Genetic Diversity and Spatial Genetic Structure in Isolated Scots Pine (Pinus sylvestris L.) Populations Native to Eastern and Southern Carpathians
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Populations
2.2. DNA Extraction, Amplification, and Sizing
2.3. Genetic Data Analysis
2.4. Spatial Genetic Structure (SGS)
3. Results
3.1. Genetic Diversity
3.2. Population Genetic Structure
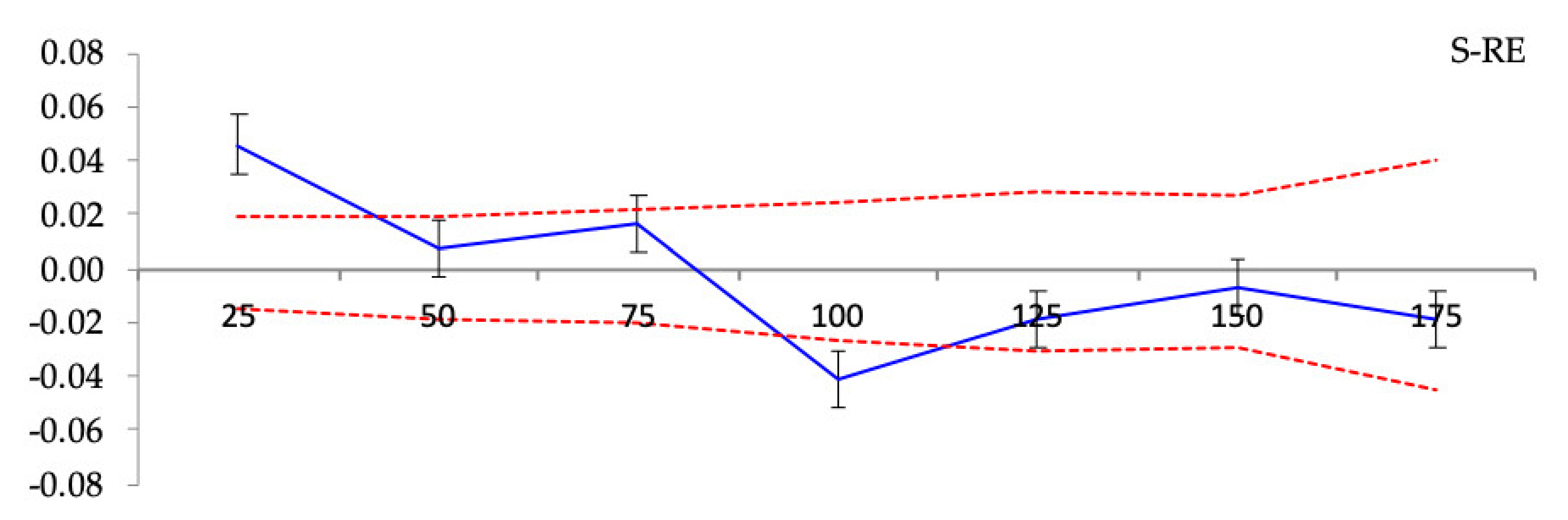
3.3. Spatial Genetic Structure
4. Discussion
Spatial Genetic Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Habel, J.C.; Assmann, T. Relict Species: Phylogeography and Conservation Biology; Springer: Berlin/Heidelberg, Germany, 2010; ISBN 9783540921608. [Google Scholar]
- Hampe, A.; Petit, R.J. Conserving biodiversity under climate change: The rear edge matters. Ecol. Lett. 2005, 8, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Fady, B.; Aravanopoulos, F.A.; Alizoti, P.; Mátyás, C.; von Wühlisch, G.; Westergren, M.; Belletti, P.; Cvjetkovic, B.; Ducci, F.; Huber, G.; et al. Evolution-based approach needed for the conservation and silviculture of peripheral forest tree populations. For. Ecol. Manag. 2016, 375, 66–75. [Google Scholar] [CrossRef]
- Alberto, F.J.; Aitken, S.N.; Alía, R.; González-Martínez, S.C.; Hänninen, H.; Kremer, A.; Lefèvre, F.; Lenormand, T.; Yeaman, S.; Whetten, R.; et al. Potential for evolutionary responses to climate change—Evidence from tree populations. Glob. Chang. Biol. 2013, 19, 1645–1661. [Google Scholar] [CrossRef] [PubMed]
- Eckert, C.G.; Samis, K.E.; Lougheed, S.C. Genetic variation across species’ geographical ranges: The central–marginal hypothesis and beyond. Mol. Ecol. 2008, 17, 1170–1188. [Google Scholar] [CrossRef] [PubMed]
- De-Lucas, A.I.; GonzÁlez-MartÍnez, S.C.; Vendramin, G.G.; Hidalgo, E.; Heuertz, M. Spatial genetic structure in continuous and fragmented populations of Pinus pinaster Aiton. Mol. Ecol. 2009, 18, 4564–4576. [Google Scholar] [CrossRef]
- Epperson, B.K. Spatial Structure of Genetic Variation within Populations of Forest Trees. In Population Genetics of Forest Trees; Springer: Dordrecht, The Netherlands, 1992; pp. 257–278. [Google Scholar] [CrossRef]
- Piotti, A.; Leonardi, S.; Heuertz, M.; Buiteveld, J.; Geburek, T.; Gerber, S.; Kramer, K.; Vettori, C.; Vendramin, G.G. Within-population genetic structure in beech (Fagus sylvatica L.) stands characterized by different disturbance histories: Does forest management simplify population substructure? PLoS ONE 2013, 8, e73391. [Google Scholar] [CrossRef]
- Vekemans, X.; Hardy, O.J. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. Ecol. 2004, 13, 921–935. [Google Scholar] [CrossRef]
- Budde, K.B.; González-Martínez, S.C.; Navascués, M.; Burgarella, C.; Mosca, E.; Lorenzo, Z.; Zabal-Aguirre, M.; Vendramin, G.G.; Verdú, M.; Pausas, J.G.; et al. Increased fire frequency promotes stronger spatial genetic structure and natural selection at regional and local scales in Pinus halepensis Mill. Ann. Bot. 2017, 119, 1061–1072. [Google Scholar] [CrossRef]
- Morton, N.E. Isolation by Distance. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Elsevier Science: San Diego, CA, USA, 2013; Volume 28, p. 139. ISBN 9780080961569. [Google Scholar]
- Valbuena-Carabana, M.; Gonzalez-Martinez, S.C.; Hardy, O.J.; Gil, L. Fine-scale spatial genetic structure in mixed oak stands with different levels of hybridization. Mol. Ecol. 2007, 16, 1207–1219. [Google Scholar] [CrossRef]
- Mosca, E.; Di Pierro, E.A.; Budde, K.B.; Neale, D.B.; González-Martínez, S.C. Environmental effects on fine-scale spatial genetic structure in four Alpine keystone forest tree species. Mol. Ecol. 2018, 27, 647–658. [Google Scholar] [CrossRef]
- Curtu, A.L.; Craciunesc, I.; Enescu, C.M.; Vidalis, A.; Sofletea, N. Fine-scale spatial genetic structure in a multi-oak-species (Quercus spp.) forest. IForest 2015, 8, 324–332. [Google Scholar] [CrossRef]
- González-Díaz, P.; Jump, A.S.; Perry, A.; Wachowiak, W.; Lapshina, E.; Cavers, S. Ecology and management history drive spatial genetic structure in Scots pine. For. Ecol. Manag. 2017, 400, 68–76. [Google Scholar] [CrossRef]
- Naydenov, K.; Senneville, S.; Beaulieu, J.; Tremblay, F.; Bousquet, J. Glacial vicariance in Eurasia: Mitochondrial DNA evidence from Scots pine for a complex heritage involving genetically distinct refugia at mid-northern latitudes and in Asia Minor. BMC Evol. Biol. 2007, 7, 233. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, F.; Pinzauti, F.; Kujala, S.T.; González-Martínez, S.C.; Vendramin, G.G. Novel polymorphic nuclear microsatellite markers for Pinus sylvestris L. Conserv. Genet. Resour. 2012, 4, 231–234. [Google Scholar] [CrossRef]
- Curt, T.; Prévosto, B. Rooting strategy of naturally regenerated beech in Silver birch and Scots pine woodlands. Plant Soil 2003, 255, 265–279. [Google Scholar] [CrossRef]
- Mátyaás, C.; Ackzell, L.; Samuel, C.J.A. EUFORGEN Technical Guidelines for Genetic Conservation and Use for Scots Pine (Pinus sylvestris); International Plant Genetic Resources Institute: Rome, Italy, 2004; 6. [Google Scholar]
- Picon-Cochard, C.; Coll, L.; Balandier, P. The role of below-ground competition during early stages of secondary succession: The case of 3-year-old Scots pine (Pinus sylvestris L.) seedlings in an abandoned grassland. Oecologia 2006. [Google Scholar] [CrossRef]
- Egnell, G. Effects of slash and stump harvesting after final felling on stand and site productivity in Scots pine and Norway spruce. For. Ecol. Manag. 2016. [Google Scholar] [CrossRef]
- Hebda, A.; Wójkiewicz, B.; Wachowiak, W. Genetic characteristics of Scots pine in Poland and reference populations based on nuclear and chloroplast microsatellite markers. Silva Fenn. 2017, 51, 1–17. [Google Scholar] [CrossRef][Green Version]
- Tóth, E.G.; Vendramin, G.G.; Bagnoli, F.; Cseke, K.; Höhn;, M. High genetic diversity and distinct origin of recently fragmented Scots pine (Pinus sylvestris L.) populations along the Carpathians and the Pannonian Basin. Tree Genet. Genomes 2017, 13. [Google Scholar] [CrossRef]
- Prus-Głowacki, W.; Urbaniak, L.; Bujas, E.; Curtu, A.L. Genetic variation of isolated and peripheral populations of Pinus sylvestris (L.) from glacial refugia. Flora Morphol. Distrib. Funct. Ecol. Plants 2012. [Google Scholar] [CrossRef]
- Tanţǎu, I.; Feurdean, A.; de Beaulieu, J.L.; Reille, M.; Fǎrcaş, S. Holocene vegetation history in the upper forest belt of the Eastern Romanian Carpathians. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2011. [Google Scholar] [CrossRef]
- San-Miguel-Ayanz, J.; de Rigo, D.; Caudullo, G.; Houston Durrant, T.; Mauri, A. European Atlas of Forest Tree Species; Publications Office of the European Union: Brussels, Belgium, 2016; ISBN 9789279367403. [Google Scholar]
- Feurdean, A.; Tanţâu, I.; Fârcaş, S. Holocene variability in the range distribution and abundance of Pinus, Picea abies, and Quercus in Romania; implications for their current status. Quat. Sci. Rev. 2011. [Google Scholar] [CrossRef]
- Şofletea, N.; Curtu, A.L. Dendrologie; Editura Universității Transilvania: Brasov, Romania, 2007; ISBN 9789736358852. [Google Scholar]
- Bernhardsson, C.; Floran, V.; Ganea, S.L.; García-gil, M.R. Forest Ecology and Management Present genetic structure is congruent with the common origin of distant Scots pine populations in its Romanian distribution. For. Ecol. Manag. 2016, 361, 131–143. [Google Scholar] [CrossRef]
- Feurdean, A.; Wohlfarth, B.; Björkman, L.; Tantau, I.; Bennike, O.; Willis, K.J.; Farcas, S.; Robertsson, A.M. The influence of refugial population on Lateglacial and early Holocene vegetational changes in Romania. Rev. Palaeobot. Palynol. 2007. [Google Scholar] [CrossRef]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Soranzo, N.; Provan, J.; Powell, W. Characterization of microsatellite loci in Pinus sylvestris L. Mol. Ecol. 1998, 7, 1260–1261. [Google Scholar]
- Van Oosterhout, C.; Weetman, D.; Hutchinson, W.F. Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol. Ecol. Notes 2006, 6, 255–256. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinform. (Oxf. Engl.) 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Weiß, C.H. StatSoft, Inc., Tulsa, OK.: STATISTICA, Version 8. Asta Adv. Stat. Anal. 2007, 91, 339–341. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Manni, F.; Guerard, E.; Heyer, E. Geographic Patterns of (Genetic, Morphologic, Linguistic) Variation: How Barriers Can Be Detected by Using Monmonier’s Algorithm. Hum. Biol. 2007, 76, 173–190. [Google Scholar] [CrossRef]
- Monmonier, M.S. Maximum-Difference Barriers: An Alternative Numerical Regionalization Method*. Geogr. Anal. 2010, 5, 245–261. [Google Scholar] [CrossRef]
- Dieringer, D.; Schlötterer, C. MICROSATELLITE ANALYSER (MSA): A platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes 2003, 3, 167–169. [Google Scholar] [CrossRef]
- Smouse, P.E.; Peakall, R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 1999, 82, 561–573. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. SPAGeDi 1.5 a Program for Spatial Pattern Analysis of Genetic Diversity User’s Manual. Mol. Ecol. Notes 2002, 2, 618–620. Available online: https://www2.ulb.ac.be/sciences/lagev/fichiers/manual_SPAGeDi.pdf (accessed on 7 November 2019). [CrossRef]
- Loiselle, B.A.; Sork, V.L.; Nason, J.; Graham, C. Spatial genetic structure of a tropical understory shrub, PSYCHOTRIA OFFICINALIS (RuBIACEAE). Am. J. Bot. 1995, 82, 1420–1425. [Google Scholar] [CrossRef]
- Ganea, S.; Ranade, S.S.; Hall, D. Development and transferability of two multiplexes nSSR in Scots pine (Pinus sylvestris L.). J. For. Res. 2015, 26, 361–368. [Google Scholar] [CrossRef]
- Scalfi, M.; Piotti, A.; Rossi, M.; Piovani, P. Genetic variability of Italian southern Scots pine (Pinus sylvestris L.) populations: The rear edge of the range. Eur. J. For. Res. 2009. [Google Scholar] [CrossRef]
- Belletti, P.; Ferrazzini, D.; Piotti, A.; Monteleone, I.; Ducci, F. Genetic variation and divergence in Scots pine (Pinus sylvestris L.) within its natural range in Italy. Eur. J. For. Res. 2012, 131, 1127–1138. [Google Scholar] [CrossRef]
- Nowakowska, J.A.; Zachara, T.; Konecka, A. Genetic variability of Scots pine (Pinus sylvestris L.) and Norway spruce (Picea abies L. Karst.) natural regeneration compared with their maternal stands. For. Res. Pap. 2014, 75, 47–54. [Google Scholar] [CrossRef]
- Pavia, I.; Mengl, M.; João Gaspar, M.; Carvalho, A.; Heinze, B.; Lima-Brito, J. Preliminary evidence of two potentialy native populations of Pinus sylvestris L. in Portugal based on nuclear and chloroplast SSR markers. Austrian J. For. Res. 2014, 1, 1–22. [Google Scholar]
- Tóth, E.G.; Bede-Fazekas, Á.; Vendramin, G.G.; Bagnoli, F.; Höhn, M. Mid-Pleistocene and Holocene demographic fluctuation of Scots pine (Pinus sylvestris L.) in the Carpathian Mountains and the Pannonian Basin: Signs of historical expansions and contractions. Quat. Int. 2019, 504, 202–213. [Google Scholar] [CrossRef]
- Gil MR, G.; Floran, V.; Östlund, L.; Gull, B.A. Genetic diversity and inbreeding in natural and managed populations of Scots pine. Tree Genet. Genomes 2015, 11, 28. [Google Scholar] [CrossRef]
- Castro, J.; Gómez, J.M.; García, D.; Zamora, R.; Hódar, J.A. Seed predation and dispersal in relict Scots pine forests in southern Spain. Plant Ecol. 1999, 145, 115–123. [Google Scholar] [CrossRef]
- Marquardt, P.E.; Echt, C.S.; Epperson, B.K.; Pubanz, D.M. Genetic structure, diversity, and inbreeding of eastern white pine under different management conditions. Can. J. For. Res. 2007, 37, 2652–2662. [Google Scholar] [CrossRef]
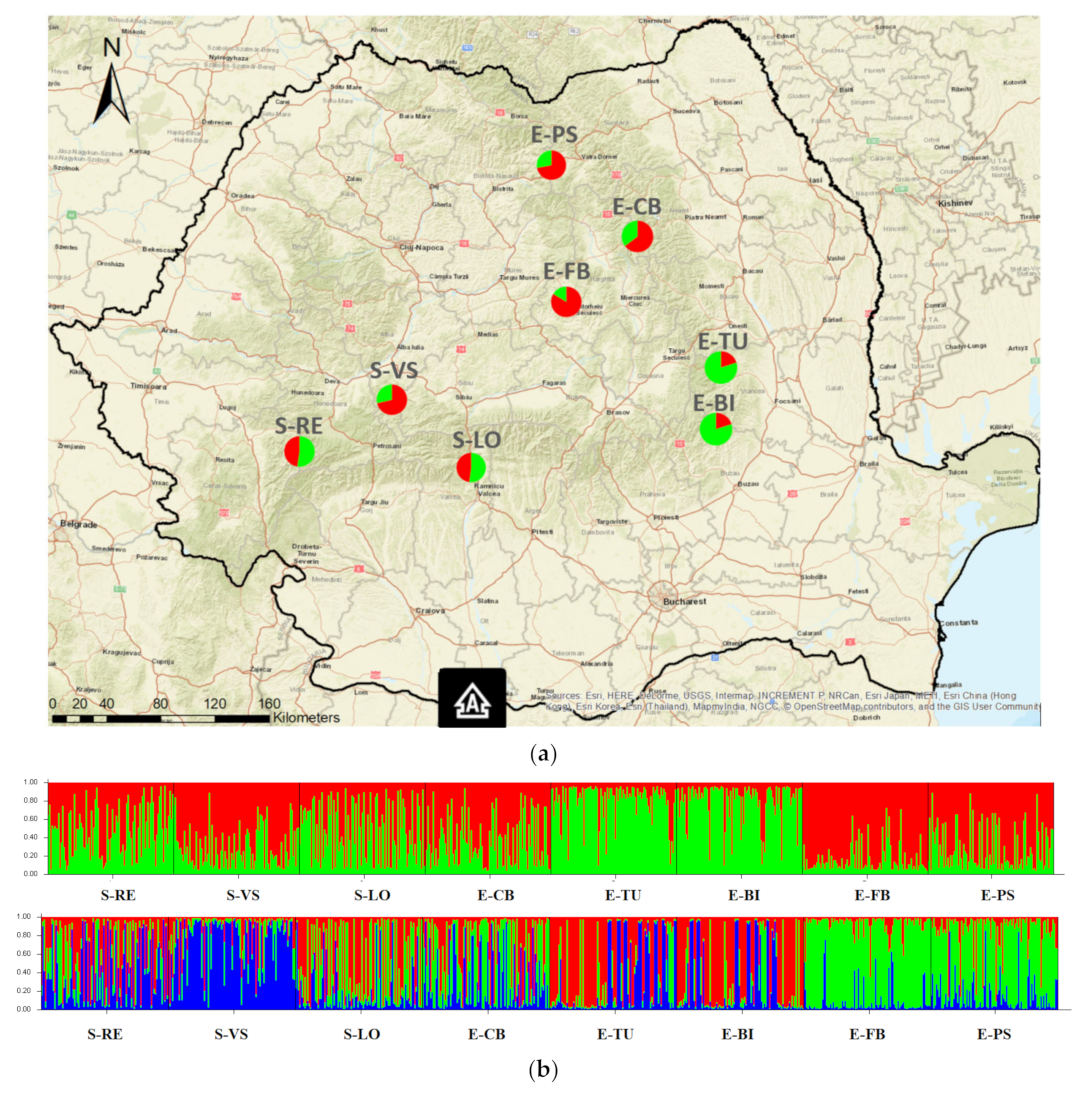



| No. | Population | Acronym | Ecotype | Sample Size | Geographic Location | ||
|---|---|---|---|---|---|---|---|
| Latitude | Longitude | Altitude (m) | |||||
| 1. | Retezat | S-RE | R | 96 | 45°26′ | 22°46′ | 680–750 |
| 45°24′ | 22°46′ | 890–925 | |||||
| 2. | Valea Sebeșului | S-VS | R | 96 | 45°42′ | 23°36′ | 750–1070 |
| 45°42′ | 23°35′ | 730–780 | |||||
| 3. | Lotrișor | S-LO | R | 96 | 45°18′ | 24°16′ | 340–510 |
| 4. | Cheile Bicazului | E-CB | R | 96 | 46°49′ | 25°49′ | 1060–1110 |
| 5. | Tulnici | E-TU | A | 96 | 45°55′ | 26°36′ | 580–610 |
| 6. | Bisoca | E-BI | A | 96 | 45°33′ | 26°40′ | 930–950 |
| 7. | Fântâna Brazilor | E-FB | PB | 96 | 46°30′ | 25°15′ | 950–960 |
| 8. | Poiana Stampei | E-PS | PB | 96 | 47°18′ | 25°07′ | 920 |
| Population | Ecotype | Na | Ne | He | FIS | Pa | |
|---|---|---|---|---|---|---|---|
| S-RE | R | Mean | 9.750 | 4.553 | 0.724 | 0.162 | 3 |
| SE | 0.977 | 0.698 | 0.062 | 0.104 | |||
| S-VS | R | Mean | 10.500 | 5.165 | 0.733 | 0.049 | 4 |
| SE | 2.062 | 1.114 | 0.057 | 0.090 | |||
| S-LO | R | Mean | 11.750 | 6.226 | 0.731 | 0.168 * | 4 |
| SE | 2.289 | 1.590 | 0.078 | 0.061 | |||
| E-CB | R | Mean | 10.750 | 5.222 | 0.672 | 0.154 | 5 |
| SE | 2.250 | 1.468 | 0.087 | 0.088 | |||
| E-TU | A | Mean | 10.375 | 5.290 | 0.710 | 0.164 | 0 |
| SE | 1.936 | 1.136 | 0.088 | 0.080 | |||
| E-BI | A | Mean | 10.250 | 5.246 | 0.711 | 0.138 | 0 |
| SE | 1.980 | 1.096 | 0.088 | 0.083 | |||
| E-FB | PB | Mean | 8.750 | 3.488 | 0.658 | −0.046 | 2 |
| SE | 1.934 | 0.463 | 0.065 | 0.073 | |||
| E-PS | PB | Mean | 9.375 | 4.214 | 0.635 | 0.187 * | 0 |
| SE | 2.299 | 1.093 | 0.094 | 0.066 | |||
| Total | Mean | 10.188 | 4.925 | 0.697 | 0.122 | 18 | |
| SE | 0.677 | 0.391 | 0.027 | 0.029 | |||
| Source | Degrees of Freedom | Sum of Squares | Mean Squares | Estimated Variation | Percent of Variation |
|---|---|---|---|---|---|
| Among population | 7 | 385.255 | 55.036 | 0.495 | 6% |
| Within population | 760 | 5711.833 | 7.516 | 7.516 | 94% |
| Total | 767 | 6097.089 | 8.011 | 100% |
| Population | F1 | bF (b-log) (±SE) | Sp = −bF/(1 − F1) |
|---|---|---|---|
| (95% CI) | |||
| S-RE | 0.0137 * | −0.0036 ± 0.0041 *** | 0.0036 |
| (0.0004–0.0081) | |||
| S-VS | 0.0875 ** | −0.0046 ± 0.0041 *** | 0.0049 |
| (0.0021–0.0087) | |||
| S-LO | 0.0187 ** | −0.0071 ± 0.0047 *** | 0.0072 |
| (0.0026–0.0168) | |||
| S-TU | 0.0277 * | −0.0201 ± 0.0493 ** | 0.0207 |
| (0.0021–0.0239) | |||
| S-BI | 0.0227 * | −0.0136 ± 0.0102 ** | 0.0139 |
| (0.0031–0.0356) | |||
| S-FB | 0.0446 ** | −0.0175 ± 0.0102 ** | 0.0183 |
| (0.0036–0.0303) | |||
| S-PS | −0.0007 | −0.0011 ± 0.0052 *** | 0.0011 |
| (−0.0033–0.0131) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Șofletea, N.; Mihai, G.; Ciocîrlan, E.; Curtu, A.L. Genetic Diversity and Spatial Genetic Structure in Isolated Scots Pine (Pinus sylvestris L.) Populations Native to Eastern and Southern Carpathians. Forests 2020, 11, 1047. https://doi.org/10.3390/f11101047
Șofletea N, Mihai G, Ciocîrlan E, Curtu AL. Genetic Diversity and Spatial Genetic Structure in Isolated Scots Pine (Pinus sylvestris L.) Populations Native to Eastern and Southern Carpathians. Forests. 2020; 11(10):1047. https://doi.org/10.3390/f11101047
Chicago/Turabian StyleȘofletea, Nicolae, Georgeta Mihai, Elena Ciocîrlan, and Alexandru Lucian Curtu. 2020. "Genetic Diversity and Spatial Genetic Structure in Isolated Scots Pine (Pinus sylvestris L.) Populations Native to Eastern and Southern Carpathians" Forests 11, no. 10: 1047. https://doi.org/10.3390/f11101047
APA StyleȘofletea, N., Mihai, G., Ciocîrlan, E., & Curtu, A. L. (2020). Genetic Diversity and Spatial Genetic Structure in Isolated Scots Pine (Pinus sylvestris L.) Populations Native to Eastern and Southern Carpathians. Forests, 11(10), 1047. https://doi.org/10.3390/f11101047





