transformants were selected on LB agar containing 10 µg/mL erythromycin. Plasmid pNF8 contains gfp mut1 (F64L and S65T), which is expressed from the constitutive Listeria monocytogenes dlt operon promoter [27,29].
2.3. Determination of Bacterial Generation Time and Plasmid Stability In Vitro
To determine if the presence of the plasmid affected the growth rate of the transformed bacterial strains, the generation time was measured in a short-term growth assay. Absorbance at 600 nm and fluorescence of GFP were measured every hour for a total of 9 h and then as a final point at 19 h. Bacterial overnight cultures were prepared as described above, diluted 100-fold in 25 mL LB containing tetracycline or erythromycin as appropriate, and incubated at 20 °C with shaking; three biological replicates and three technical replicates were measured, and generation times (g) were calculated for each replicate growth curve using the equation: g = 0.693/k, where k (growth rate constant) is determined from the slope of the exponential trendline [30]. Differences in generation times between the wild type and transformed strains were determined from an independent sample t-test using R software version 3.2.3 [31]. Values of p < 0.05 were considered significant. GFP production was measured in a SpectraMax M5 Microplate Reader (Molecular Devices, San Jose, CA, USA) at excitation wavelength of 485 nm and emission wavelength of 538 nm with a cut off at 530 nm.
A plasmid stability assay [25] was carried out for 120 h. Cultures of four biological replicates per bacterial strain, grown in the presence of antibiotics, were adjusted to an OD600nm of 0.5 and 100 µL of inocula were transferred to 3 mL LB without antibiotics. This was considered time 0. The cultures were incubated at 20 °C with shaking. Every 8 h, 100 µL of bacterial culture were subcultured in 3 mL LB to maintain the culture in the logarithmic phase of growth. OD600nm and fluorescence of GFP were measured as described above, and 100 µL of bacterial culture were plated on LB agar and incubated at 30 °C overnight for viable cell counts. Bacterial colonies were then replica plated on LB with the appropriate antibiotic to detect plasmid-containing colonies. Number of generations (n) was calculated from the count of plasmid-containing colonies using the equation n = 3.3 log (Nf/Ni), where Ni and Nf are the number of colonies at the beginning and end of the time interval, respectively [32].
2.4. Determination of Bacterial Colonization Patterns on P. radiata Seedling Roots
P. radiata seeds (Sheffield’s Seed Co. Inc., NY, USA) were surface sterilized as described by Wenny and Dumroese [33]. Seeds were soaked in sterile water for 24 h and stratified at 4 °C in wet sterilized vermiculite. After 15 days, they were placed in autoclaved sand Turface substrate (69% silica sand, 29% Turface, 2% MgCO3) in 12 cm × 3 cm pots (one seed per pot) and grown under controlled conditions (16 h photoperiod, day/night temperature of 23/17 °C) [34]. For each bacterial treatment, 24 three-month-old P. radiata seedlings were watered once with bacterial suspension (5 mL/pot) that was prepared by washing overnight cultures twice with 0.03 M MgSO4 and adjusting the final concentration to an OD600nm of 0.5 with the same solution. Sixteen seedlings were also treated with a control solution of 0.03 M MgSO4. Three seedlings per bacterial treatment and two control seedlings were harvested 1, 3, 5, 9, 12, 16, 24 and 31 days after inoculation (DAI). At each time point, roots were weighed and bacterial colonization assessed by epifluorescence microscopy and/or confocal microscopy. Epifluorescent imaging of P. radiata roots was done using a Leica M205FA stereo microscope with a DFC360 FX camera and GFP excitation/emission filters. Selected root areas were imaged using a Leica SP2 confocal microscope (Leica-microsystems, Concord, ON, Canada) fitted with 20 × 0.7 NA and 63 × 1.4 NA water immersion lenses, with GFP fluorescence data collected in one channel, and the endogenous red autofluorescence of the plant tissue collected in a second channel to provide anatomical context. Addition of scale bars, and minor adjustments to brightness/contrast were done using Fiji [35], and high-resolution composite images assembled using the pair-wise stitching plug-in.
For the quantification of GFP by sandwich enzyme-linked immunosorbent assay (ELISA), each seedling root was cut into about 10 pieces, and placed in a microfuge tube with 1 mL of phosphate buffered saline. Samples were vortexed for 2 min at medium speed before being macerated with a micropestle and placed in a sonication bath (FSH14H, Fisher Scientific, Waltham, MA, USA) for 2 min in order to separate bacterial clusters into single cells without causing cell lysis. Optimal sonication time was previously established by sonicating bacterial suspensions for different time periods and plating to quantify viable cells. In triplicate, 100 µL of 10-fold diluted samples were measured using a GFP PicoKine™ ELISA Kit (BosterBio, Pleasanton, CA, USA) according to the manufacturer’s instructions. Roots of the seedlings treated with 0.03 M MgSO4 were prepared in the same manner and used as negative controls. As positive controls B. simplex/pNF8 and E. billingiae/pNmI suspensions of known cell concentration were used. Quantity of GFP was normalized with the corresponding sample root weight. The entire experiment was repeated in plants harvested 1, 4, 9 and 16 DAI. In addition, serial dilutions of 100 µL macerate were plated on LB agar, grown overnight at 30 °C, and then replica plated on LB with the appropriate antibiotic to detect plasmid-containing colonies. The bacterial colonies were counted and fluorescence production was visualized using ChemiDoc™ MP Imaging System (Bio-Rad, Hercules, CA, USA). Simple linear regression was carried out to investigate the relationship between the GFP quantity, determined by ELISA from root samples inoculated with E. billingiae/pNmI or B. simplex/pNF8, and DAI. Differences were analyzed by Welch’s ANOVA with Games–Howell post-hoc test using R software. GFP concentration values were logarithmically transformed prior analysis.
3. Results
3.1. Bacterial Generation Time and Plasmid Stability
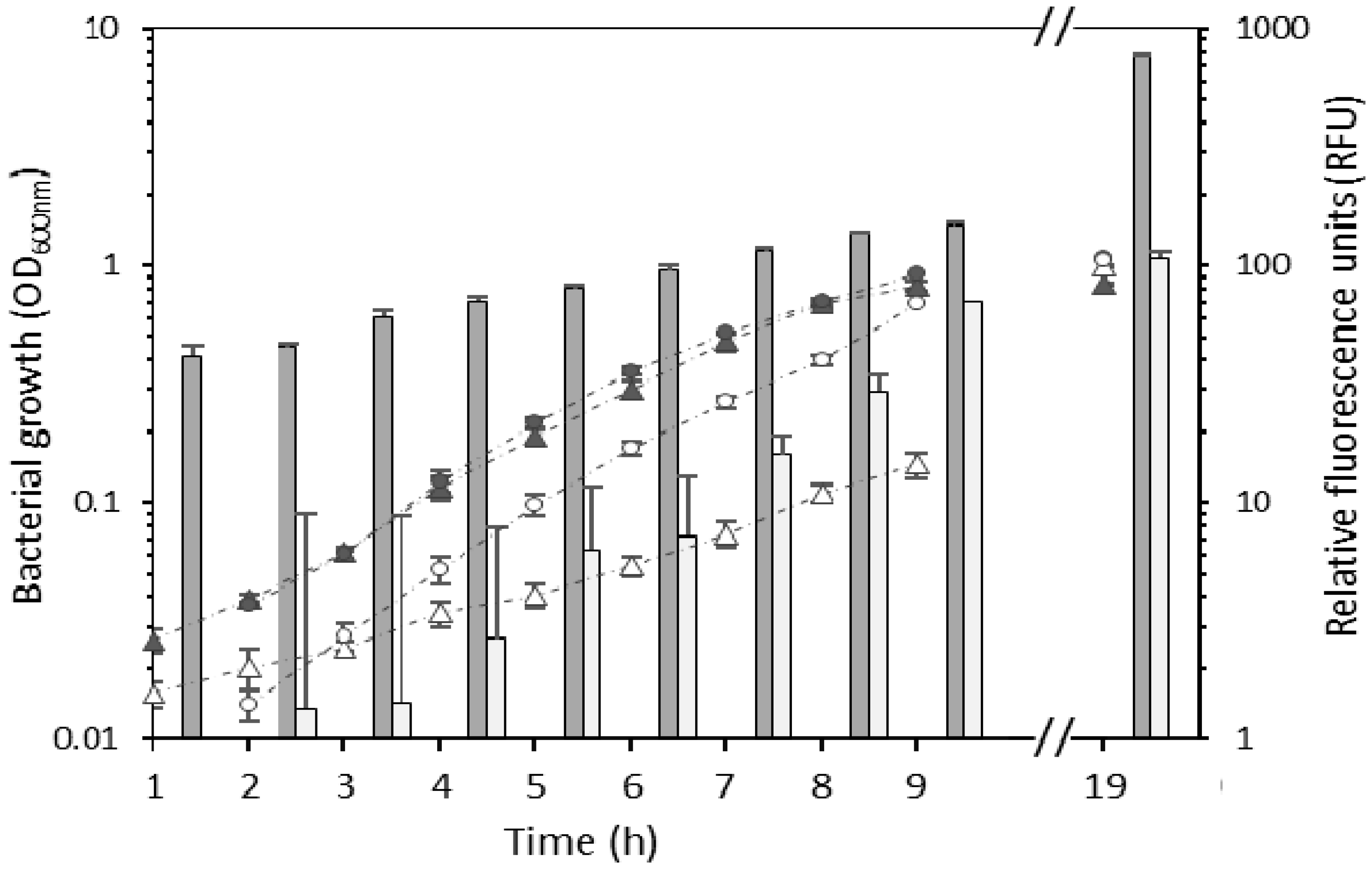
The biocontrol bacterial strains E. billingiae S31R1 and B. simplex S11R41 were successfully transformed with plasmids pNmI and pNF8, respectively. The presence of the plasmid significantly impacted the generation time of B. simplex (149.9 ± 8.1 min) compared to untransformed B. simplex (83 ± 3.8 min); t(16) = −20.7, p < 0.001 (Figure 1). However, by 19 h, both strains achieved similar densities. Generation times of wild type and transformed E. billingiae were not significantly different (69.1 ± 2.2 min and 66.3 ± 2 min, respectively) (Figure 1). Fluorescence of the transformed strains increased with culture density as expected (Figure 1). GFP fluorescence was higher in B. simplex/pNF8 than in E. billingiae/pNmI varying from 47.8-fold (at the 3 h time point) to 9.8-fold (at the 9 h time point) (Figure 1). This difference is likely due to differences in the strengths of the promoters used to drive gfp transcription in each strain, plasmid copy numbers (pNF8 is derived from pAT18, a high copy number plasmid [28], while pNmI is derived from pRK415, a low copy number plasmid [24]), and GFP expression and/or activity in heterologous hosts.
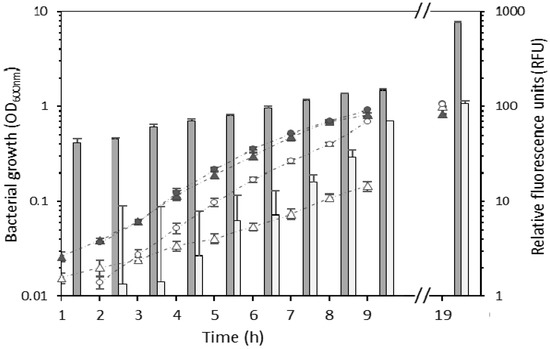
Figure 1.
Growth of wild type Erwinia billingiae S31R1 (grey circles), wild type Bacillus simplex S11R41 (grey triangles), E. billingiae/pNmI (white circles) and B. simplex/pNF8 (white triangles). Bars represent the mean of the relative fluorescence at each time point for B. simplex/pNF8 (dark grey) or E. billingiae/pNmI (light grey). Error bars indicate the pooled standard deviation of the three biological replicates and three technical replicates. Measurements were not taken in the time period corresponding to the horizontal axis break.
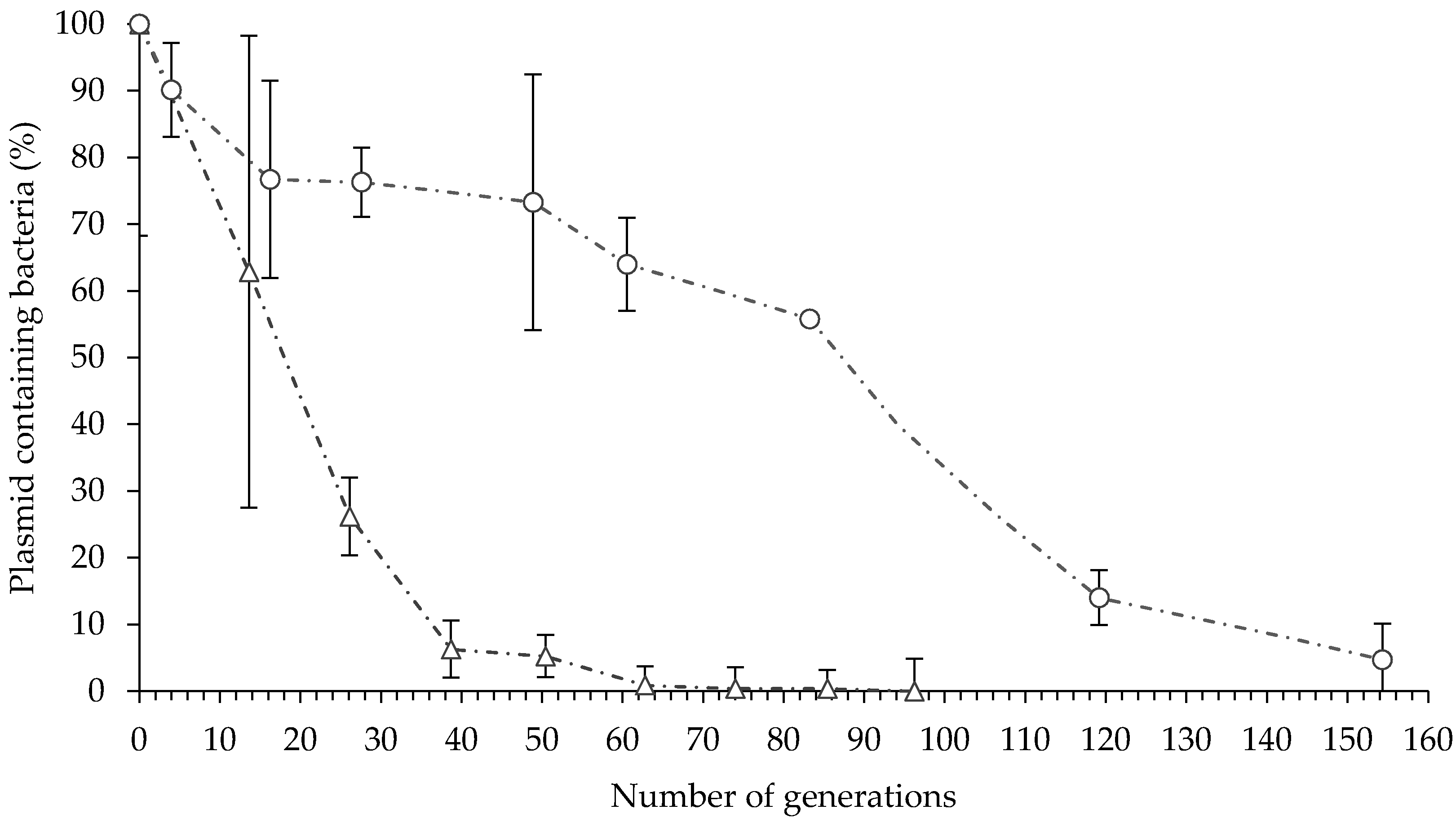
Plasmid-containing E. billingiae were detected after 154 generations in vitro in the absence of selective pressure, and after 88 generations, 50% of the transformed E. billingiae maintained the plasmid containing the gfp gene (Figure 2). In contrast, plasmid-containing B. simplex decreased sharply to 50% after 18 generations in the absence of selective antibiotics (Figure 2). Plasmid-containing B. simplex were not detected after 85 generations.
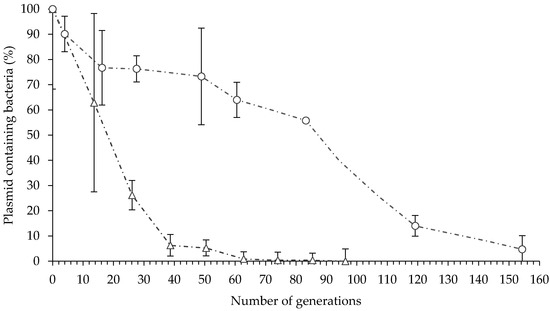
Figure 2.
In vitro plasmid maintenance in E. billingiae/pNmI (white circles) and B. simplex/pNF8 (white triangles). Error bars indicate the standard deviation of four biological replicates.
3.2. Bacterial Colonization Patterns on P. radiata Seedling Roots
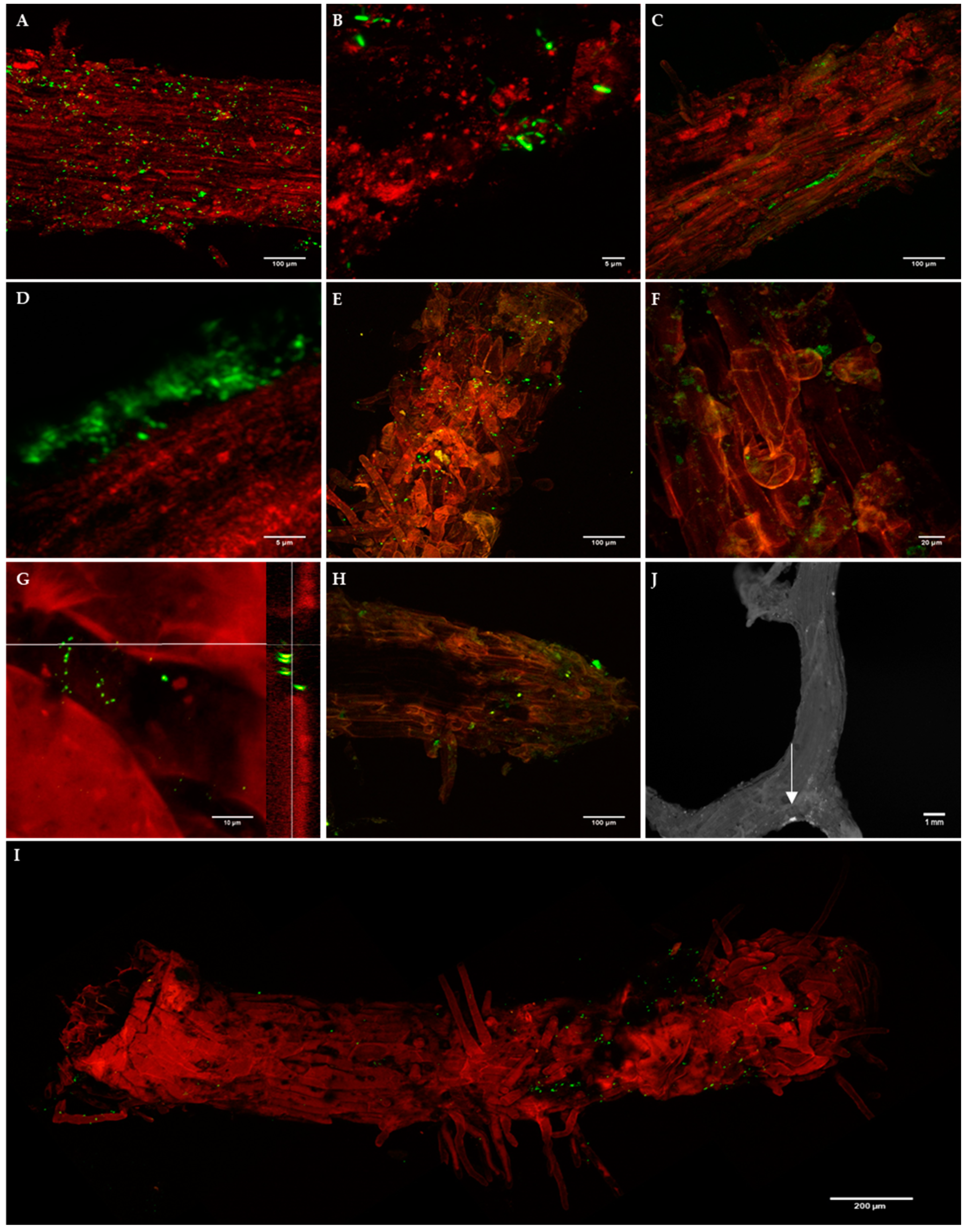
The presence of both biocontrol bacterial strains on P. radiata roots was detected by epifluorescence microscopy and/or confocal microscopy, although colonization patterns of E. billingiae and B. simplex were different (Figure 3). B. simplex was found from 1 DAI to 31 DAI, initially mainly on the surface of older root tissues, distributed randomly and sometimes in clusters (Figure 3A,B,E). An abundance of B. simplex was also noted on the substrate surrounding P. radiata roots. At a later colonization stage (24 DAI and 31 DAI), bacterial clusters appeared on new tissues and at the base of emerging lateral roots (Figure 3G,I,J). E. billingiae was found from 1 DAI to 16 DAI colonizing the intercellular spaces within the epidermal tissue and appeared in clusters of high density in deeper layers (Figure 3C,D,F,H). There was a distinct presence of the bacteria on root hairs. In general, both bacterial strains were more abundant on the upper portions of the roots although they were present all along the roots.
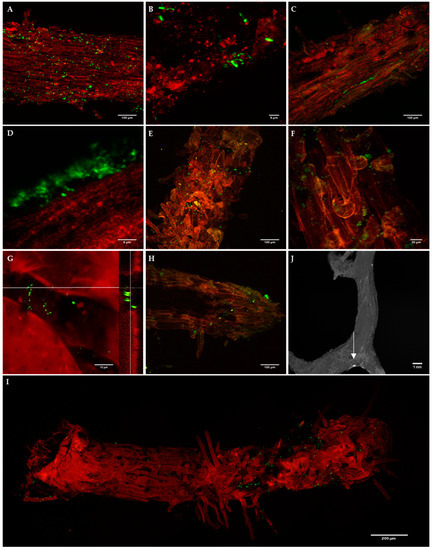
Figure 3.
Confocal images of B. simplex/pNF8 (A,B,E,G,J) and E. billingiae/pNmI (C,D,F,H) on Pinus radiata root epidermis at 1 days after inoculation (DAI) (A–D), 9 DAI (E,F) and 24 DAI (G,H,J). Epifluorescence image of B. simplex/pNF8 31 DAI (I); the white arrow points a bacterial cluster. G: Confocal image of B. simplex/pNF8 cluster and its orthogonal view at 24 DAI. Bacteria, green fluorescence; plant tissue, red autofluorescence to provide anatomical context. At 1 DAI B. simplex/pNF8 were found on the surface of primary and secondary roots forming small clusters or individually (A,B) without showing preferences for any root areas, after 9 DAI their presence was concentrated at areas abundant in root hairs (E,I) and at the base of emerging lateral roots (J). High density clusters of E. billingiae/pNmI appeared from 1 DAI (C,D,F,H) which also were present at root tips at 24 DAI (H). From 1 DAI presence of E. billingiae was also detected on root hairs (C,H).
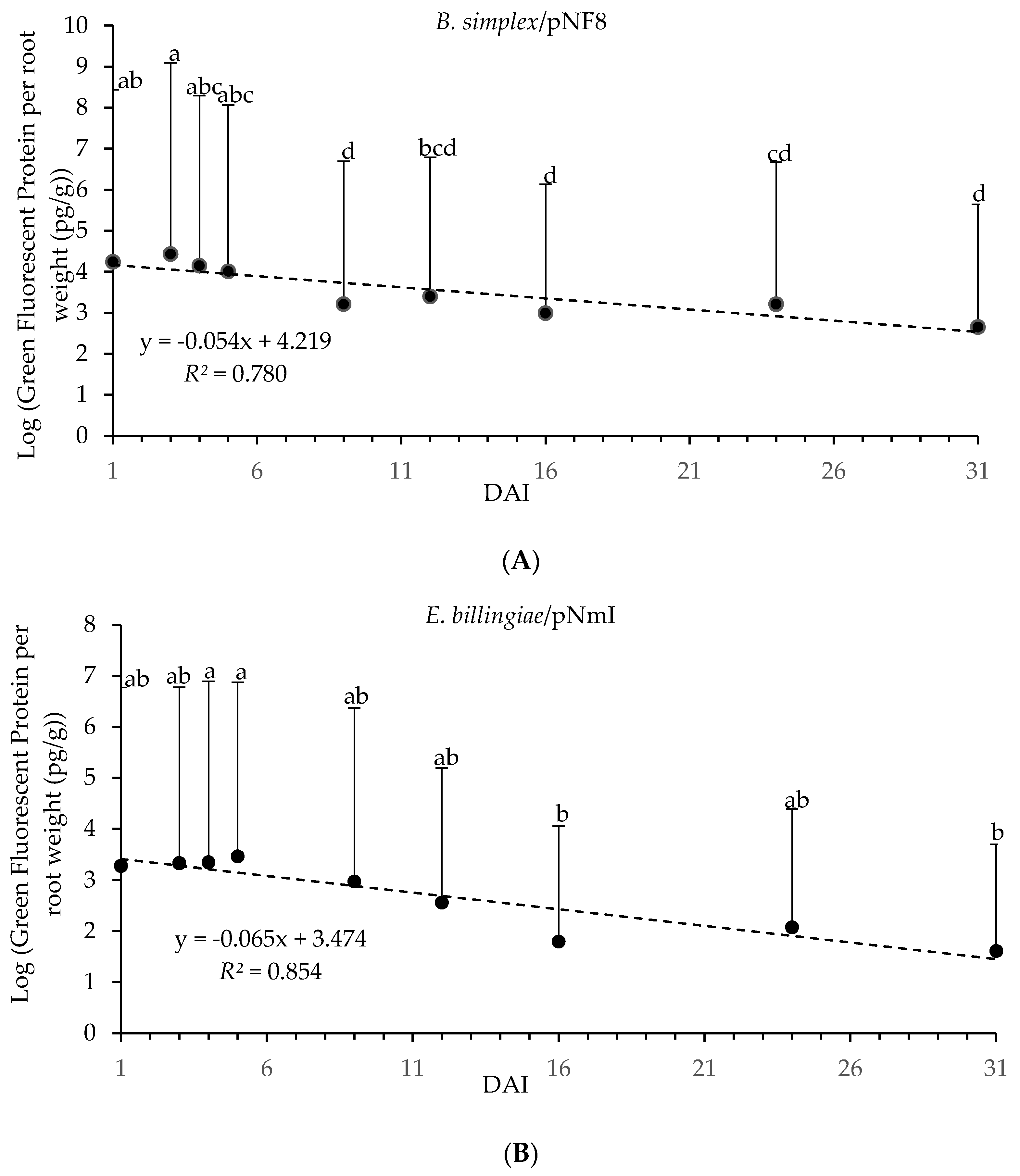
GFP from E. billingiae/pNmI and B. simplex/pNF8 in P. radiata seedling roots, quantified by ELISA, was detected throughout 31 DAI, and for both bacterial strains GFP content over time followed a similar trend (Figure 4). A significant regression equation obtained between GFP content and DAI was found for roots inoculated with E. billingiae/pNmI, (F(1,7) = 40.883, p < 0.01); and for roots inoculated with B. simplex/pNF8, (F(1,7) = 24.862, p < 0.01). DAI and GFP content were negatively correlated for E. billingiae/pNmI (β = −0.924, p < 0.001) and for B. simplex/pNF8 (β = −0.883, p < 0.01). Significant differences were found among GFP content related to DAI for plants inoculated with E. billingiae/pNmI (Welch’s F(8, 33.5) = 5.09, p < 0.01) and B. simplex/pNF8 (Welch’s F(8, 33.16) = 14.14, p < 0.001). Maximum significant values of GFP from E. billingiae/pNmI and B. simplex/pNF8 were observed at 4 and 5 DAI, and 3 DAI, respectively. The lowest values of GFP from E. billingiae/pNmI were detected at 16 and 31 DAI and from B. simplex/pNF8 at 9, 16 and 31 DAI (Figure 4). Plate count method showed a similar trend (data not shown).
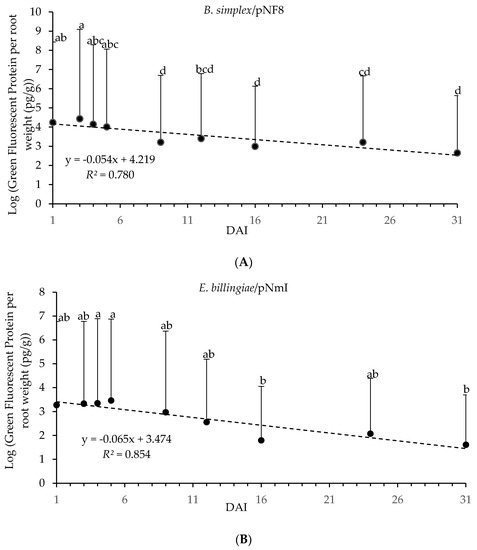
Figure 4.
Log transformed GFP content (pg) from E. billingiae/pNmI (B) and B. simplex/pNF8 (A) per root weight (g) over 31 days determined by ELISA. Error bars indicate the pooled standard deviation calculated from the standard deviation of the measured GFP concentration of the replicates and the standard deviation obtained for the plant weights. Statistically significant differences of p < 0.05 in GFP content in E. billingiae/pNmI (B) or B. simplex/pNF8 (A) over time are presented with lowercase letters.
4. Discussion
We report the colonization dynamics on P. radiata roots of two GFP-expressing biocontrol bacterial strains, E. billingiae S31R1 and B. simplex S11R41, using confocal microscopy and ELISAs. Both bacteria were labeled by transformation with plasmids containing the gfp gene, and bacterial growth rates and plasmid stability were assessed in vitro. Although integration of the gfp gene into the bacterial chromosome may increase its stability, fluorescence emitted by GFP expressed from a single chromosomal copy of gfp is often insufficient to visualize individual cells and chromosome insertion may disrupt the physiology and/or ecology of these natural isolates that are not well characterized at the molecular level.
E. billingiae S31R1 was transformed with the broad host range, low copy number vector pRK415 [24] containing the par locus which stabilized the plasmid in a number of bacterial hosts [36,37]. Plasmids containing the par locus were maintained in the absence of selective pressure in Gram-negative bacterial strains Escherichia coli, Agrobacterium tumefaciens, Azotobacter vinelandii, Acinetobacter calcoaceticus, Caulobacter crescentus, Rhizobium etli CE3, Sinorhizobium meliloti, and Pseudomonas spp. [36,38,39,40,41,42,43]. In the present study, 50% of the transformed E. billingiae maintained the plasmid after 88 generations in vitro; however, fewer than 10% of the E. billingiae cells retained the plasmid after 154 generations.
Compared to E. billingiae S31R1/pNmI cultures, B. simplex S11R41 carrying the high copy number pNF8 plasmid, which does not encode stabilization functions, was rapidly outgrown in log phase cultures by cells that did not retain the plasmid; 50% of the transformed bacteria did not carry the plasmid after 18 generations and plasmids were not detected after 85 generations. Maintenance of the plasmid imposed a metabolic burden on B. simplex as evidenced by the significant increase in generation time of the transformed strain compared to the wild type strain. Differences in growth rates between transformed and non-transformed bacterial strains can lead to plasmid loss from the population in long-term assays [44]. Cells unburdened by the plasmid grow faster and over time become the dominant strain in a culture grown in the absence of selective pressure for the plasmid. The rapid rate of pNF8 loss from the population was unexpected as Andersen et al. [45] reported that 30% of Listeria monocytogenes transformed with pNF8 retained the plasmid after 100 generations in vitro, and 100% of L. monocytogenes recovered from mice tissues after 48 h maintained the plasmid [28]. For Bacillus spp., 80% of a Bacillus cereus strain VA1 population lost the plasmid after four days of growth in log phase [46]; however, Hassen and Labuschagne [47] detected transformed B. simplex strains in tomato roots 10 DAI. Thus, the stability of pNF8 may depend on the bacterial host and growth conditions.
Fluorescent E. billingiae cells were visualized on P. radiata seedling roots up to 24 DAI by confocal microscopy and GFP was detectable by ELISA to the end of the experiment at 31 DAI, although levels were reduced 70-fold compared to the maximum levels detected at 5 DAI. Despite the observation that pNF8 in B. simplex was more unstable than pNmI in E. billingiae in vitro, transformed B. simplex S11R41 were observed on P. radiata roots over 31 DAI and GFP levels decreased 60-fold from the highest levels obtained at 3 DAI. The increase in GFP concentration on roots in the first days following soil application may be explained by colonization of motile cells and proliferation of attached cells [48]. The observed decrease in both bacterial populations on the seedling roots after 5 days is consistent with other studies and may be due to exhaustion of nutrients, deposition of excreted products, and insufficient growth of roots due to space limitations in small containers [43]. Considering that bacterial generation times are generally much longer on roots (7.2 h on average for bacteria on P. radiata roots [49]) than in vitro, and that there was no significant difference in the generation times of E. billingiae S31R1/pNmI and the wild type strain in vitro, it is unlikely that E. billingiae cells without pNmI would rapidly overtake the root population, although plasmid loss may also have contributed to the decrease in GFP contents, especially in the later measurements (24 and 31 DAI). Bacillus spp. also generally do not proliferate rapidly in the rhizosphere; generation time on P. radiata roots was 39 h in one study [49]. However, the instability of pNF8 observed in B. simplex might also have affected GFP content, especially at later timepoints (24 and 31 DAI).
E. billingiae is primarily considered to be an epiphyte, having been isolated from leaves, fruits, or trunks of pear, apple, chestnut and hawthorn, often as a secondary colonizer of diseased tissues [50,51,52,53]. Here, we observed E. billingiae S31R1 microcolonies on P. radiata roots as early as 1 DAI and up to 24 DAI, which suggests that this bacterium rapidly colonizes and proliferates in pine roots. The lower resolution of confocal images of E. billingiae clusters compared to those for B. simplex may be due to enclosure of microcolonies in a mucoid-like layer. Bacteria have been found in small, discontinuous biofilms on the surface of many plant roots including pine (e.g., [54]). In some cases, E. billingiae S31R1 was positioned in intercellular spaces suggesting that this strain may be able to colonize pine root endophytically. At a later colonization stage (24 DAI), E. billingiae was detected on root hairs and root tips which are sites with high nutrient levels due to release of exudates, secretions and lysates [55].
B. simplex has been isolated from the rhizosphere of a variety of plants including wheat, grass, apple and raspberry, and can colonize the roots of several crops enhancing their growth [47,56,57,58]. B. simplex KBSIF-3 cells were visualized along the length of tomato roots 7 DAI [47]. The colonization pattern of B. simplex S11R41 on P. radiata roots was also initially scattered along the surface of the roots but then became localized in clusters of cells at the base of emerging lateral roots and in the root elongation zone and root hairs by 31 DAI. Some of the cells were attached by contact of the cell pole with the root surface, which is believed to be transient [59], and others were attached along their long axis, a more permanent interaction. As observed in other root colonization studies [60], B. simplex did not appear to form biofilms.
5. Conclusions
Although fluorescent B. simplex S11R41/pNF8 and E. billingiae S31R1/pNmI populations decreased over time on P. radiata roots, bacterial microcolony formation and localization in specific plant tissues indicates that both bacterial strains can stably colonize roots of P. radiata seedlings. Plasmid pNmI containing the par locus was more stable than plasmid pNF8, however this could also be related to differences in host compatibility. The detection of GFP by ELISA over the duration of the experiment suggests that it can be used as a complementary tool to confocal microscopy for detection of gfp-transformed bacterial strains in planta over time.
Author Contributions
Conceptualization, N.M. and C.L.P.; formal analysis, N.M., E.I. and C.L.P.; investigation, N.M., B.D.C., T.J.D.C., E.I. and C.L.P.; methodology, N.M., B.D.C., T.J.D.C., E.I. and C.L.P.; visualization, N.M. and B.D.C.; writing—original draft, N.M., B.D.C., T.J.D.C., E.I. and C.L.P.; writing—review and editing, N.M., B.D.C., T.J.D.C., E.I. and C.L.P.
Funding
This research was funded by the Canadian Tree Fund: Jack Kimmel research grant, the Project: Healthy Forest: LIFE14 ENV/ES/000179 and the Project: RTA 2017-00063-CO4-03INIA.
Acknowledgments
We thank Michelle Perley for the construction of plasmid pRK415par, and Matthew Methven, Greenhouse Tech/Facilities Manager, and Rossella Calvaruso at UNB (Fredericton, NB, Canada), and Ion Ortueta, María Eugenia Sanz and Raquel Marquinez at NEIKER-Tecnalia (Arkaute, Álava, Spain) for their technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mead, D.J. Sustainable Management of Pinus radiata Plantations; FAO Forestry Paper No. 170; FAO: Rome, Italy, 2013. [Google Scholar]
- HAZI. El Bosque Vasco en Cifras. Available online: http://www.nasdap.ejgv.euskadi.eus/contenidos/informacion/inventario_forestal_2011/es_agripes/adjuntos/El%20bosque%20vasco%20en%20cifras%202011_2012.pdf (accessed on 29 May 2019).
- Murua, J.R.; Albiac, J.; Astorkiza, I.; Eguía, B.; Ferrero, A.; Moreno, J. Libro Blanco del Sector de la Madera: Actividad Forestal e Industria de Transformación de la Madera: Evolución Reciente y Perspectivas en Euskadi, 1st ed.; Eusko Jaurlaritzaren Argitalpen Zerbitzu Nagusia: Vitoria-Gasteiz, Spain, 2016. [Google Scholar]
- MacKenzie, M. Infection changes and volume loss in a 19-year-old Pinus radiata stand affected by Armillaria root rot. N. Z. J. For. Res. 1987, 17, 100–108. [Google Scholar]
- Hood, I.A.; Sandberg, C.J. Armillaria populations in a Pinus radiata plantation on a former indigenous rainforest site. N. Z. J. For. Sci. 1993, 23, 62–77. [Google Scholar]
- Mesanza, N.; Iturritxa, E. Root and butt rot caused by Heterobasidion annosum in atlantic coniferous ecosystems of Spain. For. Pathol. 2012, 42, 514–520. [Google Scholar] [CrossRef]
- Doğmuş-Lehtijärvi, H.T.; Erdoğan, R.C.; Lehtijärvi, A.; Woodward, S.; Aday Kaya, A.G. Pathogenicity of Heterobasidion annosum (Fr.) Bref. sensu stricto on coniferous tree species in Turkey. For. Pathol. 2016, 46, 22–28. [Google Scholar] [CrossRef]
- Mesanza, N.; Patten, C.L.; Iturritxa, E. Distribution and characterization of Armillaria complex in Atlantic forest ecosystems of Spain. Forests 2017, 8, 235. [Google Scholar] [CrossRef]
- Garbelotto, M.; Gonthier, P. Biology, epidemiology, and control of Heterobasidion species worldwide. Annu. Rev. Phytopathol. 2013, 51, 39–59. [Google Scholar] [CrossRef]
- Pratt, J.E.; Lloyd, J.D. The use of disodium octaborate tetrahydrate to control conifer butt rot caused by Heterobasidion annosum. Proc. Crop Prot. North. Br. 1996, 207–212. [Google Scholar]
- Johansson, S.M.; Pratt, J.E.; Asiegbu, F.O. Treatment of Norway spruce and Scots pine stumps with urea against the root and butt rot fungus Heterobasidion annosum-possible modes of action. For. Ecol. Manag. 2002, 157, 87–100. [Google Scholar] [CrossRef]
- Asiegbu, F.O.; Adomas, A.; Stenlid, J. Conifer root and butt rot caused by Heterobasidion annosum (Fr.) Bref. s.l. Mol. Plant Pathol. 2005, 6, 395–409. [Google Scholar] [CrossRef]
- Baumgartner, K.; Coetzee, M.P.A.; Hoffmeister, D. Secrets of the subterranean pathosystem of Armillaria. Mol. Plant Pathol. 2011, 12, 515–534. [Google Scholar] [CrossRef]
- Gonthier, P.; Thor, M. Annosus root and butt rots. In Infectious Forest Diseases; Gonthier, P., Nicolotti, G., Eds.; CAB International: Wallingford, UK, 2013; pp. 128–158. [Google Scholar]
- Shaw III, C.G.; Roth, L.F. Control of Armillaria root rot in managed coniferous forests: A literature review. Eur. J. For. Pathol. 1978, 8, 163–174. [Google Scholar] [CrossRef]
- Mesanza, N.; Iturritxa, E.; Patten, C.L. Native rhizobacteria as biocontrol agents of Heterobasidion annosum s.s. and Armillaria mellea infection of Pinus radiata. Biol. Control 2016, 101, 8–16. [Google Scholar] [CrossRef]
- Lam, S.T.; Ellis, D.M.; Ligon, J.M. Genetic approaches for studying rhizosphere colonisation. Plant Soil 1990, 129, 11–18. [Google Scholar] [CrossRef]
- Chin-A-Woeng, T.F.C.; de Priester, W.; van der Bij, A.J.; Lugtenberg, B.J. Description of the colonization of a gnotobiotic tomato rhizosphere by Pseudomonas fluorescens biocontrol strain WCS365, using scanning electron microscopy. Mol. Plant-Microbe Interact. 1997, 10, 79–86. [Google Scholar] [CrossRef]
- Prieto, P.; Mercado-Blanco, J. Endophytic behaviour of biocontrol strain Pseudomonas fluorescens PICF7 in olive roots using confocal microscopy. FEMS Microbiol Ecol. 2008, 64, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Palacio-Bielsa, A.; Roselló, M.; Llop, P.; López, M.M. Erwinia spp. from pome fruit trees: Similarities and differences among pathogenic and non-pathogenic species. Trees 2012, 26, 13–29. [Google Scholar] [CrossRef]
- Köberl, M.; Ramadan, E.M.; Adam, M.; Cardinale, M.; Hallmann, J.; Heuer, H.; Smalla, K.; Berg, G. Bacillus and Streptomyces were selected as broad-spectrum antagonists against soilborne pathogens from arid areas in Egypt. FEMS Microbiol. Lett. 2013, 342, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, R.; Luu, V.T.; Weinhold, A.; Goldberg, J.; Oh, Y.; Baldwin, I.T. Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc. Natl. Acad. Sci. USA 2015, 112, E5013–E5020. [Google Scholar] [CrossRef] [PubMed]
- Yanisch-Perron, C.; Vieira, J.; Messing, J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985, 33, 103–119. [Google Scholar] [CrossRef]
- Keen, N.T.; Tamaki, S.; Kobayashi, D.Y.; Trollinger, D.J. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 1988, 70, 191–197. [Google Scholar] [CrossRef]
- Weinstein, M.; Roberts, R.C.; Helinski, D.R. A region of the broad-host-range plasmid RK2 causes stable in planta inheritance of plasmids in Rhizobium meliloti cells isolated from alfalfa root nodules. J. Bacteriol. 1992, 174, 7486–7489. [Google Scholar] [CrossRef] [PubMed]
- Kalogeraki, V.S.; Winans, S.C. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 1997, 188, 69–75. [Google Scholar] [CrossRef]
- Fortineau, N.; Trieu-Cuot, P.; Gaillot, O.; Pellegrini, E.; Berche, P.; Gaillard, J.L. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res. Microbiol. 2000, 151, 353–360. [Google Scholar] [CrossRef]
- Romero, D.; Pérez-García, A.; Veening, J.W.; de Vicente, A.; Kuipers, O.P. Transformation of undomesticated strains of Bacillus subtilis by protoplast electroporation. J. Microbiol. Meth. 2006, 66, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, G.; Doyle, M.P. Green Fluorescent Protein Labeling of Listeria, Salmonella, and Escherichia coli O157:H7 for Safety-Related Studies. PLoS ONE 2011, 6, e18083. [Google Scholar] [CrossRef] [PubMed]
- White, D. The Physiology and Biochemistry of Prokaryotes, 3rd ed.; Oxford University Press: New York, NY, USA, 2007. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Boyd, C.E. Water Quality: An Introduction; Kluwer Academic Publishers: Boston, MA, USA, 2000. [Google Scholar]
- Wenny, D.L.; Dumroese, R.K. Germination of conifer seeds surface sterilized with bleach. Tree Planters’ Notes 1987, 38, 18–21. [Google Scholar]
- Chanway, C.P.; Radley, R.A.; Hall, F.B. Inoculation of conifer seeds with plant growth promoting Bacillus strains causes increased seedlings emergence and biomass. Soil Biol. Biochem. 1991, 23, 575–590. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.C.; Burioni, R.; Helinski, D.R. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. J. Bacteriol. 1990, 172, 6204–6216. [Google Scholar] [CrossRef]
- Sobecky, P.A.; Easter, C.L.; Bear, P.D.; Helinski, D.R. Characterization of the stable maintenance properties of the par region of broad-Host-range plasmid RK2. J. Bacteriol. 1996, 178, 2086–2093. [Google Scholar] [CrossRef][Green Version]
- Davis, T.L.; Helinski, D.R.; Roberts, R.C. Transcription and autoregulation of the stabilizing functions of broad-host-range plasmid RK2 in Escherichia coli, Agrobacterium tumefaciens, and Pseudomonas aeruginosa. Mol. Microbiol. 1992, 6, 1981–1994. [Google Scholar] [CrossRef]
- Sia, E.A.; Roberts, R.C.; Easter, C.; Helinski, D.R.; Figurski, D.H. Different relative importances of the par operons and the effect of conjugal transfer on the maintenance of intact promiscuous plasmid RK2. J. Bacteriol. 1995, 177, 2789–2797. [Google Scholar] [CrossRef]
- Taté, R.; Riccio, A.; Caputo, E.; Cermola, M.; Favre, R.; Patriarca, E.J. The Rhizobium etli trpB gene is essential for an effective symbiotic interaction with Phaseolus vulgaris. Mol. Plant-Microbe Interact. 1999, 12, 926–933. [Google Scholar] [CrossRef]
- Dombrecht, B.; Vanderleyden, J.; Michiels, J. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol. Plant-Microbe Interact. 2001, 14, 426–430. [Google Scholar] [CrossRef]
- Deng, W.L.; Rehm, A.H.; Charkowski, A.O.; Rojas, C.M.; Collmer, A. Pseudomonas syringae exchangeable effector loci: Sequence diversity in representative pathovars and virulence function in P. syringae pv. syringae B728a. J. Bacteriol. 2003, 8, 2592–2602. [Google Scholar] [CrossRef]
- Rincon, A.; Ruiz-Diez, B.; Garcia-Fraile, S.; Garcia, J.A.; Fernandez-Pascual, M.; Pueyo, J.J.; de Felipe, M.R. Colonisation of Pinus halepensis roots by Pseudomonas fluorescens and interaction with the ectomycorrhizal fungus Suillus granulatus. FEMS Microbiol. Ecol. 2005, 51, 303–311. [Google Scholar] [CrossRef]
- Lau, B.T.C.; Malkus, P.; Paulsson, J. New quantitative methods for measuring plasmid loss rates reveal unexpected stability. Plasmid 2013, 70, 353–361. [Google Scholar] [CrossRef]
- Andersen, J.B.; Roldgaard, B.B.; Lindner, A.B.; Christensen, B.B.; Licht, T.R. Construction of a multiple fluorescence labelling system for use in co-invasion studies of Listeria monocytogenes. BMC Microbiol. 2006, 6, 86. [Google Scholar] [CrossRef]
- Artursson, V.; Jansson, J.K. Use of Bromodeoxyuridine Immunocapture To Identify Active Bacteria Associated with Arbuscular Mycorrhizal Hyphae. Appl Environ. Microbiol. 2003, 69, 6208–6215. [Google Scholar] [CrossRef]
- Hassen, A.I.; Labuschagne, N. Root colonization and growth enhancement in wheat and tomato by rhizobacterial isolates from the rhizoplane of grasses. World J. Microbiol. Biotechnol. 2010, 26, 1837–1848. [Google Scholar] [CrossRef]
- Espinosa-Urgel, M.; Kolter, R.; Ramos, J.L. Root colonisation by Pseudomonas putida: Love at first sight. Microbiology 2002, 148, 341–343. [Google Scholar] [CrossRef]
- Bowen, G.D.; Rovira, A.D. Microbial colonization of plant roots. Ann. Rev. Phytopalhol. 1976, 14, 121–144. [Google Scholar] [CrossRef]
- Wensing, A.; Gernold, M.; Geider, K. Detection of Erwinia species from the apple and pear flora by mass spectroscopy of whole cells and with novel PCR primers. J. Appl. Microbiol. 2011, 112, 147–158. [Google Scholar] [CrossRef]
- Gehring, I.; Geider, K. Identification of Erwinia species isolated from apples and pears by differential PCR. J. Microbiol. Methods 2012, 89, 57–62. [Google Scholar] [CrossRef]
- Klein, J.M.; Bennett, R.W.; MacFarland, L.; Abranches Da Silva, M.E.; Meza-Turner, B.M.; Dark, P.M.; Frey, M.E.; Wellappili, D.P.; Beugli, A.D.; Jue, H.J.; et al. Draft genome sequence of Erwinia billingiae OSU19-1, isolated from a pear tree canker. Genome Announc. 2015, 3, e01119-15. [Google Scholar] [CrossRef]
- McEvoy, A.; O’Regan, F.; Fleming, C.C.; Moreland, B.P.; Pollock, J.A.; McGuinness, B.W.; Hodkinson, T.R. Bleeding canker of horse chestnut (Aesculus hippocastanum) in Ireland: Incidence, severity and characterization using DNA sequences and real-time PCR. Plant Pathol. 2016, 65, 1419–1429. [Google Scholar] [CrossRef]
- Nurmiaho-Lassila, E.L.; Timonen, S.; Haahtela, K.; Sen, R. Bacterial colonization patterns of intact Pinus sylvestris mycorrhizospheres in dry pine forest soil: An electron microscopy study. Can. J. Microbiol. 1997, 43, 1017–1035. [Google Scholar] [CrossRef]
- Bent, E.; Breuil, C.; Enebak, S.; Chanway, C.P. Surface colonization of lodgepole pine (Pinus contorta var. latifolia [Dougl. Engelm.]) roots by Pseudomonas fluorescens and Paenibacillus polymyxa under gnotobiotic conditions. Plant Soil 2002, 241, 187–196. [Google Scholar] [CrossRef]
- Erturk, Y.; Ercisli, S.; Cakmakci, R. Yield and growth response of strawberry to plant growth-promoting rhizobacteria inoculation. J. Plant Nutr. 2012, 35, 817–826. [Google Scholar] [CrossRef]
- Urquiza, C.C.; Hernández, I.A.; Medina, J.A.C.; López, M.A.R.; Cruz, J.P.; Zarazúa, R.L.R.; Casas, Z.G.M.; Arteaga, D.L.J.; García, A.A.C.; Pérez, J.C.; et al. Identification by MALDI-TOF mass spectrometry of mercury-resistant bacteria associated with the rhizosphere of an apple orchard. Geomicrobiol. J. 2017, 34, 176–182. [Google Scholar] [CrossRef]
- Akinrinlola, R.J.; Yuen, G.Y.; Drijber, R.A.; Adesemoye, A.O. Evaluation of Bacillus strains for plant growth promotion and predictability of efficacy by in vitro physiological traits. Int. J. Microbiol. 2018, 11. [Google Scholar] [CrossRef]
- Hinsa, S.M.; O’Toole, G.A. Identification and characterization of the LapD protein: LapD modulates the secretion of LapA. Microbiology 2006, 152, 1375–1383. [Google Scholar] [CrossRef]
- Qi, J.; Aiuchi, D.; Tani, M.; Asano, S.; Koike, M. Potential of entomopathogenic Bacillus thuringiensis as plant growth promoting rhizobacteria and biological control agents for tomato Fusarium wilt. IJOEAR 2016, 2, 55–63. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).