Groundwater Depth Overrides Tree-Species Effects on the Structure of Soil Microbial Communities Involved in Nitrogen Cycling in Plantation Forests
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site Description and Sample Collection
2.2. Soil Chemical Analysis
2.3. DNA Extraction
2.4. Length-heterogeneity PCR (LH-PCR)
2.5. Denaturing Gradient Gel Electrophoresis (DGGE) Analysis
2.6. Quantitative PCR (Q-PCR)
2.7. Statistical Analysis
3. Results
3.1. Soil Chemical Characteristics
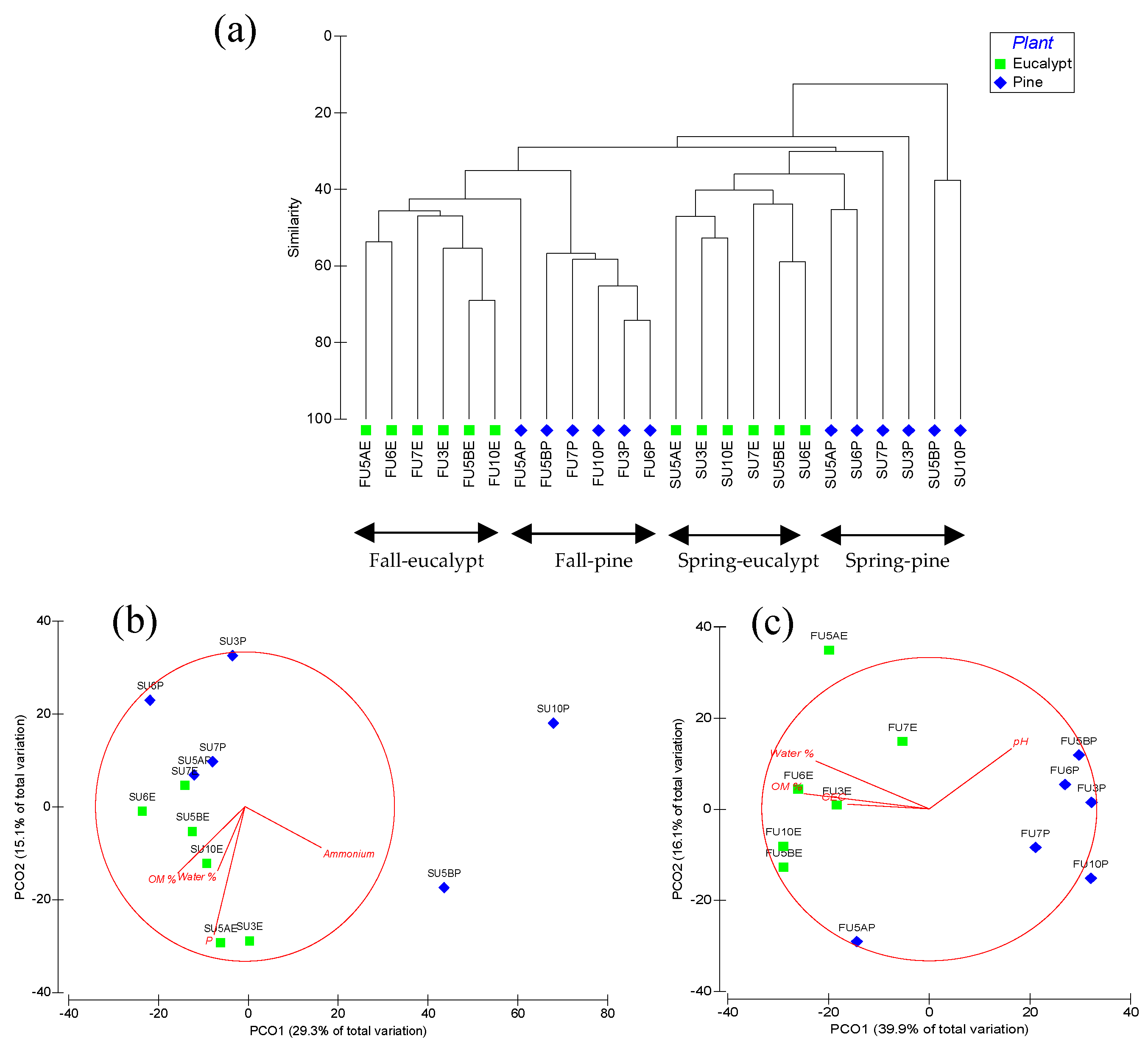
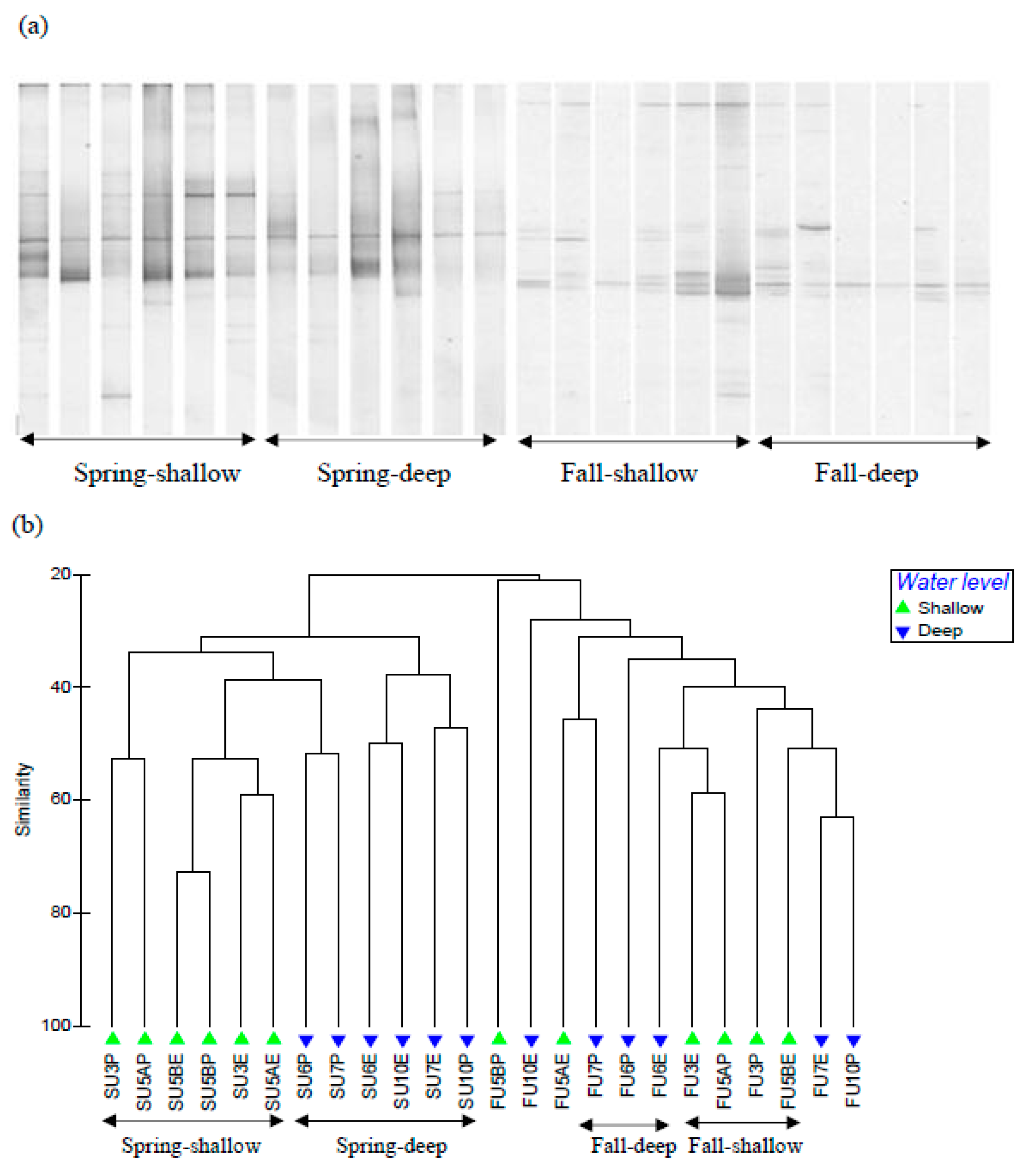
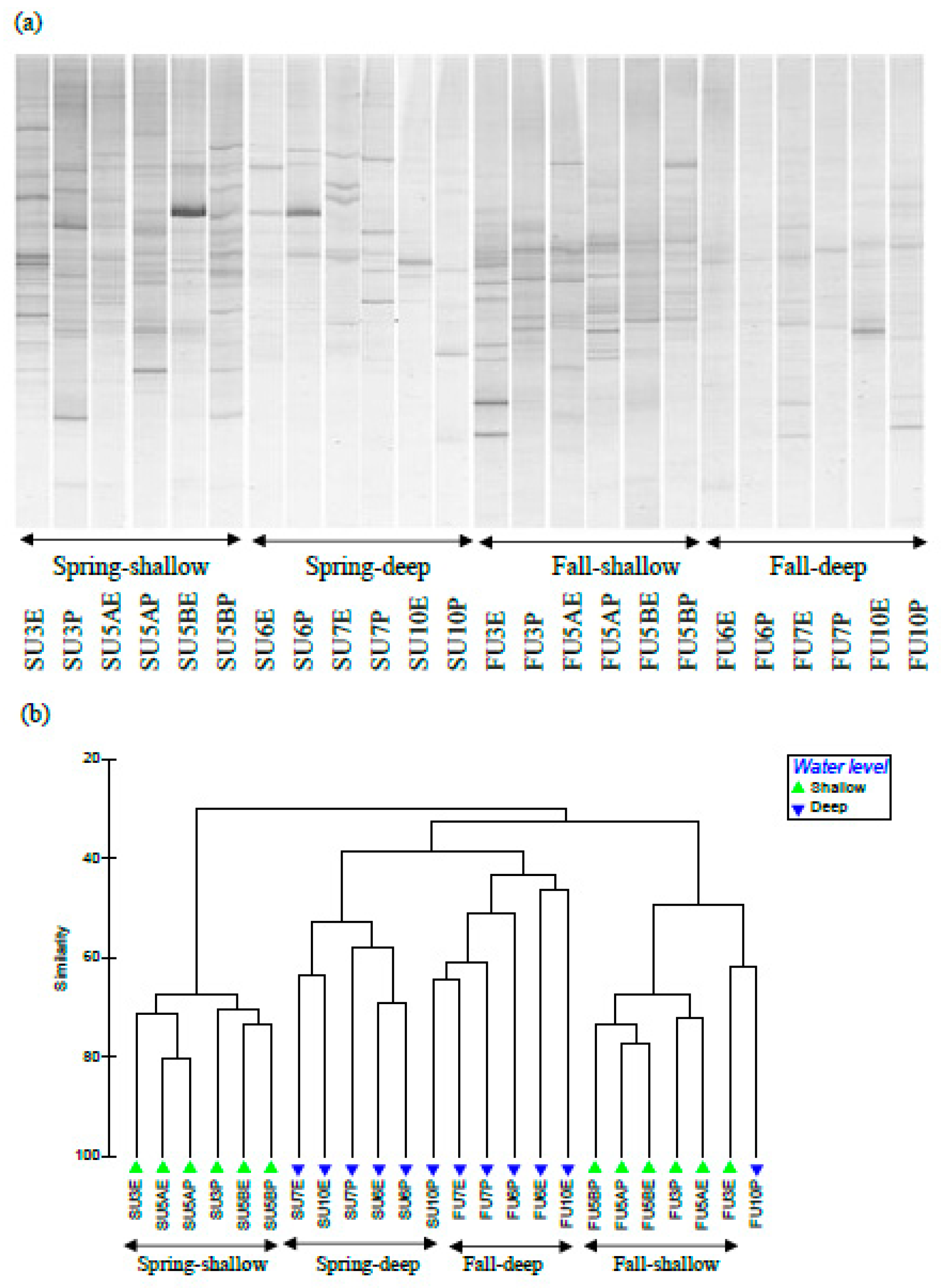
3.2. Soil fungal Communities
3.3. Soil Bacterial and Archaeal Communities
3.4. Soil Nitrification Archaea and Bacteria
3.5. Soil Denitrification Bacteria
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hattenschwiler, S.; Tiunov, A.V.; Scheu, S. Biodiversity and litter decomposition interrestrial ecosystems. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 191–218. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 1980, 286, 885–886. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Berg, G. Plant–microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Van Der Wal, A.; Geydan, T.D.; Kuyper, T.W.; De Boer, W. A thready affair: Linking fungal diversity and community dynamics to terrestrial decomposition processes. FEMS Microbiol. Rev. 2013, 37, 477–494. [Google Scholar] [CrossRef]
- Bossuyt, H.; Denef, K.; Six, J.; Frey, S.; Merckx, R.; Paustian, K. Influence of microbial populations and residue quality on aggregate stability. Appl. Soil Ecol. 2001, 16, 195–208. [Google Scholar] [CrossRef]
- Denef, K.; Six, J.; Paustian, K.; Merckx, R. Importance of macroaggregate dynamics in controlling soil carbon stabilization: Short-term effects of physical disturbance induced by dry–wet cycles. Soil Boil. Biochem. 2001, 33, 2145–2153. [Google Scholar] [CrossRef]
- Levy-Booth, D.J.; Prescott, C.; Grayston, S.J. Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Boil. Biochem. 2014, 75, 11–25. [Google Scholar] [CrossRef]
- Schleper, C.; Holben, W.; Klenk, H.P. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl. Environ. Microbiol. 1997, 63, 321–323. [Google Scholar] [CrossRef] [Green Version]
- Elshahed, M.S.; Najar, F.Z.; Roe, B.A.; Oren, A.; Dewers, T.A.; Krumholz, L.R. Survey of Archaeal Diversity Reveals an Abundance of Halophilic Archaea in a Low-Salt, Sulfide- and Sulfur-Rich Spring. Appl. Environ. Microbiol. 2004, 70, 2230–2239. [Google Scholar] [CrossRef] [Green Version]
- Buckley, D.H.; Graber, J.R.; Schmidt, T.M. Phylogenetic Analysis of Nonthermophilic Members of the Kingdom Crenarchaeota and Their Diversity and Abundance in Soils. Appl. Environ. Microbiol. 1998, 64, 4333–4339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oline, D.K.; Schmidt, S.K.; Grant, M.C. Biogeography and Landscape-Scale Diversity of the Dominant Crenarchaeota of Soil. Microb. Ecol. 2006, 52, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Joulian, C.; Ollivier, B.; Patel, B.K.C.; Roger, P.A. Phenotypic and phylogenetic characterization of dominant culturable methanogens isolated from ricefield soils. Fems Microbiol. Ecol. 1998, 25, 135–145. [Google Scholar] [CrossRef]
- Leininger, S.; Urich, T.; Schloter, M.; Schwark, L.; Qi, J.; Nicol, G.W.; Prosser, J.I.; Schuster, S.C.; Schleper, C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 2006, 442, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Kemnitz, D.; Kolb, S.; Conrad, R. High abundance of Crenarchaeota in a temperate acidic forest soil. FEMS Microbiol. Ecol. 2007, 60, 442–448. [Google Scholar] [CrossRef] [Green Version]
- Jurgens, G.; Lindstrom, K.; Saano, A. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl. Environ. Microbiol. 1997, 63, 803–805. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, L.; Bricio, C.; Blesa, A.; Hidalgo, A.; Berenguer, J. Transferable Denitrification Capability of Thermus thermophilus. Appl. Environ. Microbiol. 2013, 80, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Francis, C.; Roberts, K.J.; Beman, J.M.; Santoro, A.; Oakley, B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 2005, 102, 14683–14688. [Google Scholar] [CrossRef] [Green Version]
- Jia, Z.J.; Conrad, R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 2009, 11, 1658–1671. [Google Scholar] [CrossRef]
- Gubry-Rangin, C.; Nicol, G.W.; Prosser, J.I. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol. Ecol. 2010, 74, 566–574. [Google Scholar] [CrossRef] [Green Version]
- Erguder, T.H.; Boon, N.; Wittebolle, L.; Marzorati, M.; Verstraete, W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol. Rev. 2009, 33, 855–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szukics, U.; Hackl, E.; Zechmeister-Boltenstern, S.; Sessitsch, A. Rapid and dissimilar response of ammonia oxidizing archaea and bacteria to nitrogen and water amendment in two temperate forest soils. Microbiol. Res. 2011, 167, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Lücker, S.; Wagner, M. A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria. Trends Microbiol. 2016, 24, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Wertz, S.; Poly, F.; Le Roux, X.; Degrange, V.; Degrange, V. Development and application of a PCR-denaturing gradient gel electrophoresis tool to study the diversity of Nitrobacter-like nxrA sequences in soil. FEMS Microbiol. Ecol. 2008, 63, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Broeckling, C.D.; Broz, A.K.; Bergelson, J.; Manter, D.K.; Vivanco, J.M. Root Exudates Regulate Soil Fungal Community Composition and Diversity▿. Appl. Environ. Microbiol. 2007, 74, 738–744. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Chen, C.; Xu, Z.; Liu, Y. Effects of single and mixed species forest ecosystems on diversity and function of soil microbial community in subtropical China. J. Soils Sediments 2011, 12, 228–240. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.; Holden, P. Influence of Drying-Rewetting Frequency on Soil Bacterial Community Structure. Microb. Ecol. 2003, 45, 63–71. [Google Scholar] [CrossRef]
- Kieft, T.L.; Amy, P.S.; Brockman, F.J.; Fredrickson, J.K.; Bjornstad, B.N.; Rosacker, L.L. Microbial abundance and activities in relation to water potential in the vadose zones of arid and semiarid sites. Microb. Ecol. 1993, 26, 59–78. [Google Scholar] [CrossRef]
- Schimel, J.P.; Gulledge, J.; Clein-Curley, J.S.; Lindstrom, J.E.; Braddock, J.F. Moisture effects on microbial activity and community structure in decomposing birch litter in the Alaskan taiga. Soil Boil. Biochem. 1999, 31, 831–838. [Google Scholar] [CrossRef]
- Wilkinson, S.; Anderson, J.M.; Scardelis, S.; Tisiafouli, M.; Taylor, A.; Wolters, V. PLFA profiles of microbial communities in decomposing conifer litters subject to moisture stress. Soil Boil. Biochem. 2002, 34, 189–200. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P.; Holden, P.A. Variations in microbial community composition through two soil depth profiles. Soil Boil. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef]
- Felsmann, K.; Baudis, M.; Gimbel, K.; Kayler, Z.E.; Ellerbrock, R.; Bruehlheide, H.; Bruckhoff, J.; Welk, E.; Puhlmann, H.; Weiler, M.; et al. Soil Bacterial Community Structure Responses to Precipitation Reduction and Forest Management in Forest Ecosystems across Germany. PLoS ONE 2015, 10, e0122539. [Google Scholar] [CrossRef]
- Gleeson, D.B.; Muller, C.; Banerjee, S.; Ma, W.; Siciliano, S.D.; Murphy, D. Response of ammonia oxidizing archaea and bacteria to changing water filled pore space. Soil Boil. Biochem. 2010, 42, 1888–1891. [Google Scholar] [CrossRef]
- Gleeson, D.B.; Herrmann, A.M.; Livesley, S.; Murphy, D. Influence of water potential on nitrification and structure of nitrifying bacterial communities in semiarid soils. Appl. Soil Ecol. 2008, 40, 189–194. [Google Scholar] [CrossRef]
- Singh, B.K.; Dawson, L.A.; Macdonald, C.A.; Buckland, S.M. Impact of biotic and abiotic interaction on soil microbial communities and functions: A field study. Appl. Soil Ecol. 2009, 41, 239–248. [Google Scholar] [CrossRef]
- Jones, C.M.; Hallin, S. Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J. 2010, 4, 633–641. [Google Scholar] [CrossRef]
- Hamer, J.; Veneklaas, E.J.; Renton, M.; Poot, P. Links between soil texture and root architecture of Eucalyptus species may limit distribution ranges under future climates. Plant Soil 2015, 403, 217–229. [Google Scholar] [CrossRef]
- Coyle, D.R.; Coleman, M.D.; Aubrey, D.P. Above- and below-ground biomass accumulation, production, and distribution of sweetgum and loblolly pine grown with irrigation and fertilization. Can. J. For. Res. 2008, 38, 1335–1348. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.W. Determination of ammonium in KCl extracts of soils by the salicylate method. Commun. Soil Sci. Plant Anal. 1983, 14, 1051–1062. [Google Scholar] [CrossRef]
- Mikkonen, A.; Santalahti, M.E.; Lappi, K.; Pulkkinen, A.-M.; Montonen, L.; Suominen, L. Bacterial and archaeal communities in long-term contaminated surface and subsurface soil evaluated through coextracted RNA and DNA. FEMS Microbiol. Ecol. 2014, 90, 103–114. [Google Scholar] [CrossRef]
- Wu, T.; Chellemi, D.O.; Graham, J.H.; Martin, K.; Rosskopf, E.N. Comparison of Soil Bacterial Communities Under Diverse Agricultural Land Management and Crop Production Practices. Microb. Ecol. 2007, 55, 293–310. [Google Scholar] [CrossRef] [PubMed]
- Mills, D.; Fitzgerald, K.; Litchfield, C.D.; Gillevet, P.M. A comparison of DNA profiling techniques for monitoring nutrient impact on microbial community composition during bioremediation of petroleum-contaminated soils. J. Microbiol. Methods 2003, 54, 57–74. [Google Scholar] [CrossRef]
- Amann, R.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casamayor, E.O.; Schäfer, H.; Bañeras, L.; Pedrós-Alió, C.; Muyzer, G. Identification of and Spatio-Temporal Differences between Microbial Assemblages from Two Neighboring Sulfurous Lakes: Comparison by Microscopy and Denaturing Gradient Gel Electrophoresis. Appl. Environ. Microbiol. 2000, 66, 499–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997, 63, 4704–4712. [Google Scholar] [CrossRef] [Green Version]
- Throback, I.N.; Enwall, K.; Jarvis, A.; Hallin, S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbio. Ecol. 2004, 49, 401–417. [Google Scholar] [CrossRef]
- Michotey, V.; Méjean, V.; Bonin, P. Comparison of Methods for Quantification of Cytochrome cd1-Denitrifying Bacteria in Environmental Marine Samples. Appl. Environ. Microbiol. 2000, 66, 1564–1571. [Google Scholar] [CrossRef] [Green Version]
- Pastorelli, R.; Piccolo, R.; Simoncini, S.; Landi, S. New Primers for Denaturing Gradient Gel Electrophoresis Analysis of Nitrate-Reducing Bacterial Community in Soil. Pedosphere 2013, 23, 340–349. [Google Scholar] [CrossRef]
- Kutvonen, H.; Rajala, P.; Carpén, L.; Bomberg, M. Nitrate and ammonia as nitrogen sources for deep subsurface microorganisms. Front. Microbiol. 2015, 6, 98. [Google Scholar] [CrossRef] [Green Version]
- Hallin, S.; Lindgren, P.E. PCR detection of genes encoding nitrile reductase in denitrifying bacteria. Appl. Environ. Microbiol. 1999, 65, 1652–1657. [Google Scholar] [CrossRef] [Green Version]
- Goldmann, K.; Schröter, K.; Pena, R.; Schöning, I.; Schrumpf, M.; Buscot, F.; Polle, A.; Wubet, T. Divergent habitat filtering of root and soil fungal communities in temperate beech forests. Sci. Rep. 2016, 6, 31439. [Google Scholar] [CrossRef] [PubMed]
- Barbi, F.; Prudent, E.; Vallon, L.; Buée, M.; Dubost, A.; Legout, A.; Marmeisse, R.; Fraissinet-Tachet, L.; Luis, P. Tree species select diverse soil fungal communities expressing different sets of lignocellulolytic enzyme-encoding genes. Soil Boil. Biochem. 2016, 100, 149–159. [Google Scholar] [CrossRef]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Boil. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Zumsteg, A.; Bååth, E.; Stierli, B.; Zeyer, J.; Frey, B. Bacterial and fungal community responses to reciprocal soil transfer along a temperature and soil moisture gradient in a glacier forefield. Soil Boil. Biochem. 2013, 61, 121–132. [Google Scholar] [CrossRef]
- Peltoniemi, K.; Fritze, H.; Laiho, R. Response of fungal and actinobacterial communities to water-level drawdown in boreal peatland sites. Soil Boil. Biochem. 2009, 41, 1902–1914. [Google Scholar] [CrossRef] [Green Version]
- Sahrawat, K.L. Factors Affecting Nitrification in Soils. Commun. Soil Sci. Plant Anal. 2008, 39, 1436–1446. [Google Scholar] [CrossRef] [Green Version]
- Bowles, M.; Nigro, L.M.; Teske, A.P.; Joye, S. Denitrification and environmental factors influencing nitrate removal in Guaymas Basin hydrothermally altered sediments. Front. Microbiol. 2012, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Manis, E.; Royer, T.V.; Johnson, L.T.; Leff, L. Denitrification in Agriculturally Impacted Streams: Seasonal Changes in Structure and Function of the Bacterial Community. PLOS ONE 2014, 9, e105149. [Google Scholar] [CrossRef]
- Zhang, X.; Barberán, A.; Zhu, X.; Zhang, G.; Han, X. Water Content Differences Have Stronger Effects than Plant Functional Groups on Soil Bacteria in a Steppe Ecosystem. PLOS ONE 2014, 9, e115798. [Google Scholar] [CrossRef]
- Szukics, U.; Abell, G.C.; Hödl, V.; Mitter, B.; Sessitsch, A.; Hackl, E.; Zechmeister-Boltenstern, S. Nitrifiers and denitrifiers respond rapidly to changed moisture and increasing temperature in a pristine forest soil. FEMS Microbiol. Ecol. 2010, 72, 395–406. [Google Scholar] [CrossRef]
- Morales, S.; Cosart, T.; Holben, W.E. Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J. 2010, 4, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Cavigelli, M.A.; Robertson, G.P. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology 2000, 81, 1402–1414. [Google Scholar] [CrossRef]
- Huhe; Borjigin, S.; Buhebaoyin; Wu, Y.; Li, M.; Cheng, Y. Microbial Nitrogen-Cycle Gene Abundance in Soil of Cropland Abandoned for Different Periods. PLOS ONE 2016, 11, e0154697. [Google Scholar] [CrossRef] [PubMed]
- Zogg, G.P.; Zak, D.; Ringelberg, D.B.; White, D.C.; Macdonald, N.W.; Pregitzer, K.S. Compositional and Functional Shifts in Microbial Communities Due to Soil Warming. Soil Sci. Soc. Am. J. 1997, 61, 475. [Google Scholar] [CrossRef] [Green Version]
- Smit, E.; Leeflang, P.; Gommans, S.; Broek, J.V.D.; Van Mil, S.; Wernars, K. Diversity and Seasonal Fluctuations of the Dominant Members of the Bacterial Soil Community in a Wheat Field as Determined by Cultivation and Molecular Methods. Appl. Environ. Microbiol. 2001, 67, 2284–2291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.; Yeom, J.; Han, J.; Kim, J.; Park, W. Seasonal changes in nitrogen-cycle gene abundances and in bacterial communities in acidic forest soils. J. Microbiol. 2012, 50, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Bru, D.; Ramette, A.; Saby, N.P.A.; Dequiedt, S.; Ranjard, L.; Jolivet, C.; Arrouays, D.; Philippot, L. Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 2010, 5, 532–542. [Google Scholar] [CrossRef] [Green Version]
- Ferroni, G.D.; Kaminski, J.S. Psychrophiles, psychrotrophs, and mesophiles in an environment which experiences seasonal temperature fluctuations. Can. J. Microbiol. 1980, 26, 1184–1191. [Google Scholar] [CrossRef]




| Gene | Primer | Primer Sequences | References |
|---|---|---|---|
| amoA AOA | Arch-amoAF * | 5′-(C/G)TAATGGTCTGGCTTAGACG-3′ | [18] |
| Arch-amoAR | 5′-GCGGCCATCCATCTGTATGT-3′ | ||
| amoA AOB | amoA-1F * | 5’-GGGGTTTCTACTGGTGGT-3′ | [45] |
| amoA-2R | 5’-CCCCTC(G/T)G(C/G)AAAGCCTTCTTC-3′ | ||
| nrxA | F1370 F1 nrxA | 5’-CAGACCGACGTGTGCGAAAG-3’ | [24] |
| F2843 R2 nrxA * | 5’-TCCACAAGGAACGGAAGGTC-3’ | ||
| narG | narG2179F | 5’-TAC(A/T)T(C/G)CT(C/G)AAGTACCT(C/G)CT-3’ | [48] |
| narG2488R * | 5’-C(C/G)TTGTAGATCTCCCA(A/G)TC-3’ | [49] | |
| nirK | F1aCu | 5’-ATCATGGT(C/G)CTGCCGCG-3’ | [50] |
| R3Cu * | 5’-GCCTCGATCAG(A/G)TTGTGGTT-3’ | ||
| nirS | cd3aF | 5’-GT(C/G) AAC GT(C/G)AAG GA(A/G)AC(C/G)GG-3’ | [47] |
| R3cd * | 5’-GA(C/G)TTCGG(A/G)TG(C/G)GTCTTGA-3’ | ||
| nosZ | nosZ-F | 5’-CG(C/T)TGT TC(A/C)TCGACAGCCAG-3’ | [46] |
| nosZ1622R * | 5’-CGC (G/A)A(C/G)GGCAA(C/G)AAG GT(C/G)CG-3’ |
| Effects and Interactions | Soil Organic Matter (%) * | Soil pH ** | Soil Water (%) | Ammonium (ppm) | Nitrate (ppm) | Phosphorus (ppm) | Potassium (ppm) | Magnesium (ppm) | Calcium (ppm) | CEC (meq/100 g) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Groundwater | Deep | 2.0 ± 0.3 a | 4.2 ± 0.2 a | 11.6 ± 4.3 b | 0.6 ± 0.2 a | 26.9 ± 7.8 a | 80.5 ± 11.2 a | 34.4 ± 4.9 a | 34.2 ± 9.1 a | 116.6 ± 94.2 a | 4.0 ± 0.8 a |
| Shallow | 2.4 ± 0.3 a | 3.8 ± 0.1 b | 17.9 ± 6.5 a | 1.0 ± 0.5 a | 33.0 ± 9.5 a | 96.0 ± 18.2a | 37.8 ± 8.5a | 42.4 ± 11.9 a | 300.0 ± 134.7 a | 5.1 ± 1.2 a | |
| Tree | Eucalypt | 2.9 ± 0.3 a | 4.1 ± 0.2 a | 18.3 ±5.9 a | 0.6 ± 0.1 a | 29.3 ± 7.7 a | 111.1 ±18.7 a | 42.8 ± 8.9 a | 54.5 ± 13.0 a | 319.9 ± 133.9 a | 6.3 ± 1.3 a |
| Pine | 1.5 ± 0.1 b | 3.9 ± 0.1 a | 11.3 ± 4.4 b | 1.0 ± 0.5 a | 30.7 ± 9.7 a | 65.4 ± 4.9 b | 29.3 ± 3.0 a | 22.1 ± 3.4 b | 96.7 ± 14.9 a | 2.9 ± 0.4 b | |
| Season | Spring | 2.4 ± 0.2 a | 4.1 ± 0.2 a | 14.3 ± 4.9 a | 0.7 ± 0.2 a | 47.6 ± 9.7 a | 65.0 ± 6.1 b | 47.2 ± 8.3 a | 57.2 ± 12.5 a | 327.8 ± 132.9 a | 6.3 ± 1.3 a |
| Fall | 2.1 ± 0.3 a | 3.9 ± 0.1 a | 15.7 ± 7.6 a | 0.9 ± 0.5 a | 12.3 ± 1.6 b | 111.5 ± 18.3 a | 25.0 ± 8.2 b | 19.4 ± 2.9 b | 88.8 ±12.4 a | 2.9 ± 0.4 b | |
| Significance level | |||||||||||
| Groundwater | 0.2736 | 0.0443 | 0.0012 | 0.4336 | 0.5830 | 0.2249 | 0.6894 | 0.4339 | 0.1300 | 0.2929 | |
| Tree | 0.0006 | 0.6223 | 0.0004 | 0.4495 | 0.8978 | 0.0078 | 0.1180 | 0.0058 | 0.0696 | 0.0045 | |
| Season | 0.3406 | 0.5369 | 0.5731 | 0.7148 | 0.0048 | 0.0089 | 0.0150 | 0.0019 | 0.0537 | 0.0043 | |
| Groundwater × Tree | 0.5380 | 0.9121 | 0.5244 | 0.7722 | 0.8150 | 0.1814 | 0.6039 | 0.3410 | 0.2485 | 0.3923 | |
| Groundwater × Season | 0.8624 | 0.0976 | 0.7863 | 0.3169 | 0.4149 | 0.0704 | 0.5500 | 0.4810 | 0.1479 | 0.3363 | |
| Tree × Season | 0.4465 | 0.3769 | 0.0555 | 0.3183 | 0.8267 | 0.0083 | 0.0651 | 0.0398 | 0.1323 | 0.0519 | |
Gene | Season | Water Level | Plant Species | |||
|---|---|---|---|---|---|---|
| Spring | Fall | Spring | Fall | |||
| Nitrification | amoA AOA | 0.01 * | 0.02 * | 0.58 | 0.68 | 0.86 |
| amoA AOB | nd | nd | nd | nd | nd | |
| nrxA | nd | nd | nd | nd | nd | |
| Denitrification | narG | nd | nd | nd | nd | nd |
| nirK | 0.01 * | 0.04 * | 0.22 | 0.46 | 0.63 | |
| nirS | nd | nd | nd | nd | nd | |
| nosZ | 0.01 * | 0.83 | 0.32 | 0.61 | 0.88 | |
| Genes | Variables | Correlation * |
|---|---|---|
| amoA AOA | Nitrate (ppm) | 17.4% |
| Mg (ppm) | 11.5% | |
| Ca (ppm) | 10.4% | |
| nosZ | All less than 10.0% | |
| nirK | Nitrate (ppm) | 16.6% |
| Mg (ppm) | 14.0% | |
| K (ppm) | 13.1% | |
| Water (%) | 10.7% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, T.; Gray, A.; Liu, G.; Kaminski, H.; Osi Efa, B.; Aubrey, D.P. Groundwater Depth Overrides Tree-Species Effects on the Structure of Soil Microbial Communities Involved in Nitrogen Cycling in Plantation Forests. Forests 2020, 11, 275. https://doi.org/10.3390/f11030275
Wu T, Gray A, Liu G, Kaminski H, Osi Efa B, Aubrey DP. Groundwater Depth Overrides Tree-Species Effects on the Structure of Soil Microbial Communities Involved in Nitrogen Cycling in Plantation Forests. Forests. 2020; 11(3):275. https://doi.org/10.3390/f11030275
Chicago/Turabian StyleWu, Tiehang, Ashley Gray, Gan Liu, Hilary Kaminski, Bolanle Osi Efa, and Doug P. Aubrey. 2020. "Groundwater Depth Overrides Tree-Species Effects on the Structure of Soil Microbial Communities Involved in Nitrogen Cycling in Plantation Forests" Forests 11, no. 3: 275. https://doi.org/10.3390/f11030275
APA StyleWu, T., Gray, A., Liu, G., Kaminski, H., Osi Efa, B., & Aubrey, D. P. (2020). Groundwater Depth Overrides Tree-Species Effects on the Structure of Soil Microbial Communities Involved in Nitrogen Cycling in Plantation Forests. Forests, 11(3), 275. https://doi.org/10.3390/f11030275






