Selection of Suitable Reference Genes Based on Transcriptomic Data in Ginkgo biloba under Different Experimental Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Treatments
2.2. Total RNA Isolation and cDNA Synthesis
2.3. Gene Sequence Search and Primer Design
2.4. qRT-PCR Assays
2.5. Data Analysis
3. Results
3.1. Designing and Validation of Primers
3.2. Expression Profile of Candidate Reference Genes
3.3. geNorm Analysis
3.4. NormFinder Analysis
3.5. BestKeeper Analysis
3.6. Analysis by the ΔCt Method
3.7. RefFinder Analysis
3.8. Validation of the Stability of Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Overbergh, L.; Giulietti, A.; Valckx, D.; Decallonne, B.; Bouillon, R.; Mathieu, C. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J. Biomol. Tech. 2003, 14, 33–43. [Google Scholar] [PubMed]
- Cheng, H.; Li, L.; Xu, F.; Cheng, S.; Cao, F.; Wang, Y.; Yuan, H.; Jiang, D.; Wu, C. Expression patterns of a cinnamyl alcohol dehydrogenase gene involved in lignin biosynthesis and environmental stress in Ginkgo biloba. Mol. Biol. Rep. 2013, 40, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Li, L.; Cheng, S.; Cao, F.; Wang, Y.; Yuan, H. Molecular cloning and function assay of a chalcone isomerase gene (GbCHI) from Ginkgo biloba. Plant Cell Rep. 2011, 30, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Li, L.; Zhang, W.; Cheng, H.; Sun, N.; Cheng, S.; Wang, Y. Isolation, characterization, and function analysis of a flavonol synthase gene from Ginkgo biloba. Mol. Biol. Rep. 2011, 39, 2285–2296. [Google Scholar] [CrossRef]
- Liao, Y.L.; Shen, Y.B.; Chang, J.; Zhang, W.W.; Cheng, S.Y.; Xu, F. Isolation, expression, and promoter analysis of GbWRKY2: A novel transcription factor gene from Ginkgo biloba. Int. J. Genom. 2015, 2015, 607185. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xie, W.; Yu, X.; Zhang, Z.; Zhao, Y.; Wang, N.; Wang, Y. Selection of suitable reference genes for RT-qPCR gene expression analysis in Siberian wild rye (Elymus sibiricus) under different experimental conditions. Genes 2019, 10, 451. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Liu, Y.; Ma, X.; Shuai, Q.; Gai, J.; Li, Y. Evaluation of Reference Genes for Normalization of Gene Expression Using Quantitative RT-PCR under Aluminum, Cadmium, and Heat Stresses in Soybean. PLoS ONE 2017, 12, e0168965. [Google Scholar] [CrossRef]
- Galli, V.; Borowski, J.M.; Perin, E.C.; Da Silva Messias, R.; Labonde, J.; Dos Santos Pereira, I.; Silva, S.D.; Rombaldi, C.V. Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses. Gene 2015, 554, 205–214. [Google Scholar] [CrossRef]
- Yang, Q.; Yin, J.; Li, G.; Qi, L.; Yang, F.; Wang, R.; Li, G. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol. Biol. Rep. 2014, 41, 2325–2334. [Google Scholar] [CrossRef]
- Jain, M.; Tian, C.; Jiang, Q.; Wang, F.; Wang, G.L.; Xu, Z.S.; Xiong, A.S. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS ONE 2015, 10, e0117569. [Google Scholar] [CrossRef] [Green Version]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef] [Green Version]
- Cao, F.; Cheng, H.; Cheng, S.; Li, L.; Xu, F.; Yu, W.; Yuan, H. Expression of selected Ginkgo biloba heat shock protein genes after cold treatment could be induced by other abiotic stress. Int. J. Mol. Sci. 2012, 13, 5768–5788. [Google Scholar] [CrossRef]
- Yao, X.; Zhou, G.S.; Tang, Y.P.; Qian, Y.F.; Guan, H.L.; Pang, H.; Zhu, S.; Mo, X.; Su, S.L.; Jin, C.; et al. Simultaneous quantification of flavonol glycosides, terpene lactones, biflavones, proanthocyanidins, and ginkgolic acids in Ginkgo biloba leaves from fruit cultivars by ultrahigh-performance liquid chromatography coupled with triple quadrupole mass spectrometry. Biomed. Res. Int. 2013, 2013, 582591. [Google Scholar] [CrossRef] [Green Version]
- Ude, C.; Schubert-Zsilavecz, M.; Wurglics, M. Ginkgo biloba extracts: A review of the pharmacokinetics of the active ingredients. Clin. Pharmacokinet. 2013, 52, 727–749. [Google Scholar] [CrossRef]
- Kressmann, S.; Müller, W.; Blume, H. Pharmaceutical quality of different Ginkgo biloba brands. J. Pharm. Pharmacol. 2010, 54, 661–669. [Google Scholar] [CrossRef] [Green Version]
- Mohanta, T.K.; Tamboli, Y.; Zubaidha, P.K. Phytochemical and medicinal importance of Ginkgo biloba L. Nat. Prod. Res. 2014, 28, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, P.; Kong, Q.; Xing, S.; Liu, X.; Sun, L. The contents of terpene trilactone and flavonoid in leaves of seedlings from ancient female Ginkgo trees in China. Hortic. Plant J. 2017, 3, 165–171. [Google Scholar] [CrossRef]
- Yao, X.; Shang, E.; Zhou, G.; Tang, Y.; Guo, S.; Su, S.; Jin, C.; Qian, D.; Qin, Y.; Duan, J.A. Comparative characterization of total flavonol glycosides and terpene lactones at different ages, from different cultivation sources and genders of Ginkgo biloba leaves. Int. J. Mol. Sci. 2012, 13, 10305–10315. [Google Scholar] [CrossRef]
- Lobstein, A.; Rietsch-Jako, L.; Haag-Berrurier, M.; Anton, R. Seasonal variations of the flavonoid content from Ginkgo biloba leaves. Planta Med. 1991, 57, 430–433. [Google Scholar] [CrossRef] [Green Version]
- Chiu, K.L.; Cheng, Y.C.; Chen, J.H.; Chang, C.J.; Yang, P.W. Supercritical fluids extraction of Ginkgo ginkgolides and flavonoids. J. Supercrit. Fluids 2002, 24, 77–87. [Google Scholar] [CrossRef]
- Miao, S.F.; Yu, J.P.; Du, Z.; Guan, Y.X.; Yao, S.J.; Zhu, Z.Q. Supercritical fluid extraction and micronization of Ginkgo flavonoids from Ginkgo biloba leaves. Ind. Eng. Chem. Res. 2010, 49, 5461–5466. [Google Scholar] [CrossRef]
- Chen, S.; Xing, X.H.; Huang, J.J.; Xu, M.S. Enzyme-assisted extraction of flavonoids from Ginkgo biloba leaves: Improvement effect of flavonol transglycosylation catalyzed by Penicillium decumbens cellulase. Enzyme Microb. Technol. 2011, 48, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Sati, P.; Dhyani, P.; Bhatt, I.D.; Pandey, A. Ginkgo biloba flavonoid glycosides in antimicrobial perspective with reference to extraction method. J. Tradit. Complement. Med. 2019, 9, 15–23. [Google Scholar] [CrossRef]
- Han, S.; Wu, Z.; Jin, Y.; Yang, W.; Shi, H. RNA-Seq analysis for transcriptome assembly, gene identification, and SSR mining in Ginkgo (Ginkgo biloba L.). Tree Genet. Genomes 2015, 11, 37. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, J.; Zhou, Q.; Xin, Y.; Wang, G.; Xu, L.A. De novo transcriptome analysis revealed genes involved in flavonoid biosynthesis, transport and regulation in Ginkgo biloba. Ind. Crops Prod. 2018, 124, 226–235. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, Y.; Kim, S.U.; Chen, Z.; Nie, G.; Cheng, S.; Ye, J.; Xu, F. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci. Rep. 2020, 10, 13723. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Meng, X.; Liao, Y.; Yu, T.; Cao, J.; Tan, J.; Xu, F.; Cheng, S. Characterization and functional analysis of two novel 3-hydroxy-3-methylglutaryl-coenzyme A reductase genes (GbHMGR2 and GbHMGR3) from Ginkgo biloba. Sci. Rep. 2019, 9, 14109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagano, A.J.; Kawagoe, T.; Sugisaka, J.; Honjo, M.N.; Iwayama, K.; Kudoh, H. Annual transcriptome dynamics in natural environments reveals plant seasonal adaptation. Nat. Plants 2019, 5, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, C.; Wang, Y.; Ding, Z.; Zhao, L. Global transcriptional analysis reveals the complex relationship between tea quality, leaf senescence and the responses to cold-drought combined stress in Camellia sinensis. Front. Plant Sci. 2016, 7, 1858. [Google Scholar] [CrossRef] [Green Version]
- Migocka, M.; Papierniak, A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breed. 2010, 28, 343–357. [Google Scholar] [CrossRef]
- Niu, X.; Chen, M.; Huang, X.; Chen, H.; Tao, A.; Xu, J.; Qi, J. Reference gene selection for qRT-PCR normalization analysis in kenaf (Hibiscus cannabinus L.) under abiotic stress and hormonal stimuli. Front. Plant Sci. 2017, 8, 771. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Han, X.; Chen, S.; Zheng, L.; He, X.; Liu, M.; Qiao, G.; Wang, Y.; Zhuo, R. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Salix matsudana under different abiotic stresses. Sci. Rep. 2017, 7, 40290. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Shen, G.A.; Liu, C.; Liu, X.; Tan, F.; Sun, X.; Tang, K. Molecular cloning and sequence analysis of a novel chalcone synthase cDNA from Ginkgo biloba. DNA Seq. 2004, 15, 283–290. [Google Scholar] [CrossRef]
- Ye, J.; Cheng, S.; Zhou, X.; Chen, Z.; Kim, S.U.; Tan, J.; Zheng, J.; Xu, F.; Zhang, W.; Liao, Y. A global survey of full-length transcriptome of Ginkgo biloba reveals transcript variants involved in flavonoid biosynthesis. Ind. Crops Prod. 2019, 139, 111547. [Google Scholar] [CrossRef]
- Narsai, R.; Ivanova, A.; Ng, S.; Whelan, J. Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol. 2010, 10, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, H.; Fu, Y.; He, W.; Wang, L.; Wei, Y. Selection of appropriate reference genes for quantitative real-time PCR in Oxytropis ochrocephala Bunge using transcriptome datasets under abiotic stress treatments. Front. Plant Sci. 2015, 6, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Cong, P.; Tian, Y.; Zhu, Y. Using RNA-seq data to select reference genes for normalizing gene expression in apple roots. PLoS ONE 2017, 12, e0185288. [Google Scholar] [CrossRef]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dheda, K. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 118–119. [Google Scholar] [CrossRef] [Green Version]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.W.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Weinmann, S.; Roll, S.; Schwarzbach, C.; Vauth, C.; Willich, S.N. Effects of Ginkgo biloba in dementia: Systematic review and meta-analysis. BMC Geriatr. 2010, 10, 14. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.H.; Hu, X.; Tang, W.; Zheng, X.; Kim, Y.S.; Lee, B.H.; Zhu, J.K. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell Biol. 2006, 26, 9533–9543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruelland, E.; Vaultier, M.N.; Zachowski, A.; Hurry, V. Chapter 2 cold signalling and cold acclimation in plants. Adv. Bot. Res. 2009, 49, 35–150. [Google Scholar] [CrossRef]
- Xu, F.; Cheng, S.; Cheng, S.; Wang, Y.; Du, H. Time course of expression of chalcone synthase gene in Ginkgo biloba. J.Plant Physiol. Mol. Biol. 2007, 33, 309. [Google Scholar]
- Lo Piero, A.R.; Puglisi, I.; Rapisarda, P.; Petrone, G. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J. Agric. Food Chem. 2005, 53, 9083–9088. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, S.; Liu, X.; Shang, J.; Zhang, A.; Zhu, Z.; Zha, D. Chalcone synthase (CHS) family members analysis from eggplant (Solanum melongena L.) in the flavonoid biosynthetic pathway and expression patterns in response to heat stress. PLoS ONE 2020, 15, e0226537. [Google Scholar]
- Peng, X.; Wu, H.; Chen, H.; Zhang, Y.; Qiu, D.; Zhang, Z. Transcriptome profiling reveals candidate flavonol-related genes of Tetrastigma hemsleyanum under cold stress. BMC Genom. 2019, 20, 687. [Google Scholar] [CrossRef] [Green Version]
- Anjum, S.A.; Wang, L.; Farooq, M.; Khan, I.; Xue, L. Methyl jasmonate-induced alteration in lipid peroxidation, antioxidative defence system and yield in soybean under drought. J. Agron. Crop Sci. 2011, 197, 296–301. [Google Scholar] [CrossRef]
- Wang, J.; Qian, J.; Yao, L.; Lu, Y. Enhanced production of flavonoids by methyl jasmonate elicitation in cell suspension culture of Hypericum perforatum. Bioresour. Bioprocess. 2015, 2, 5. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.M.; Gershenzon, J.; Bohlmann, J. Induction of volatile terpene biosynthesis and diurnal emission by methyl jasmonate in foliage of Norway spruce. Plant Physiol. 2003, 132, 1586–1599. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Ma, C.; Qi, D.; Lv, H.; Yang, T.; Peng, Q.; Chen, Z.; Lin, Z. Transcriptional responses and flavor volatiles biosynthesis in methyl jasmonate-treated tea leaves. BMC Plant Biol. 2015, 15, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Zhang, X.; Fu, M.; Zeng, H.; Ye, J.; Zhang, W.; Liao, Y.; Xu, F. Effects of different stress treatments on the total terpene trilactone content and expression levels of key genes in Ginkgo biloba leaves. Plant Mol. Biol. Rep. 2020. [Google Scholar] [CrossRef]
- Awasthi, P.; Mahajan, V.; Jamwal, V.L.; Kapoor, N.; Rasool, S.; Bedi, Y.S.; Gandhi, S.G. Cloning and expression analysis of chalcone synthase gene from Coleus forskohlii. J. Genet. 2016, 95, 647–657. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Wang, R.; Xian, B.; Ren, C.; Liu, Q.; Wu, Q.; Pei, J. Integrated metabolomics and transcriptome analysis on flavonoid biosynthesis in safflower (Carthamus tinctorius L.) under MeJA treatment. BMC Plant Biol. 2020, 20, 353. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Pan, X.; Li, Y.; Cui, L.; Zhang, Y.; Zhang, Z.; Pan, G.; Yang, J.; Cao, P.; Yang, A. Identification and characterization of chalcone synthase gene family members in Nicotiana tabacum. J. Plant Growth Regul. 2017, 36, 374–384. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, Q.; Shen, W.; El Mohtar, C.A.; Zhao, X.; Gmitter, F.G., Jr. Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biol. 2018, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Luo, X.; He, J.; Zhang, C.; Liao, X.; Xu, X.; Feng, S.; Yu, C.; Jiang, Z.; Meng, Y.; et al. Bioactive compounds induced in Physalis angulata L. by methyl-jasmonate: An investigation of compound accumulation patterns and biosynthesis-related candidate genes. Plant Mol. Biol. 2020, 103, 341–354. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, M.R.; Ruedell, C.M.; Ricachenevsky, F.K.; Sperotto, R.A.; Pasquali, G.; Fett-Neto, A.G. Reference gene selection for quantitative reverse transcription-polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill. BMC Mol. Biol. 2010, 11, 73. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Liu, F.; Huang, W.; Sun, Q.; Huang, X. Identification of reliable reference genes for qRT-PCR in the ephemeral plant Arabidopsis pumila based on full-length transcriptome data. Sci. Rep. 2019, 9, 8408. [Google Scholar] [CrossRef]
- Zhang, C.; Jiang, Q.N.; Wang, Y.G.; Fu, J.X.; Dong, B.; Zhou, L.H.; Zhao, H.B. Transcriptome-based validation of proper reference genes for reverse trascription quantitative PCR analysis of Sinocalycanthus chinensis. Biol. Plant. 2020, 64, 253–257. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wu, Z.; Bao, W.; Hu, H.; Chen, M.; Chai, T.; Wang, H. Identification and evaluation of reference genes for quantitative real-time PCR analysis in Polygonum cuspidatum based on transcriptome data. BMC Plant Biol. 2019, 19, 498. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar Reddy, P.; Srinivas Reddy, D.; Sivasakthi, K.; Bhatnagar-Mathur, P.; Vadez, V.; Sharma, K.K. Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Front. Plant Sci. 2016, 7, 529. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, J.; Huang, S.; Guo, T.; Deng, L.; Hua, W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 2014, 538, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Chen, Z.; Ju, Y.; Zhang, H.; Cai, M.; Pan, H.; Zhang, Q. Reference gene selection for qRT-PCR analysis of flower development in Lagerstroemia indica and L. speciosa. PLoS ONE 2018, 13, e0195004. [Google Scholar] [CrossRef] [Green Version]
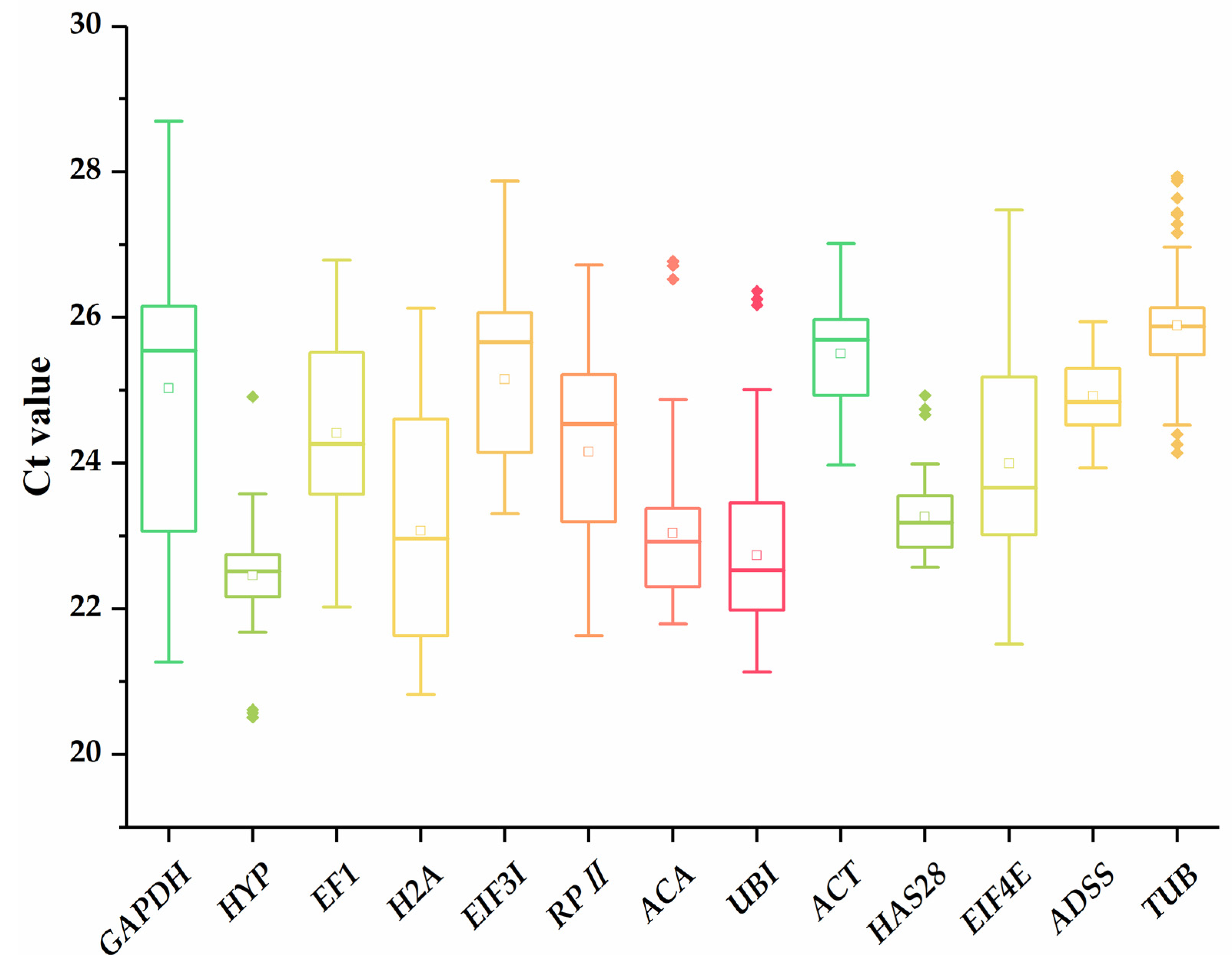
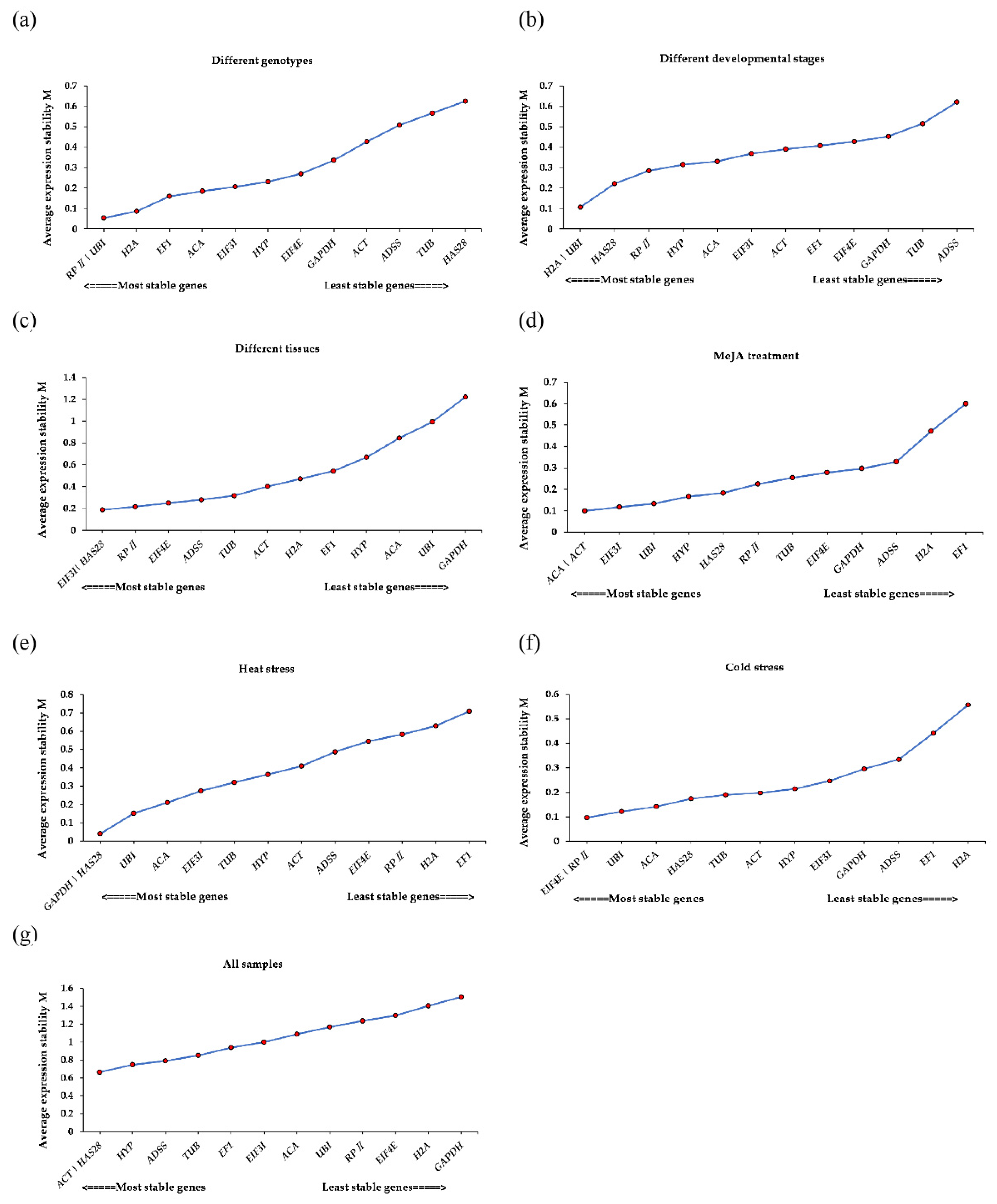
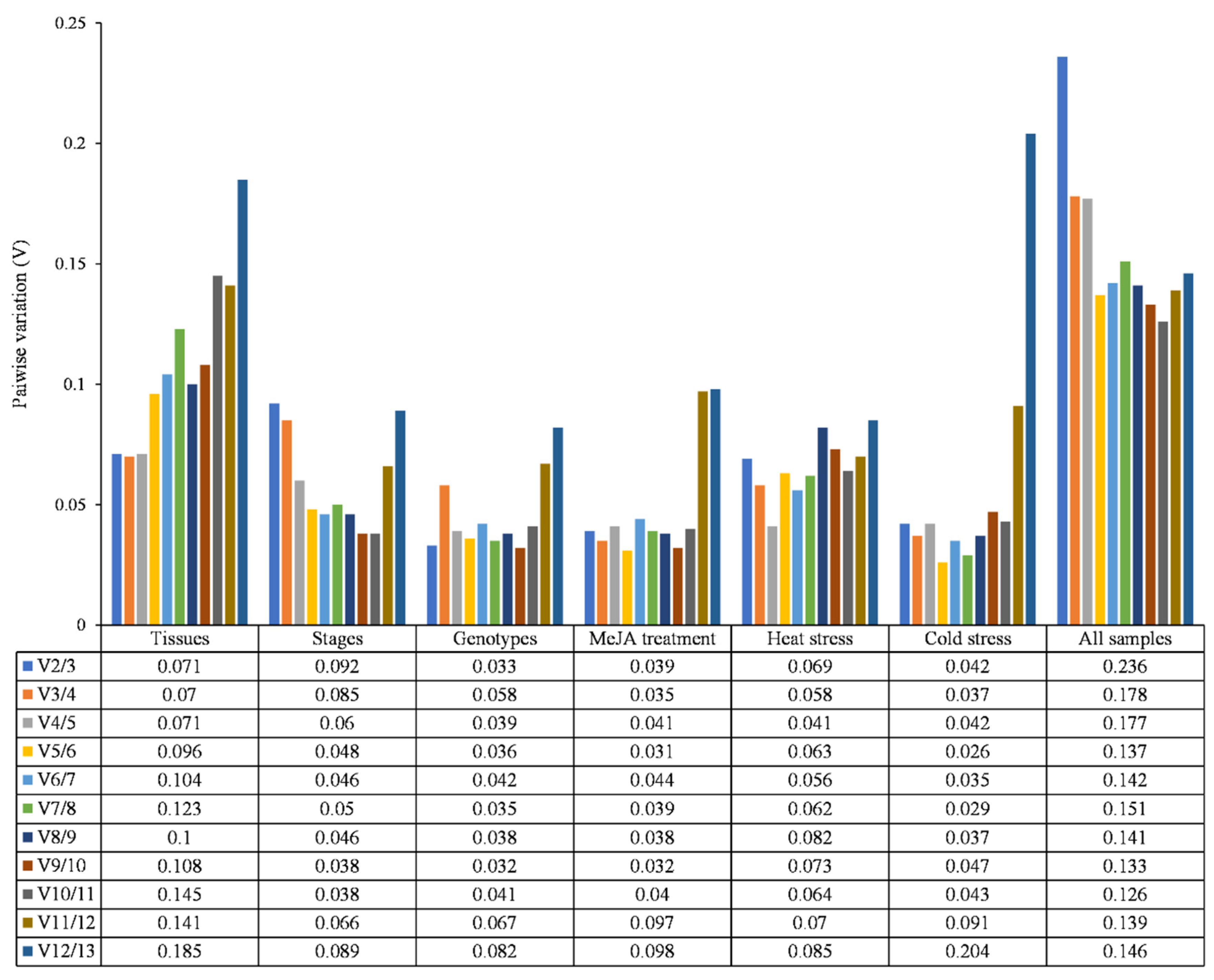


| Gene | Gene Number | Gene Description | Primer Sequence (5′-3′) Forward/Reverse | Product Size (bp) | Efficiency (%) | R2 |
|---|---|---|---|---|---|---|
| TUB | Gb_02392 | γ-Tubulin | F: TCATACAGACACCGACTCAA R: CAATCTCCACTCCTCCATC | 124 | 105.07 | 0.996 |
| EIF4E | Gb_08649 | Eukaryotic translation initiation factor 4E | F: AAGTGGGAGGACCCTAAATG R: GCTAACAAAGTGTAGAGCCAGAG | 101 | 106.25 | 0.9975 |
| EIF3I | Gb_07392 | Eukaryotic translation initiation factor 3 subunit I | F: CAAGGCAGAGCAGTGAGT R: ATCCCAGATGCGGAGAAC | 128 | 100.68 | 0.996 |
| HAS28 | Gb_13272 | 28 kDa heat- and acid-stable phosphoprotein | F: CAGAACAAGCGAGGAAAG R: CCAGACAAGGCAAGGATA | 164 | 95.76 | 0.9956 |
| UBI | Gb_24579 | Ubiquitin | F: GCCATCAGACTTGCTACG R: CACTTTCCAACCCACTCA | 112 | 103.98 | 0.996 |
| RPII | Gb_40102 | RNA polymerase II | F: TACCATGCCTAATGTGCC R: CCTGTGCTCCTCTAATCCA | 139 | 103.40 | 0.9801 |
| HYP | Gb_05998 | Hypothetical protein | F: TGTGTACCCCTCAGGAACCG R: AAGCATCAGTTTGGGCAGGA | 146 | 96.55 | 0.9989 |
| EF1 | Gb_14413 | Elongation factor 1 | F: TGGCAGAGGAAGCAACTA R: GGATGAAACCCAGATACAAG | 144 | 95.84 | 0.9912 |
| GAPDH | L26924.1 | Glyceraldehyde-3-phosphate dehydrogenase | F: ATCCACGGGAGTCTTCAC R: CTCATTCACGCCAACAAC | 121 | 103.70 | 0.9974 |
| H2A | Gb_34906 | Histone H2A.6 | F: GGATAACAAGAAGACCAGGATT R: TTTGCCAGAAGCACCAGA | 163 | 101.23 | 0.995 |
| ACT | Gb_00790 | Actin | F: GTCTCGCCAAGTGGAAAGGT R: GCACACGATGCACCACTATC | 134 | 103.31 | 0.999 |
| ACA | Gb_36873 | Acetyl-coenzyme A carboxylase | F: CAGAGGCAGCAATGAGAA R: CTGTGATGGAAGCGAGGG | 110 | 104.05 | 0.9987 |
| ADSS | Gb_32787 | Adenylosuccinate synthetase | F: TGGGGTGACGAAGGAAAGGG R: CTCCCTGACAACGAGCCACA | 114 | 107.28 | 0.9975 |
| CHS | AY496931.1 | Chalcone synthase | F: CAAGCGCATGTGCGACAAGT R: CACCTCCACCACCACCATGT | 139 | 102.04 | 0.9995 |
| Method | Rank (Better—Good—Average) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| Different genotypes | |||||||||||||
| ΔCT | UBI | EF1 | RPII | ACA | H2A | HYP | EIF3I | EIF4E | ACT | GAPDH | ADSS | TUB | HAS28 |
| BestKeeper | EF1 | ACT | ADSS | H2A | TUB | RPII | UBI | ACA | HYP | EIF3I | HAS28 | EIF4E | GAPDH |
| NormFinder | EF1 | UBI | HYP | ACA | RPII | H2A | EIF3I | EIF4E | ACT | GAPDH | ADSS | TUB | HAS28 |
| GeNorm | RPII| UBI | H2A | EF1 | ACA | EIF3I | HYP | EIF4E | GAPDH | ACT | ADSS | TUB | HAS28 | |
| RefFinder | EF1 | UBI | RPII | H2A | ACA | HYP | ACT | EIF3I | ADSS | EIF4E | TUB | GAPDH | HAS28 |
| Different developmental stages | |||||||||||||
| ΔCT | HYP | HAS28 | UBI | RPII | ACA | H2A | EIF3I | ACT | EIF4E | EF1 | GAPDH | TUB | ADSS |
| BestKeeper | H2A | UBI | RPII | HAS28 | ACA | EIF4E | GAPDH | EF1 | HYP | ACT | EIF3I | TUB | ADSS |
| NormFinder | HAS28 | HYP | UBI | RPII | H2A | ACA | EIF3I | ACT | EIF4E | EF1 | GAPDH | TUB | ADSS |
| GeNorm | H2A |UBI | HAS28 | RPII | HYP | ACA | EIF3I | ACT | EF1 | EIF4E | GAPDH | TUB | ADSS | |
| RefFinder | UBI | HAS28 | H2A | HYP | RPII | ACA | EIF3I | EIF4E | ACT | EF1 | GAPDH | TUB | ADSS |
| Different tissues | |||||||||||||
| ΔCT | EIF3I | RPII | HAS28 | ADSS | EIF4E | TUB | H2A | ACT | EF1 | HYP | ACA | UBI | GAPDH |
| BestKeeper | EIF3I | RPII | ADSS | HAS28 | EIF4E | ACT | H2A | TUB | HYP | EF1 | ACA | UBI | GAPDH |
| NormFinder | EIF3I | RPII | ADSS | HAS28 | EIF4E | TUB | H2A | ACT | EF1 | HYP | ACA | UBI | GAPDH |
| GeNorm | EIF3I | HAS28 | RPII | EIF4E | ADSS | TUB | ACT | H2A | EF1 | HYP | ACA | UBI | GAPDH | |
| RefFinder | EIF3I | RPII | HAS28 | ADSS | EIF4E | TUB | ACT | H2A | EF1 | HYP | ACA | UBI | GAPDH |
| MeJA treatment | |||||||||||||
| ΔCT | ACA | ACT | HAS28 | HYP | RPII | UBI | EIF3I | TUB | EIF4E | GAPDH | ADSS | H2A | EF1 |
| BestKeeper | EIF3I | ACA | ACT | HAS28 | UBI | HYP | ADSS | RPII | GAPDH | EIF4E | TUB | H2A | EF1 |
| NormFinder | RPII | ACA | TUB | HAS28 | EIF4E | ACT | HYP | EIF3I | UBI | GAPDH | ADSS | H2A | EF1 |
| GeNorm | ACA | ACT | EIF3I | UBI | HYP | HAS28 | RPII | TUB | EIF4E | GAPDH | ADSS | H2A | EF1 | |
| RefFinder | ACA | ACT | EIF3I | RPII | HAS28 | HYP | UBI | TUB | EIF4E | GAPDH | ADSS | H2A | EF1 |
| Cold stress | |||||||||||||
| ΔCT | UBI | RPII | ACA | EIF4E | ACT | HAS28 | HYP | TUB | EIF3I | GAPDH | ADSS | EF1 | H2A |
| BestKeeper | GAPDH | RPII | ACA | HYP | ADSS | EIF4E | UBI | EIF3I | ACT | HAS28 | TUB | H2A | EF1 |
| NormFinder | EIF4E | RPII | UBI | ACA | ACT | HYP | HAS28 | TUB | EIF3I | GAPDH | ADSS | EF1 | H2A |
| GeNorm | EIF4E | RPII | UBI | ACA | HAS28 | TUB | ACT | HYP | EIF3I | GAPDH | ADSS | EF1 | H2A | |
| RefFinder | RPII | EIF4E | UBI | ACA | GAPDH | HYP | ACT | HAS28 | TUB | EIF3I | ADSS | EF1 | H2A |
| Heat stress | |||||||||||||
| ΔCT | HAS28 | GAPDH | ACA | UBI | HYP | EIF3I | TUB | EIF4E | ACT | ADSS | RPII | H2A | EF1 |
| BestKeeper | ACA | HYP | UBI | HAS28 | GAPDH | EIF3I | RPII | TUB | EIF4E | ADSS | H2A | ACT | EF1 |
| NormFinder | HAS28 | GAPDH | ACA | UBI | HYP | EIF3I | TUB | ADSS | EIF4E | ACT | RPII | H2A | EF1 |
| GeNorm | GAPDH | HAS28 | UBI | ACA | EIF3I | TUB | HYP | ACT | ADSS | EIF4E | RPII | H2A | EF1 | |
| RefFinder | HAS28 | GAPDH | ACA | UBI | HYP | EIF3I | TUB | EIF4E | ADSS | ACT | RPII | H2A | EF1 |
| All samples | |||||||||||||
| ΔCT | HAS28 | HYP | ADSS | ACT | TUB | EF1 | ACA | EIF3I | UBI | RPII | EIF4E | H2A | GAPDH |
| BestKeeper | HAS28 | HYP | ADSS | TUB | ACT | ACA | UBI | EF1 | EIF3I | RPII | EIF4E | H2A | GAPDH |
| NormFinder | HYP | HAS28 | ADSS | ACT | TUB | EF1 | ACA | EIF3I | UBI | RPII | EIF4E | H2A | GAPDH |
| GeNorm | ACT | HAS28 | HYP | ADSS | TUB | EF1 | EIF3I | ACA | UBI | RPII | EIF4E | H2A | GAPDH | |
| RefFinder | HAS28 | HYP | ACT | ADSS | TUB | EF1 | ACA | EIF3I | UBI | RPII | EIF4E | H2A | GAPDH |
| Gene name | UBI | EF1 | RPII | HYP | HAS28 | H2A | EIF3I | ACA | ACT | GAPDH | EIF4E | TUB | ADSS |
| Number of times the best gene was identified | 5 | 3 | 4 | 2 | 10 | 2 | 6 | 4 | 3 | 2 | 2 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, T.; Yang, X.; Fu, F.; Wang, G.; Cao, F. Selection of Suitable Reference Genes Based on Transcriptomic Data in Ginkgo biloba under Different Experimental Conditions. Forests 2020, 11, 1217. https://doi.org/10.3390/f11111217
Zhou T, Yang X, Fu F, Wang G, Cao F. Selection of Suitable Reference Genes Based on Transcriptomic Data in Ginkgo biloba under Different Experimental Conditions. Forests. 2020; 11(11):1217. https://doi.org/10.3390/f11111217
Chicago/Turabian StyleZhou, Tingting, Xiaoming Yang, Fangfang Fu, Guibin Wang, and Fuliang Cao. 2020. "Selection of Suitable Reference Genes Based on Transcriptomic Data in Ginkgo biloba under Different Experimental Conditions" Forests 11, no. 11: 1217. https://doi.org/10.3390/f11111217
APA StyleZhou, T., Yang, X., Fu, F., Wang, G., & Cao, F. (2020). Selection of Suitable Reference Genes Based on Transcriptomic Data in Ginkgo biloba under Different Experimental Conditions. Forests, 11(11), 1217. https://doi.org/10.3390/f11111217






