An Automatic UAV Based Segmentation Approach for Pruning Biomass Estimation in Irregularly Spaced Chestnut Orchards
Abstract
1. Introduction
2. Materials and Methods
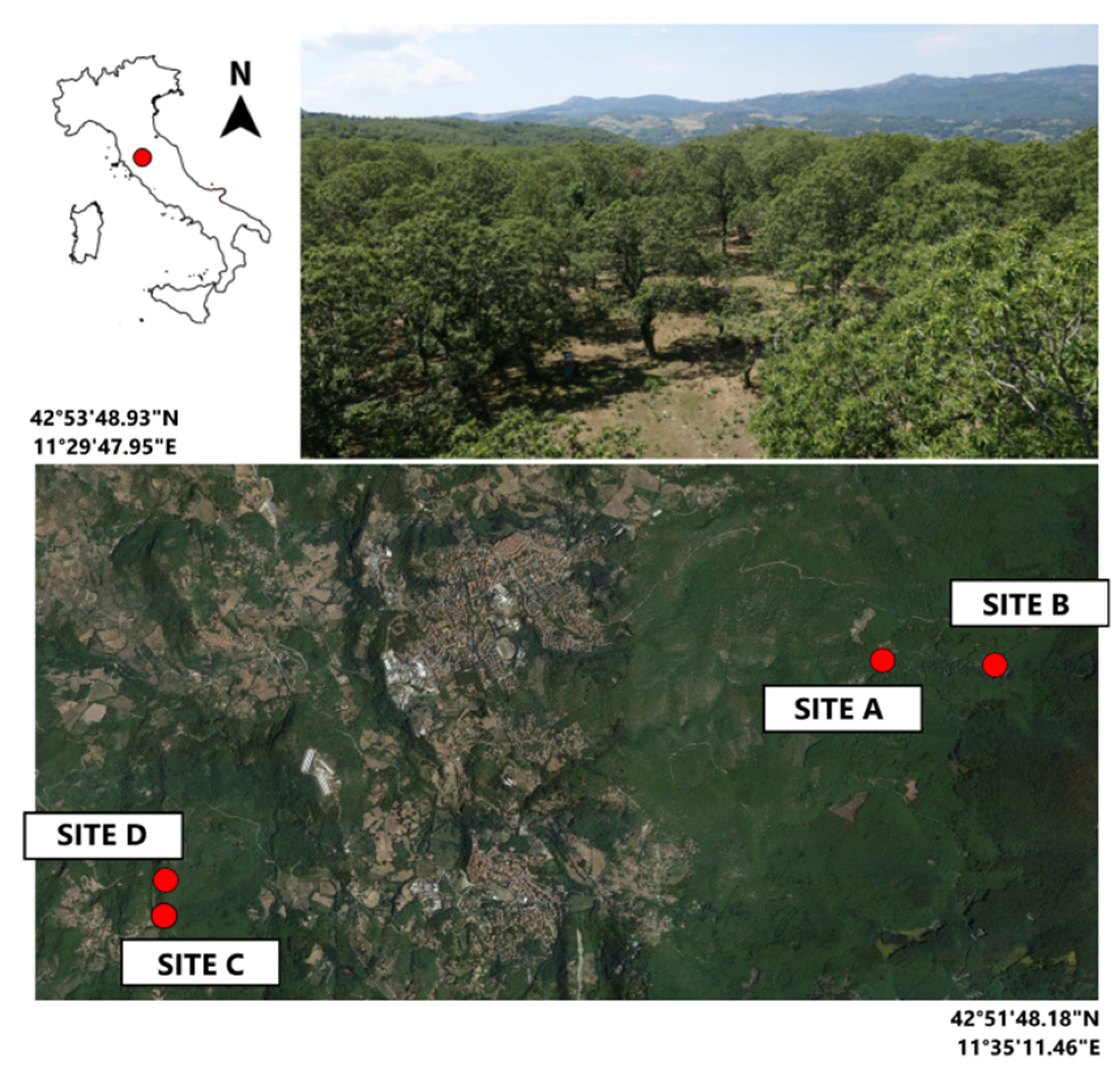
2.1. Experimental Sites
2.2. Pruning Wood Biomass Ground Measurement
2.3. UAV Platform and Data Processing
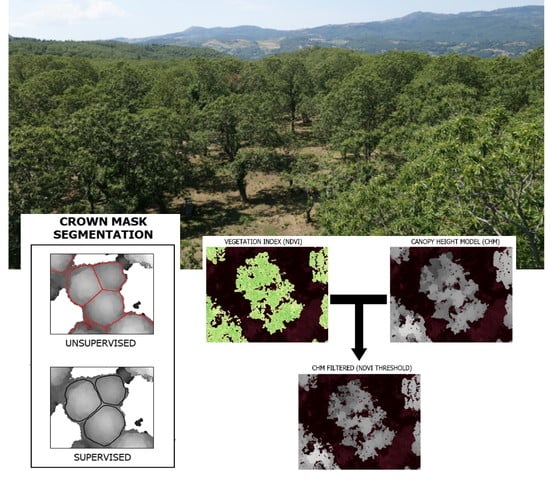
2.4. Double Filtering Approach
2.5. Puning Wood Biomass Estimation
3. Results
3.1. Wpw Ground Measurement
3.2. Supervised Data Extraction
3.3. Unsupervised Data Extraction
3.4. Geometric Data Comparison between the Supervised and the Unsupervised Approach
3.5. Wpw Estimation
4. Discussion
5. Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gallo, R.; Grigolato, S.; Cavalli, R.; Mazzetto, F. GNSS-based operational monitoring devices for forest logging operation chains. J. Agric. Eng. 2013, 44, 140–144. [Google Scholar] [CrossRef]
- Rossi, F.C.; Fritz, A.; Becker, G. Combining satellite and UAV imagery to delineate forest cover and basal area after mixed-severity fires. Sustainability 2018, 10, 2227. [Google Scholar] [CrossRef]
- Peña, M.A.; Altmann, S.H. Use of satellite-derived hyperspectral indices to identify stress symptoms in an Austrocedrus chilensis forest infested by the aphid Cinara cupressi. Int. J. Pest Manag. 2009, 55, 197–206. [Google Scholar] [CrossRef]
- Paiva, P.F.P.R.; Ruivo, M.D.L.P.; Júnior, O.M.D.S.; Maciel, M.D.N.M.; Braga, T.G.M.; De Andrade, M.M.N.; Junior, P.C.D.S.; Da Rocha, E.S.; De Freitas, T.P.M.; Leite, T.V.D.S.; et al. Deforestation in protect areas in the Amazon: A threat to biodiversity. Biodivers. Conserv. 2019, 29, 19–38. [Google Scholar] [CrossRef]
- Dash, J.P.; Pearse, G.D.; Watt, M.S. UAV multispectral imagery can complement satellite data for monitoring forest health. Remote Sens. 2018, 10, 1216. [Google Scholar] [CrossRef]
- Matese, A.; Toscano, P.; Di Gennaro, S.F.; Genesio, L.; Vaccari, F.P.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B. Intercomparison of UAV, aircraft and satellite remote sensing platforms for precision viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef]
- Yu, L.; Huang, J.; Zong, S.; Huang, H.; Luo, Y. Detecting shoot beetle damage on Yunnan pine using Landsat time-series data. Forests 2018, 9, 39. [Google Scholar]
- Puliti, S.; Ene, L.T.; Gobakken, T.; Næsset, E. Use of partial-coverage UAV data in sampling for large scale forest inventories. Remote Sens. Environ. 2017, 194, 115–126. [Google Scholar] [CrossRef]
- Fankhauser, K.E.; Strigul, N.S.; Gatziolis, D. Augmentation of traditional forest inventory and Airborne laser scanning with unmanned aerial systems and photogrammetry for forest monitoring. Remote Sens. 2018, 10, 1562. [Google Scholar] [CrossRef]
- Clarivate. Web of Science (All Databases). Available online: http://clarivate.com (accessed on 23 December 2019).
- Safonova, A.; Tabik, S.; Alcaraz-Segura, D.; Rubtsov, A.; Maglinets, Y.; Herrera, F. Detection of Fir Trees (Abies sibirica) Damaged by the Bark Beetle in Unmanned Aerial Vehicle Images with Deep Learning. Remote Sens. 2019, 11, 643. [Google Scholar] [CrossRef]
- Swinfield, T.; Lindsell, J.A.; Williams, J.V.; Harrison, R.D.; Agustiono; Habibi; Gemita, E.; Schönlieb, C.B.; Coomes, D.A. Accurate Measurement of Tropical Forest Canopy Heights and Aboveground Carbon Using Structure from Motion. Remote Sens. 2019, 11, 928. [Google Scholar] [CrossRef]
- Roşca, S.; Suomalainen, J.; Bartholomeus, H.; Herold, M. Comparing terrestrial laser scanning and unmanned aerial vehicle structure from motion to assess top of canopy structure in tropical forests. Interface Focus 2018, 8, 20170038. [Google Scholar] [CrossRef]
- Liang, X.; Wang, Y.; Pyörälä, J.; Lehtomäki, M.; Yu, X.; Kaartinen, H.; Kukko, A.; Honkavaara, E.; Issaoui, A.E.I.; Nevalainen, O.; et al. Forest in situ observations using unmanned aerial vehicle as an alternative of terrestrial measurements. For. Ecosyst. 2019, 6, 20. [Google Scholar] [CrossRef]
- Maes, W.H.; Huete, A.R.; Avino, M.; Boer, M.M.; Dehaan, R.; Pendall, E.; Griebel, A.; Steppe, K. Can UAV-based infrared thermography be used to study plant-parasite interactions between mistletoe and Eucalypt trees? Remote Sens. 2018, 10, 2062. [Google Scholar] [CrossRef]
- Grznárová, A.; Mokroš, M.; Surový, P.; Slavík, M.; Pondelík, M.; Mergani, J. The crown diameter estimation from fixed wing type of uav imagery. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. ISPRS Arch. 2019, 42, 337–341. [Google Scholar] [CrossRef]
- Pádua, L.; Adão, T.; Guimarães, N.; Sousa, A.; Peres, E.; Sousa, J.J. Post-fire forestry recovery monitoring using high-resolution multispectral imagery from unmanned aerial vehicles. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. ISPRS Arch. 2019, 42, 301–305. [Google Scholar] [CrossRef]
- Puliti, S.; Saarela, S.; Gobakken, T.; Ståhl, G.; Næsset, E. Combining UAV and Sentinel-2 auxiliary data for forest growing stock volume estimation through hierarchical model-based inference. Remote Sens. Environ. 2018, 204, 485–497. [Google Scholar] [CrossRef]
- Aguilar, F.J.; Rivas, J.R.; Nemmaoui, A.; Peñalver, A.; Aguilar, M.A. UAV-based digital terrain model generation under leaf-off conditions to support teak plantations inventories in tropical dry forests. A case of the coastal region of Ecuador. Sensors 2019, 19, 1934. [Google Scholar] [CrossRef]
- Iizuka, K.; Kato, T.; Silsigia, S.; Soufiningrum, A.Y.; Kozan, O. Estimating and examining the sensitivity of different vegetation indices to fractions of vegetation cover at different scaling Grids for Early Stage Acacia Plantation Forests Using a Fixed-Wing UAS. Remote Sens. 2019, 11, 1816. [Google Scholar] [CrossRef]
- Zou, X.; Liang, A.; Wu, B.; Su, J.; Zheng, R.; Li, J. UAV-based high-throughput approach for fast growing Cunninghamia lanceolata (Lamb.) cultivar screening by machine learning. Forests 2019, 10, 815. [Google Scholar] [CrossRef]
- Frey, J.; Kovach, K.; Stemmler, S.; Koch, B. UAV photogrammetry of forests as a vulnerable process. A sensitivity analysis for a structure from motion RGB-image pipeline. Remote Sens. 2018, 10, 912. [Google Scholar] [CrossRef]
- Hentz, Â.M.K.; Silva, C.A.; Dalla Corte, A.P.; Netto, S.P.; Strager, M.P.; Klauberg, C. Estimating forest uniformity in Eucalyptus spp. and Pinus taeda L. stands using field measurements and structure from motion point clouds generated from unmanned aerial vehicle (UAV) data collection. For. Syst. 2018, 27, 1–17. [Google Scholar] [CrossRef]
- Liu, K.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Estimating forest structural attributes using UAV-LiDAR data in Ginkgo plantations. ISPRS J. Photogramm. Remote Sens. 2018, 146, 465–482. [Google Scholar] [CrossRef]
- Giannetti, F.; Chirici, G.; Gobakken, T.; Næsset, E.; Travaglini, D.; Puliti, S. A new approach with DTM-independent metrics for forest growing stock prediction using UAV photogrammetric data. Remote Sens. Environ. 2018, 213, 195–205. [Google Scholar] [CrossRef]
- Fraser, B.T.; Congalton, R.G. Evaluating the effectiveness of Unmanned Aerial Systems (UAS) for collecting thematic map accuracy assessment reference data in New England forests. Forests 2019, 10, 24. [Google Scholar] [CrossRef]
- Liebermann, H.; Strager, M.P. Using Unmanned Aerial Systems for Deriving Forest Stand Characteristics in Mixed Hardwoods of West Virginia Using. J. Geospatial Appl. Nat. Resour. 2018, 2, 1–31. [Google Scholar]
- Kattenborn, T.; Lopatin, J.; Förster, M.; Braun, A.C.; Fassnacht, F.E. UAV data as alternative to field sampling to map woody invasive species based on combined Sentinel-1 and Sentinel-2 data. Remote Sens. Environ. 2019, 227, 61–73. [Google Scholar] [CrossRef]
- Tuominen, S.; Balazs, A.; Honkavaara, E.; Pölönen, I.; Saari, H.; Hakala, T.; Viljanen, N. Hyperspectral UAV-Imagery and photogrammetric canopy height model in estimating forest stand variables. Silva Fenn. 2017, 51, 1–21. [Google Scholar] [CrossRef]
- Park, J.Y.; Muller-Landau, H.C.; Lichstein, J.W.; Rifai, S.W.; Dandois, J.P.; Bohlman, S.A. Quantifying leaf phenology of individual trees and species in a tropical forest using unmanned aerial vehicle (UAV) images. Remote Sens. 2019, 11, 1534. [Google Scholar] [CrossRef]
- Chung, C.H.; Wang, C.H.; Hsieh, H.C.; Huang, C.Y. Comparison of forest canopy height profiles in a mountainous region of Taiwan derived from airborne lidar and unmanned aerial vehicle imagery. GISci. Remote Sens. 2019, 56, 1289–1304. [Google Scholar] [CrossRef]
- Carr, J.C.; Slyder, J.B. Individual tree segmentation from a leaf-off photogrammetric point cloud. Int. J. Remote Sens. 2018, 39, 5195–5210. [Google Scholar] [CrossRef]
- Jayathunga, S.; Owari, T.; Tsuyuki, S. Digital Aerial Photogrammetry for Uneven-Aged Forest Management: Assessing the Potential to Reconstruct Canopy Structure and Estimate Living Biomass. Remote Sens. 2019, 11, 338. [Google Scholar] [CrossRef]
- Vaglio Laurin, G.; Ding, J.; Disney, M.; Bartholomeus, H.; Herold, M.; Papale, D.; Valentini, R. Tree height in tropical forest as measured by different ground, proximal, and remote sensing instruments, and impacts on above ground biomass estimates. Int. J. Appl. Earth Obs. Geoinf. 2019, 82, 101899. [Google Scholar] [CrossRef]
- De Luca, G.; Silva, J.M.N.; Cerasoli, S.; Araújo, J.; Campos, J.; Di Fazio, S.; Modica, G. Object-based land cover classification of cork oak woodlands using UAV imagery and Orfeo Toolbox. Remote Sens. 2019, 11, 1238. [Google Scholar] [CrossRef]
- Carvajal-Ramírez, F.; Serrano, J.M.P.R.; Agüera-Vega, F.; Martínez-Carricondo, P. A comparative analysis of phytovolume estimation methods based on UAV-photogrammetry and multispectral imagery in a mediterranean forest. Remote Sens. 2019, 11, 2579. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, J.; Ni, W.; Sun, G.; Zhang, Z.; Liu, Q.; Wang, Q. Estimation of Forest Leaf Area Index Using Height and Canopy Cover Information Extracted From Unmanned Aerial Vehicle Stereo Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 471–481. [Google Scholar] [CrossRef]
- McClelland, M.P.; van Aardt, J.; Hale, D. Manned aircraft versus small unmanned aerial system—Forestry remote sensing comparison utilizing lidar and structure-from-motion for forest carbon modeling and disturbance detection. J. Appl. Remote Sens. 2019, 14, 1. [Google Scholar] [CrossRef]
- MacDicken, K.G. Global Forest Resources Assessment 2015: What, why and how? For. Ecol. Manag. 2015, 352, 3–8. [Google Scholar] [CrossRef]
- Waite, C.E.; van der Heijden, G.M.F.; Field, R.; Boyd, D.S. A view from above: Unmanned aerial vehicles (UAVs) provide a new tool for assessing liana infestation in tropical forest canopies. J. Appl. Ecol. 2019, 56, 902–912. [Google Scholar] [CrossRef]
- Wu, Z.; Ni, M.; Hu, Z.; Wang, J.; Li, Q.; Wu, G. Mapping invasive plant with UAV-derived 3D mesh model in mountain area—A case study in Shenzhen Coast, China. Int. J. Appl. Earth Obs. Geoinf. 2019, 77, 129–139. [Google Scholar] [CrossRef]
- Franklin, S.E.; Ahmed, O.S. Deciduous tree species classification using object-based analysis and machine learning with unmanned aerial vehicle multispectral data. Int. J. Remote Sens. 2018, 39, 5236–5245. [Google Scholar] [CrossRef]
- Komárek, J.; Klouček, T.; Prošek, J. The potential of Unmanned Aerial Systems: A tool towards precision classification of hard-to-distinguish vegetation types? Int. J. Appl. Earth Obs. Geoinf. 2018, 71, 9–19. [Google Scholar] [CrossRef]
- Sothe, C.; Dalponte, M.; de Almeida, C.M.; Schimalski, M.B.; Lima, C.L.; Liesenberg, V.; Miyoshi, G.T.; Tommaselli, A.M.G. Tree species classification in a highly diverse subtropical forest integrating UAV-based photogrammetric point cloud and hyperspectral data. Remote Sens. 2019, 11, 1338. [Google Scholar] [CrossRef]
- Dash, J.P.; Watt, M.S.; Paul, T.S.H.; Morgenroth, J.; Pearse, G.D. Early detection of invasive exotic trees using UAV and manned aircraft multispectral and LiDAR Data. Remote Sens. 2019, 11, 1812. [Google Scholar] [CrossRef]
- Wallace, L.; Bellman, C.; Hally, B.; Hernandez, J.; Jones, S.; Hillman, S. Assessing the ability of image based point clouds captured from a UAV to measure the terrain in the presence of canopy cover. Forests 2019, 10, 284. [Google Scholar] [CrossRef]
- Seifert, E.; Seifert, S.; Vogt, H.; Drew, D.; van Aardt, J.; Kunneke, A.; Seifert, T. Influence of drone altitude, image overlap, and optical sensor resolution on multi-view reconstruction of forest images. Remote Sens. 2019, 11, 1252. [Google Scholar] [CrossRef]
- Brach, M.; Chan, J.C.W.; Szymański, P. Accuracy assessment of different photogrammetric software for processing data from low-cost UAV platforms in forest conditions. IForest 2019, 12, 435–441. [Google Scholar] [CrossRef]
- Tomaštík, J.; Mokroš, M.; Surový, P.; Grznárová, A.; Merganič, J. UAV RTK/PPK method-An optimal solution for mapping inaccessible forested areas? Remote Sens. 2019, 11, 721. [Google Scholar] [CrossRef]
- Otsu, K.; Pla, M.; Vayreda, J.; Brotons, L. Calibrating the severity of forest defoliation by pine processionary moth with landsat and UAV imagery. Sensors 2018, 18, 3278. [Google Scholar] [CrossRef]
- Cardil, A.; Otsu, K.; Pla, M.; Silva, C.A.; Brotons, L. Quantifying pine processionary moth defoliation in a pine-oak mixed forest using unmanned aerial systems and multispectral imagery. PLoS ONE 2019, 14, e0213027. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, X.; Yang, G.; Zhu, C.; Huo, L.; Feng, H. Assessment of defoliation during the Dendrolimus tabulaeformis Tsai et Liu disaster outbreak using UAV-based hyperspectral images. Remote Sens. Environ. 2018, 217, 323–339. [Google Scholar] [CrossRef]
- Larrinaga, A.R.; Brotons, L. Greenness Indices from a Low-Cost UAV Imagery as Tools for Monitoring Post-Fire Forest Recovery. Drones 2019, 3, 6. [Google Scholar] [CrossRef]
- Abdollahnejad, A.; Panagiotidis, D.; Surovỳ, P. Estimation and extrapolation of tree parameters using spectral correlation between UAV and Pléiades data. Forests 2018, 9, 85. [Google Scholar] [CrossRef]
- Domingo, D.; Ørka, H.O.; Næsset, E.; Kachamba, D.; Gobakken, T. Effects of UAV image resolution, camera type, and image overlap on accuracy of biomass predictions in a tropicalwoodland. Remote Sens. 2019, 11, 948. [Google Scholar] [CrossRef]
- Guerra-Hernández, J.; Cosenza, D.N.; Cardil, A.; Silva, C.A.; Botequim, B.; Soares, P.; Silva, M.; González-Ferreiro, E.; Díaz-Varela, R.A. Predicting growing stock volume of eucalyptus plantations using 3-D point clouds derived from UAV imagery and ALS data. Forests 2019, 10, 905. [Google Scholar] [CrossRef]
- Guerra-Hernández, J.; González-Ferreiro, E.; Monleón, V.J.; Faias, S.P.; Tomé, M.; Díaz-Varela, R.A. Use of multi-temporal UAV-derived imagery for estimating individual tree growth in Pinus pinea stands. Forests 2017, 8, 300. [Google Scholar] [CrossRef]
- Kachamba, D.J.; Ørka, H.O.; Næsset, E.; Eid, T.; Gobakken, T. Influence of plot size on efficiency of biomass estimates in inventories of dry tropical forests assisted by photogrammetric data from an unmanned aircraft system. Remote Sens. 2017, 9, 610. [Google Scholar] [CrossRef]
- Li, Z.; Zan, Q.; Yang, Q.; Zhu, D.; Chen, Y.; Yu, S. Remote estimation of mangrove aboveground carbon stock at the species level using a low-cost unmanned aerial vehicle system. Remote Sens. 2019, 11, 1018. [Google Scholar] [CrossRef]
- Sandino, J.; Pegg, G.; Gonzalez, F.; Smith, G. Aerial mapping of forests affected by pathogens using UAVs, hyperspectral sensors, and artificial intelligence. Sensors 2018, 18, 944. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Varhola, A.; Vastaranta, M.; Coops, N.C.; Cook, B.D.; Pitt, D.; Woods, M. A best practices guide for generating forest inventory attributes from airborne laser scanning data using an area-based approach. For. Chron. 2013, 89, 722–723. [Google Scholar] [CrossRef]
- Lindberg, E.; Holmgren, J. Individual Tree Crown Methods for 3D Data from Remote Sensing. Curr. For. Reports 2017, 3, 19–31. [Google Scholar] [CrossRef]
- Alonzo, M.; Andersen, H.E.; Morton, D.C.; Cook, B.D. Quantifying boreal forest structure and composition using UAV structure from motion. Forests 2018, 9, 119. [Google Scholar] [CrossRef]
- Fujimoto, A.; Haga, C.; Matsui, T.; Machimura, T.; Hayashi, K.; Sugita, S.; Takagi, H. An end to end process development for UAV-SfM based forest monitoring: Individual tree detection, species classification and carbon dynamics simulation. Forests 2019, 10, 680. [Google Scholar] [CrossRef]
- Wu, X.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Assessment of individual tree detection and canopy cover estimation using unmanned aerial vehicle based light detection and ranging (UAV-LiDAR) data in planted forests. Remote Sens. 2019, 11, 908. [Google Scholar] [CrossRef]
- Tian, J.; Dai, T.; Li, H.; Liao, C.; Teng, W.; Hu, Q.; Ma, W.; Xu, Y. A novel tree height extraction approach for individual trees by combining TLS and UAV image-based point cloud integration. Forests 2019, 10, 537. [Google Scholar] [CrossRef]
- Saarinen, N.; Vastaranta, M.; Näsi, R.; Rosnell, T.; Hakala, T.; Honkavaara, E.; Wulder, M.A.; Luoma, V.; Tommaselli, A.M.G.; Imai, N.N.; et al. Assessing biodiversity in boreal forests with UAV-based photogrammetric point clouds and hyperspectral imaging. Remote Sens. 2018, 10, 338. [Google Scholar] [CrossRef]
- Brieger, F.; Herzschuh, U.; Pestryakova, L.A.; Bookhagen, B.; Zakharov, E.S.; Kruse, S. Advances in the derivation of Northeast Siberian forest metrics using high-resolution UAV-based photogrammetric point clouds. Remote Sens. 2019, 11, 1447. [Google Scholar] [CrossRef]
- Brovkina, O.; Cienciala, E.; Surový, P.; Janata, P. Unmanned aerial vehicles (UAV) for assessment of qualitative classification of Norway spruce in temperate forest stands. Geo Spatial Inf. Sci. 2018, 21, 12–20. [Google Scholar] [CrossRef]
- Morales, G.; Kemper, G.; Sevillano, G.; Arteaga, D.; Ortega, I.; Telles, J. Automatic segmentation of Mauritia flexuosa in unmanned aerial vehicle (UAV) imagery using deep learning. Forests 2018, 9, 736. [Google Scholar] [CrossRef]
- Yan, W.; Guan, H.; Cao, L.; Yu, Y.; Gao, S.; Lu, J.Y. An automated hierarchical approach for three-dimensional segmentation of single trees using UAV LiDAR data. Remote Sens. 2018, 10, 1999. [Google Scholar] [CrossRef]
- Ganz, S.; Käber, Y.; Adler, P. Measuring tree height with remote sensing-a comparison of photogrammetric and LiDAR data with different field measurements. Forests 2019, 10, 694. [Google Scholar] [CrossRef]
- Balsi, M.; Esposito, S.; Fallavollita, P.; Nardinocchi, C. Single-tree detection in high-density LiDAR data from UAV-based survey. Eur. J. Remote Sens. 2018, 51, 679–692. [Google Scholar] [CrossRef]
- Huang, H.; Li, X.; Chen, C. Individual tree crown detection and delineation from very-high-resolution UAV images based on bias field and marker-controlled watershed segmentation algorithms. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 2253–2262. [Google Scholar] [CrossRef]
- Mayr, M.J.; Malß, S.; Ofner, E.; Samimi, C. Disturbance feedbacks on the height of woody vegetation in a savannah: A multi-plot assessment using an unmanned aerial vehicle (UAV). Int. J. Remote Sens. 2018, 39, 4761–4785. [Google Scholar] [CrossRef]
- Marques, P.; Pádua, L.; Adão, T.; Hruška, J.; Peres, E.; Sousa, A.; Sousa, J.J. UAV-based automatic detection and monitoring of chestnut trees. Remote Sens. 2019, 11, 855. [Google Scholar] [CrossRef]
- Apostol, B.; Petrila, M.; Lorenţ, A.; Ciceu, A.; Gancz, V.; Badea, O. Species discrimination and individual tree detection for predicting main dendrometric characteristics in mixed temperate forests by use of airborne laser scanning and ultra-high-resolution imagery. Sci. Total Environ. 2020, 698, 134074. [Google Scholar] [CrossRef]
- Tabacchi, G.; De Natale, F.; Di Cosmo, L.; Floris, A.; Gagliano, C.; Gasparini, P.; Genchi, L.; Scrinzi, G.; Tosi, V. INFC—Le Stime di Superficie 2005, Parte Prima; CRA—Istituto Sperimentale per l’Assestamento Forestale e per l’Alpicoltura: Trento, Italy, 2007; pp. 1–389. [Google Scholar]
- Pádua, L.; Hruška, J.; Bessa, J.; Adão, T.; Martins, L.M.; Gonçalves, J.A.; Peres, E.; Sousa, A.M.R.; Castro, J.P.; Sousa, J.J. Multi-temporal analysis of forestry and coastal environments using UASs. Remote Sens. 2018, 10, 24. [Google Scholar] [CrossRef]
- Pádua, L.; Adão, T.; Hruška, J.; Sousa, J.J.; Peres, E.; Morais, R.; Sousa, A. Very high resolution aerial data to support multi-temporal precision agriculture information management. Procedia Comput. Sci. 2017, 121, 407–414. [Google Scholar] [CrossRef]
- Adão, T.; Pádua, L.; Pinho, T.M.; Hruška, J.; Sousa, A.; Sousa, J.J.; Morais, R.; Peres, E. Multi-purpose chestnut clusters detection using deep learning: A preliminary approach. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. ISPRS Arch. 2019, 42, 1–7. [Google Scholar] [CrossRef]
- Lee, C. A Study on the Use of Unmanned Aerial Vehicles for Chestnut Insect Pests Control. J. Agric. Life Sci. 2018, 52, 25–29. [Google Scholar] [CrossRef]
- Matese, A.; Di Gennaro, S.F. Practical applications of a multisensor UAV platform based on multispectral, thermal and RGB high resolution images in precision viticulture. Agriculture 2018, 8, 116. [Google Scholar] [CrossRef]
- Agisoft, L.C.C. Metashape Professional, Edition 1.5.2, St. Petersburg, Russia. Available online: https://www.agisoft.com/ (accessed on 23 December 2019).
- Costa Ferreira, A.-M.; Germain, C.; Homayouni, S.; Da Costa, J.-P.; Grenier, G.; Marguerit, E.; Roby, J.-P. Transformation of high resolution aerial images in vine vigour maps at intra-block scale by semi automatic image processing. In Proceedings of the XVth International GESCO Meeting, Porec, Croatia, 20–23 June 2007; pp. 1–7. [Google Scholar]
- Fiorillo, E.; Crisci, A.; De Filippis, T.; Di Gennaro, S.F.; Di Blasi, S.; Matese, A.; Primicerio, J.; Vaccari, F.P. Airborne high-resolution images for grape classification: Changes in correlation between technological and late maturity in a Sangiovese vineyard in Central Italy. Aust. J. Grape Wine Res. 2012, 18, 80–90. [Google Scholar] [CrossRef]
- QGIS. Noosa Version. Available online: https://www.qgis.org/it/site/ (accessed on 23 December 2019).
- Isenburg, M. LAStools—Efficient Tools for LiDAR Processing. Available online: https://www.cs.unc.edu/~isenburg/lastools/ (accessed on 23 December 2019).
- Silva, C.A.; Crookston, N.L.; Hudak, A.T.; Vierling, L.A. Package “rLiDAR”. LiDAR Data Processing and Visualization. 2017. Available online: https://cran.r-project.org/web/packages/rLiDAR/index.html (accessed on 23 December 2019).
- Torres-Sánchez, J.; López-Granados, F.; Serrano, N.; Arquero, O.; Peña, J.M. High-throughput 3-D monitoring of agricultural-tree plantations with Unmanned Aerial Vehicle (UAV) technology. PLoS ONE 2015, 10, e0130479. [Google Scholar] [CrossRef] [PubMed]
- Nati, C.; Montorselli, N.B.; Olmi, R. Wood biomass recovery from chestnut orchards: Results from a case study. Agrofor. Syst. 2018, 92, 1129–1143. [Google Scholar] [CrossRef]
- Geissdoerfer, M.; Savaget, P.; Bocken, N.M.P.; Hultink, E.J. The Circular Economy e A new sustainability paradigm? J. Clean. Prod. 2017, 143, 757–768. [Google Scholar] [CrossRef]





| ID | Site A | Site B | Site C | Site D |
|---|---|---|---|---|
| Location | 42°53′18.22″ N 11°33′41.57″ E | 42°53′17.19″ N 11°33′41.75″ E | 42°52′11.71″ N 11°30′28.55″ E | 42°52′18.59″ N 11°30′29.65″ E |
| Altitude (m ASL) | 960 | 1085 | 780 | 755 |
| Surface (ha) | 0.55 | 0.32 | 0.36 | 0.32 |
| Chestnut variety | Cecio | Cecio | Bastarda Rossa | Bastarda Rossa |
| Density (trees ha−1) | 72.57 | 114.60 | 110.10 | 111.73 |
| Canopy cover (%) | 82.50 | 86.47 | 90.19 | 87.80 |
| SiteTitle | DBH (cm) | Wpw per Tree (kgdw) | Wpw per Site (kgdw) | Wpw per Surface (Mgdw ha−1) |
|---|---|---|---|---|
| A | 84.21 ± 26.55 | 625.01 ± 590.48 | 18750.28 | 24.92 |
| B | 63.24 ± 11.97 | 243.69 ± 90.42 | 7310.85 | 8.94 |
| C | 49.77 ± 11.75 | 97.37 ± 63.99 | 3115.84 | 3.86 |
| D | 53.38 ± 15.5 | 113 ± 76.39 | 3390.00 | 4.05 |
| Year | Site | Tree Height (m) | Crown Mean Height (m) | Crown Area (m2) | Crown Projected Volume (m3) | Crown Area per Site (m2) | Crown Projected Volume per Site (m3) |
|---|---|---|---|---|---|---|---|
| 2017 | A | 18.07 ± 2.8 | 14.69 ± 2.67 | 93.97 ± 49.56 | 1401.66 ± 807.53 | 3006.89 | 44,853.18 |
| B | 15.19 ± 1.38 | 12.90 ± 1.09 | 59.44 ± 20.32 | 767.61 ± 267.9 | 1902.10 | 24,563.55 | |
| C | 9.63 ± 0.98 | 7.65 ± 0.94 | 61.19 ± 18.69 | 472.7 ± 170.34 | 1958.17 | 15,126.38 | |
| D | 11.23 ± 1.36 | 8.91 ± 1.15 | 63.06 ± 18.70 | 575.36 ± 216.96 | 1954.86 | 17,836.20 | |
| 2018 | A | 17.63 ± 2.66 | 13.40 ± 2.58 | 79.74 ± 45.82 | 1103.71 ± 751.49 | 2551.80 | 35,318.81 |
| B | 14.63 ± 1.36 | 11.07 ± 1.56 | 55.57 ± 20.56 | 620.82 ± 248.39 | 1778.20 | 19,866.23 | |
| C | 9.44 ± 1.12 | 6.81 ± 1.04 | 53.16 ± 19.55 | 371.31 ± 164.53 | 1700.97 | 11,882.00 | |
| D | 11.08 ± 1.68 | 7.88 ± 1.33 | 55.62 ± 22.97 | 460.28 ± 226.76 | 1724.20 | 14,268.58 |
| Site | Reference Crowns | Matched | Split | Merged | Missed |
|---|---|---|---|---|---|
| A | 30 | 46.7% | 33.3% | 20.0% | 0.0% |
| B | 30 | 83.3% | 3.3% | 10.0% | 3.3% |
| C | 30 | 63.3% | 26.7% | 6.7% | 3.3% |
| D | 30 | 76.7% | 3.3% | 20.0% | 0.0% |
| Dataset | 30 | 67.5% | 16.7% | 14.2% | 1.7% |
| Segmentation | Site | Equation | R2 |
|---|---|---|---|
| Supervised | A | y = 1.2566x − 201.4442 | 0.78 |
| B | y = 0.2729x + 143.1937 | 0.60 | |
| C | y = 0.3549x + 25.1030 | 0.71 | |
| D | y = 0.2028x + 64.0793 | 0.69 | |
| C + D | y = 0.2393x + 53.1303 | 0.65 | |
| A + B + C + D | y = 0.6664x + 56.446 | 0.33 |
| Site | R2 | adjR2 | RMSE | rRMSE (%) | PBias (%) |
|---|---|---|---|---|---|
| A | 0.83 | 0.82 | 221.26 | 44.10 | 12.60 |
| B | 0.53 | 0.50 | 46.99 | 71.10 | −2.50 |
| C | 0.54 | 0.52 | 47.38 | 66.20 | −2.30 |
| D | 0.67 | 0.65 | 43.08 | 58.10 | −4.40 |
| C + D | 0.61 | 0.60 | 45.28 | 62.70 | −3.40 |
| A + B + C + D | 0.49 | 0.48 | 217.54 | 71.60 | −1.20 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Gennaro, S.F.; Nati, C.; Dainelli, R.; Pastonchi, L.; Berton, A.; Toscano, P.; Matese, A. An Automatic UAV Based Segmentation Approach for Pruning Biomass Estimation in Irregularly Spaced Chestnut Orchards. Forests 2020, 11, 308. https://doi.org/10.3390/f11030308
Di Gennaro SF, Nati C, Dainelli R, Pastonchi L, Berton A, Toscano P, Matese A. An Automatic UAV Based Segmentation Approach for Pruning Biomass Estimation in Irregularly Spaced Chestnut Orchards. Forests. 2020; 11(3):308. https://doi.org/10.3390/f11030308
Chicago/Turabian StyleDi Gennaro, Salvatore Filippo, Carla Nati, Riccardo Dainelli, Laura Pastonchi, Andrea Berton, Piero Toscano, and Alessandro Matese. 2020. "An Automatic UAV Based Segmentation Approach for Pruning Biomass Estimation in Irregularly Spaced Chestnut Orchards" Forests 11, no. 3: 308. https://doi.org/10.3390/f11030308
APA StyleDi Gennaro, S. F., Nati, C., Dainelli, R., Pastonchi, L., Berton, A., Toscano, P., & Matese, A. (2020). An Automatic UAV Based Segmentation Approach for Pruning Biomass Estimation in Irregularly Spaced Chestnut Orchards. Forests, 11(3), 308. https://doi.org/10.3390/f11030308