Transcriptome Sequencing and Differential Expression Analysis Reveal Molecular Mechanisms for Starch Accumulation in Chestnut
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Determination of Sugar and Starch Contents
RNA Isolation and Library Construction
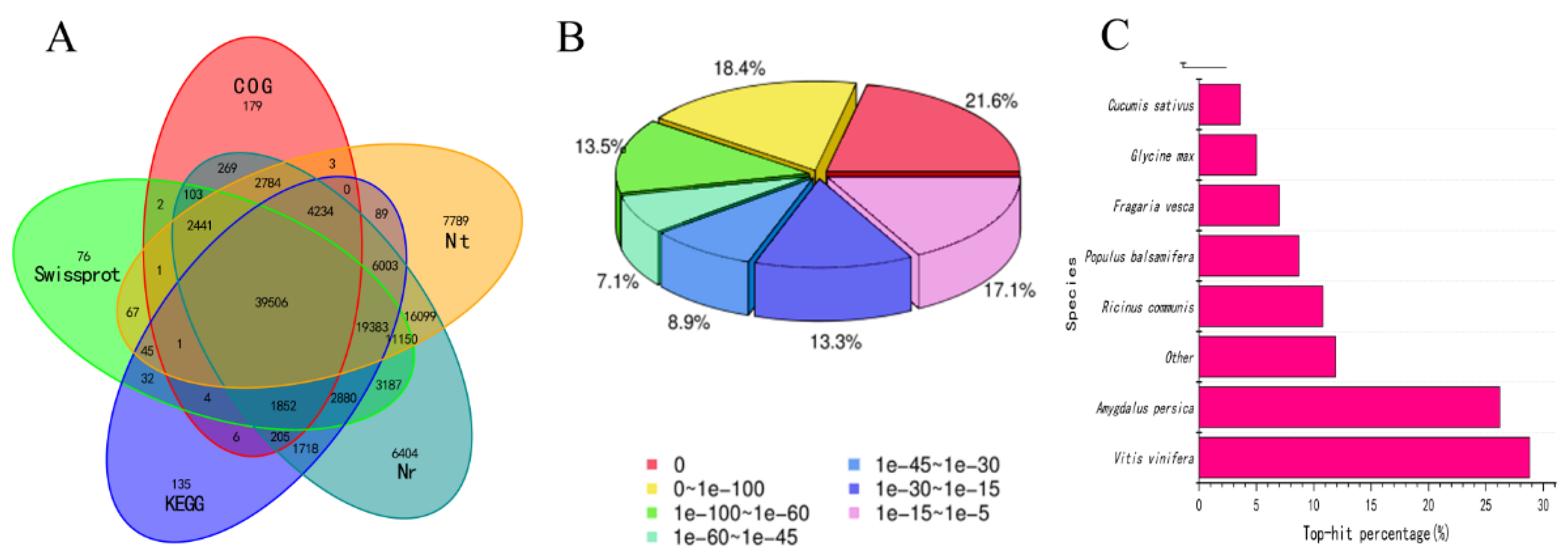
2.3. Assembly and Functional Annotation
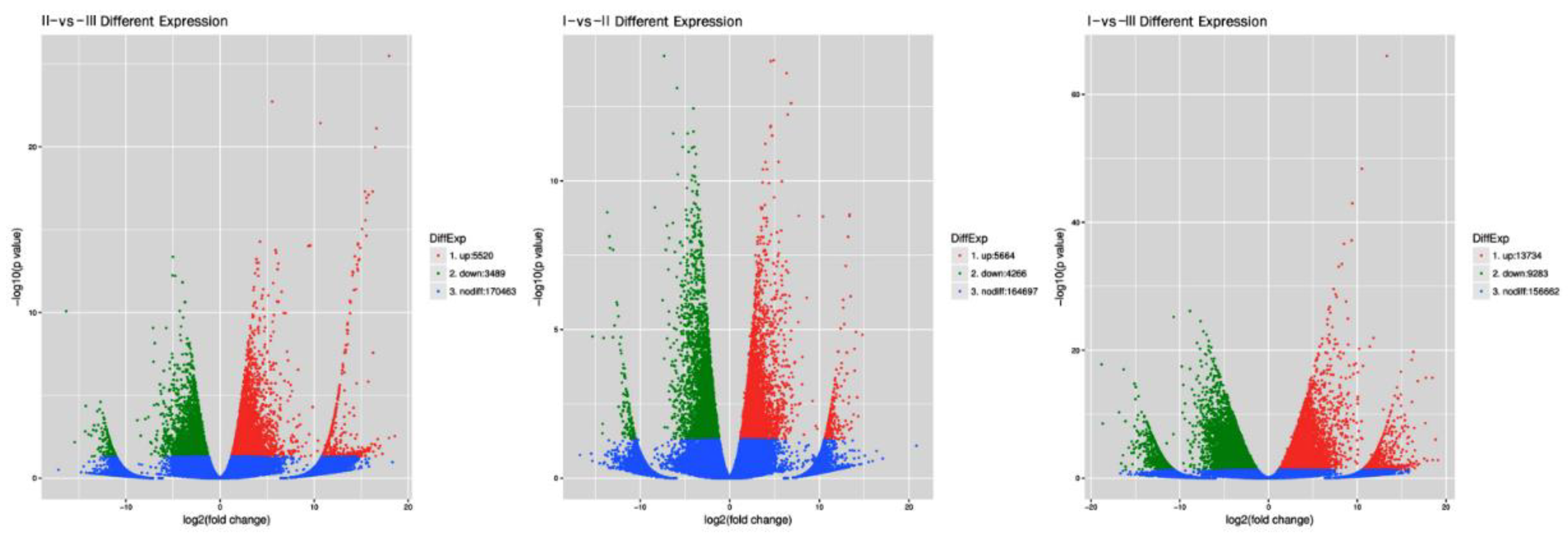
2.4. Differential Gene Expression Analysis
2.5. qRT-PCR Analysis
3. Results
3.1. Dynamics of Sugar and Starch Contents at Different Ripening Stages
3.2. Sequencing Data and DEG Statistics
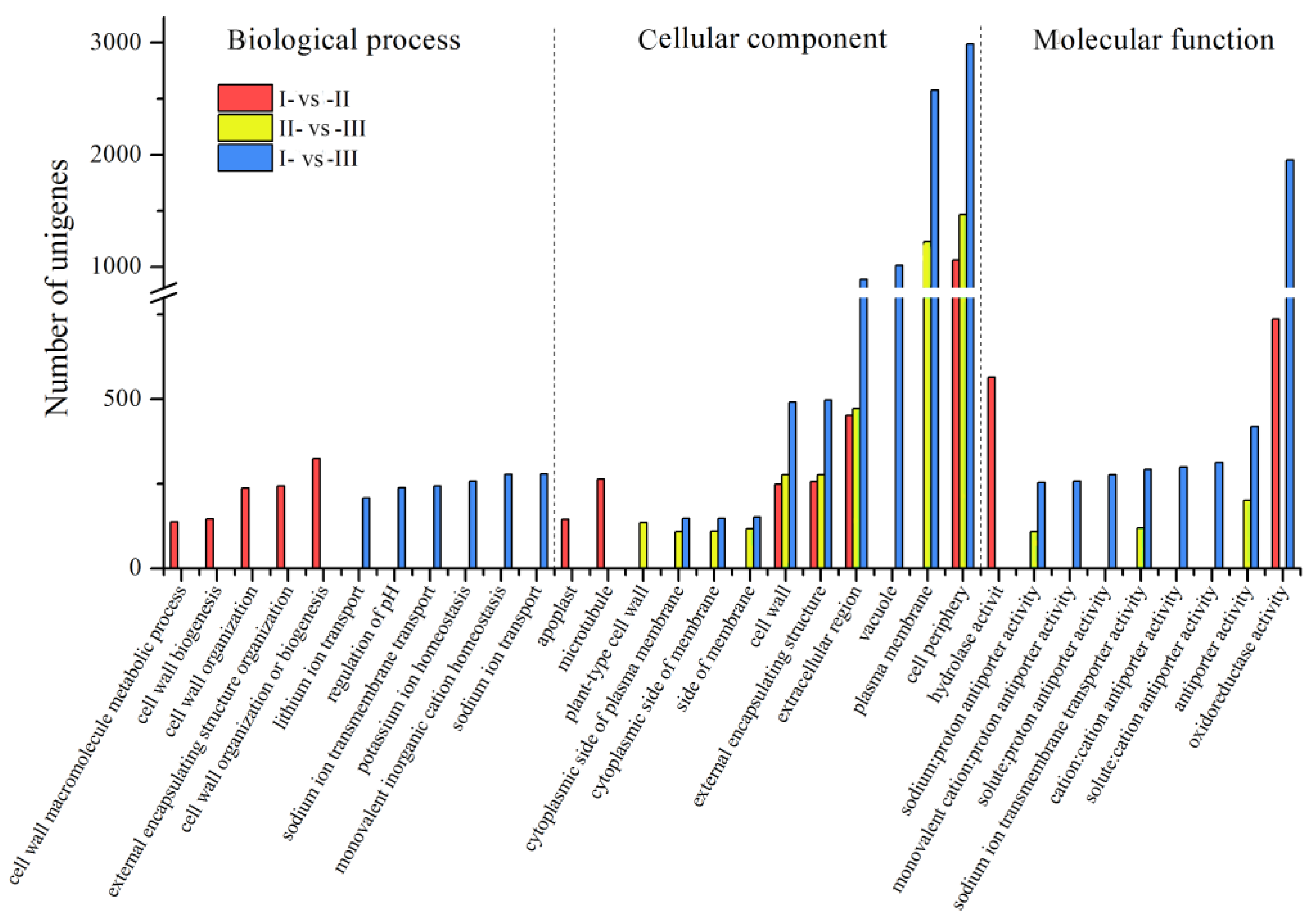
3.3. GO Annotation Analysis of DEGs
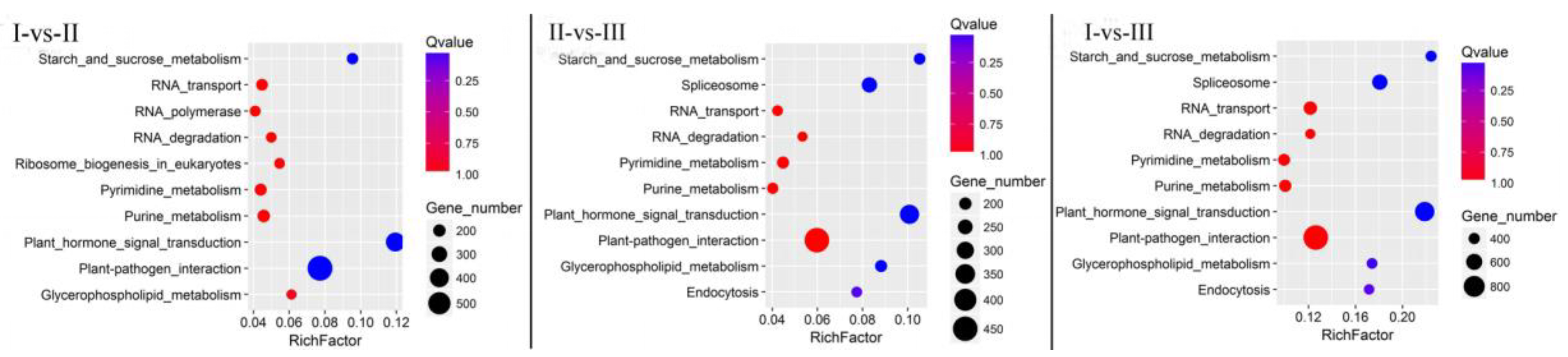
3.4. KEGG Pathway Analysis of DEGs
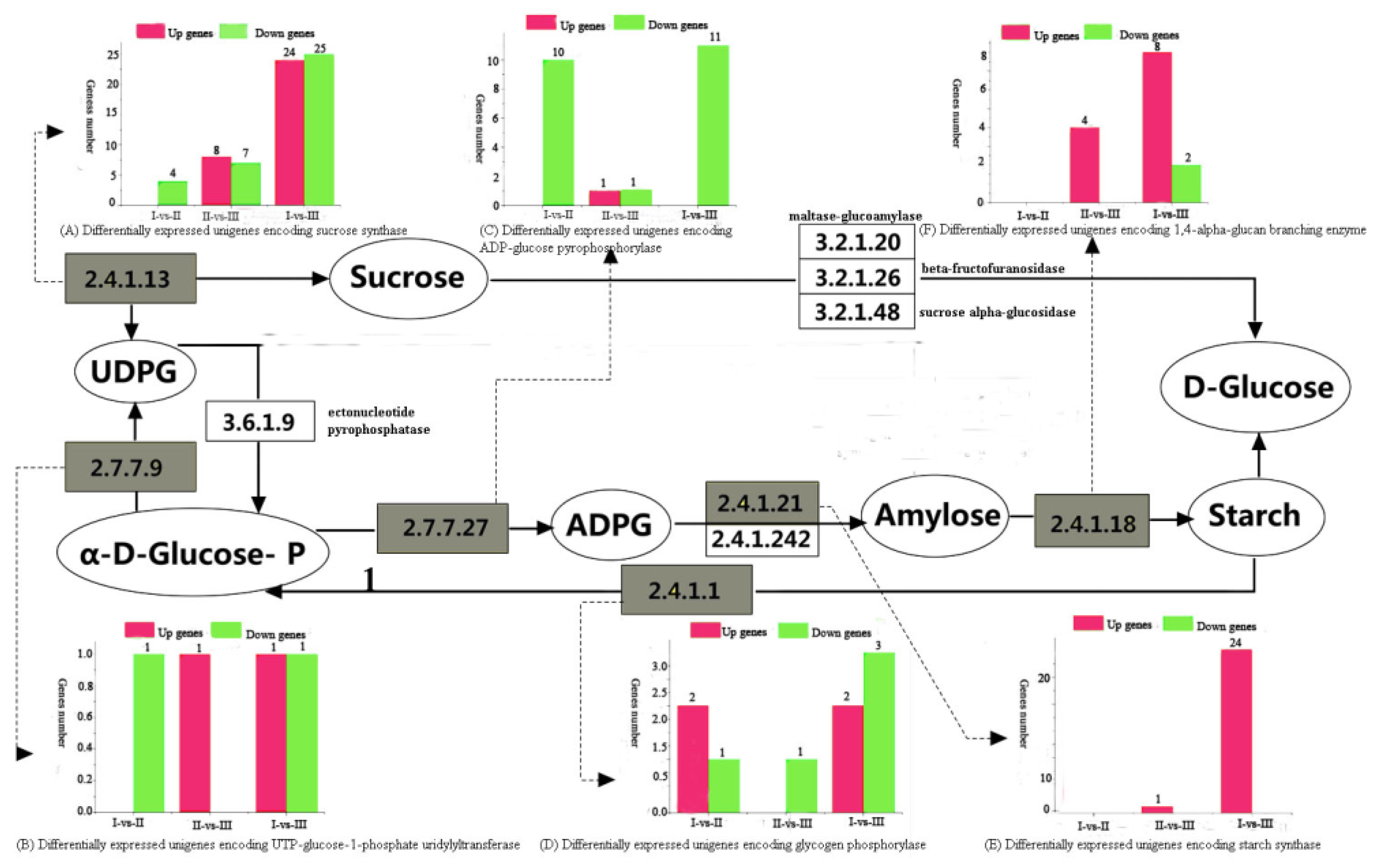
3.5. DEG Analysis of Sucrose and Starch Metabolic Pathways
3.6. Verification by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rutter, P.A.; Miller, G.; Payne, J.A. Chestnuts (Castanea). Acta Hortic. 1991, 290, 761–790. [Google Scholar] [CrossRef]
- Hou, F.; Shi, X.; Li, Q.; Xie, S.; Wu, Y.; Ouyang, J. Nutritional quality of Chinese chestnut and effect of cooking on its bioactive compounds and antioxidant activity. J. Food Process. Preserv. 2016, 40, 1383–1390. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, G.; Wu, Y.; Yang, Z.; Ouyang, J. Influence of amylose on the pasting and gel texture properties of chestnut starch during thermal processing. Food Chem. 2019, 294, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.F.; Liu, J.L.; Yang, Y.; Ren, J.; Zheng, X.Q.; Kopparapu, N.K. Effects of amylose content on the physicochemical properties of Chinese chestnut starch. Starch-Starke 2016, 68, 112–118. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, Q.; Feng, Y.; Fan, X.; Zou, F.; Zeng, X.C.H.; Cao, H.P.; Yuan, D.Y. Transcriptomic identification and expression of starch and sucrose metabolism genes in the seeds of Chinese chestnut (Castanea mollissima). J. Agric. Food Chem. 2015, 63, 929–942. [Google Scholar] [CrossRef]
- Chen, L.; Lu, D.; Wang, T.; Li, Z.; Zhao, Y.; Jiang, Y.; Zhang, Q.; Cao, Q.; Fang, K.; Xing, Y.; et al. Identification and expression analysis of starch branching enzymes involved in starch synthesis during the development of chestnut (Castanea mollissima Blume) cotyledons. PLoS ONE 2017, 12, e017779242. [Google Scholar] [CrossRef]
- Zhang, L.; Fan, X.M.; Lin, Q.; Xiang, H.; Wang, W.; Yuan, D.Y. Transcriptome analysis for developing kernel and expression analysis of starch and sucrose metabolism-related genes in Castanea henryi. J. Plant Genet. Resour. 2015, 16, 603–611. [Google Scholar]
- Guo, S.J.; Xie, P. Effects of exogenous substance on starch synthesis and related enzymes activity in Chinese chestnut. Sci. Silvae Sin. 2013, 49, 135–140. [Google Scholar]
- Barakat, A.; Staton, M.; Cheng, C.H.; Park, J.; Yassin, N.M.B.; Ficklin, S.; Yeh, C.C.; Hebard, F.; Baier, K.; Powell, W.; et al. Chestnut resistance to the blight disease: Insights from transcriptome analysis. BMC Plant Biol. 2012, 12, 38. [Google Scholar] [CrossRef] [Green Version]
- Serrazina, S.; Santos, C.; Machado, H.; Pesquita, C.; Vicentini, R.; Pais, M.S.; Sebastiana, M.; Costa, R. Castanea root transcriptome in response to Phytophthora cinnamomi challenge. Tree Genet. Genomes 2015, 11, 6. [Google Scholar] [CrossRef]
- Zhu, C.; Shi, F.; Chen, Y.; Wang, M.; Zhao, Y.; Geng, G. Transcriptome analysis of Chinese chestnut (Castanea mollissima Blume) in response to Dryocosmus kuriphilus Yasumatsu infestation. Int. J. Mol. Sci. 2019, 20, 855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Li, J.; Liu, Y.; Zhang, Q.; Gao, Y.; Fang, K.; Cao, Q.; Qin, L.; Xing, Y. Roles of the GA-mediated SPL gene family and mir156 in the floral development of Chinese chestnut (Castanea mollissima). Int. J. Mol. Sci. 2019, 20, 1577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oleksyn, J.; Zytkowiak, R.; Karolewski, P.; Reich, P.B.; Tjoelker, M.G. Genetic and environmental control of seasonal carbohydrate dynamics in trees of diverse Pinus sylvestris populations. Tree Physiol. 2000, 20, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Alexander, P.W.; Hartati, R.D.; Curtin, J. Automated potentiometric end point determination in the lane Eynon titration of reducing sugars. Electroanalysis 1989, 1, 263–269. [Google Scholar] [CrossRef]
- Jones, A.; Urban, J.; Copíková, J. A micro-analytical method for the determination of starch and amylose/amylopectin content in pea seeds. Biol. Plant. 1999, 42, 303–308. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1999, 215, 403–410. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2012, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Pan, X.C.; Wang, G.Y. China Food Composition, 2nd ed.; Peking University Medical Press: Beijing, China, 2009; p. 80. [Google Scholar]
- Li, Q.; Shi, X.; Zhao, Q.; Cui, Y.; Ouyang, J.; Xu, F. Effect of cooking methods on nutritional quality and volatile compounds of Chinese chestnut (Castanea mollissima Blume). Food Chem. 2016, 201, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Singh, R. Free sugars and their relationship with grain size and starch content in developing wheat grains. J. Sci. Food Agric. 1981, 32, 229–234. [Google Scholar] [CrossRef]
- Wang, Y.F.; Zhang, L.J.; Fan, J.J.; Ma, X.L.; Guan, Y.X. Relationship between soluble sugar Contents and starch accumulation in grain during grain-filling stage in spring maize. J. Maize Sci. 2006, 2, 81–83. [Google Scholar]
- Stein, O.; Granot, D. An overview of sucrose synthases in plants. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, Y.; Zhang, H.; Zhai, H.; Liu, Q.; He, S. A soluble starch synthase I gene, IbSSI, alters the content, composition, granule size and structure of starch in transgenic sweet potato. Sci. Rep. 2017, 7, 2315. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Ohsugi, R.; Yamagishi, T.; Sasaki, H. Effects of weak light on starch accumulation and starch synthesis enzyme activities in rice at the grain filling stage. Rice Sci. 2016, 13, 51–58. [Google Scholar]
- Shannon, J.C.; Pien, F.M.; Liu, K.C. Nucleotides and nucleotide sugars in developing maize endosperms (synthesis of ADP-glucose in brittle-1). Plant Physiol. 1996, 110, 835–843. [Google Scholar]
- Shannon, J.C.; Pien, F.M.; Cao, H.; Liu, K.C. Brittle-1, an adenylate translocator, facilitates transfer of extraplastidial synthesized ADP-glucose into amyloplasts of maize endosperms. Plant Physiol. 1998, 117, 1235–1252. [Google Scholar] [CrossRef] [Green Version]
- Zeeman, S.C.; Kossmann, J.; Smith, A.M. Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 2000, 61, 209–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Soluble Sugar (%) | Reducing Sugar (%) | Total Sugar (%) | Amylopectin (mg/g) | Amylose (mg/g) | Total Starch (mg/g) | Amylose/Amylopectin Ratio (%) | |
|---|---|---|---|---|---|---|---|
| I-YF1 | 19.27 ± 0.10b | 2.57 ± 0.90a | 21.84 ± 0.19b | 283.70 ± 2.45f | 133.12 ± 1.69a | 516.83 ± 1.77d | 46.92a |
| II-YF1 | 7.34 ± 0.06d | 1.57 ± 0.10d | 8.90 ± 0.42f | 445.36 ± 2.95c | 143.05 ± 1.75a | 588.41 ± 2.36b | 32.12c |
| III-YF1 | 15.54 ± 0.41c | 1.81 ± 0.02c | 17.35 ± 0.41d | 519.68 ± 4.66a | 102.01 ± 1.00b | 621.69 ± 5.67a | 19.62d |
| I-YRZ | 26.25 ± 0.56a | 2.15 ± 0.11b | 28.40 ± 0.10a | 237.00 ± 0.05e | 90.60 ± 0.24c | 327.60 ± 2.27e | 38.22b |
| II-YRZ | 9.25 ± 0.10d | 1.51 ± 0.51d | 10.76 ± 0.12e | 403.33 ± 1.87d | 146.24 ± 0.62a | 549.57 ± 2.46c | 36.26b |
| III-YRZ | 17.17 ± 0.55bc | 1.77 ± 0.09c | 18.9 ± 0.61c | 484.80 ± 0.1bc | 106.56 ± 0.48b | 591.36 ± 0.56b | 21.98d |
| Unigene (I-VS-II) | Unigene (II-VS-III) | Unigene (I-VS-III) |
|---|---|---|
| Unigene66368_All, Unigene51456_All, Unigene62017_All, Unigene67714_All, | Unigene40270_All, Unigene31283_All, Unigene75288_All, CL12365.Contig42_All, CL12365.Contig38_All, Unigene7815_All, CL12365.Contig34_All, CL11646.Contig2_All, Unigene12460_All, Unigene30937_All, Unigene30947_All, Unigene30932_All, Unigene80971_All, Unigene30942_All, Unigene89949_All, | Unigene31283_All, Unigene40270_All, Unigene75288_All, CL12365.Contig42_All, CL12365.Contig38_All, CL12365.Contig36_All, Unigene7815_All, CL12365.Contig37_All, CL12365.Contig19_All, CL12365.Contig33_All, CL12365.Contig12_All, CL12365.Contig3_All, CL12365.Contig21_All, CL12365.Contig25_All, CL12365.Contig23_All), CL12365.Contig24_All, CL12365.Contig14_All, CL12365.Contig41_All, CL12365.Contig4_All, CL12365.Contig34_All, CL12365.Contig5_All, CL12365.Contig43_All, CL11646.Contig2_All, CL12365.Contig9_All, Unigene51456_All, Unigene67714_All, Unigene62017_All, Unigene66368_All, Unigene12460_All, Unigene30937_All, Unigene30947_All, Unigene80971_All, Unigene30932_All, Unigene30942_All, Unigene30941_All, Unigene89949_All, Unigene30934_All, Unigene30945_All, Unigene27481_All, Unigene30946_All, Unigene80976_All, Unigene80977_All, Unigene30944_All, Unigene57545_All, Unigene80978_All, Unigene84850_All, Unigene80975_All, |
| Unigene (I-VS-II) | Unigene (II-VS-III) | Unigene (I-VS-III) |
|---|---|---|
| —— | CL2750.Contig1_All | CL12336.Contig8_All, CL12336.Contig40_All, CL12336.Contig10_All, CL12336.Contig46_All, CL12336.Contig26_All, CL12336.Contig1_All, CL12336.Contig3_All, CL12336.Contig13_All, CL12336.Contig4_All, CL12336.Contig14_All, CL12336.Contig2_All, CL12336.Contig15_All, CL12336.Contig22_All, CL12336.Contig25_All, CL12336.Contig5_All, CL12336.Contig41_All, CL12336.Contig31_All, CL12336.Contig28_All, CL12336.Contig11_All, CL12336.Contig30_All, CL12336.Contig44_All, CL2750.Contig2_All, CL2750.Contig11_All, CL2750.Contig1_All |
| YF1-vs-YRZ | Annotation | Genes |
|---|---|---|
| I | Sucrose synthetase [2.4.1.13] | Unigene23727_All |
| Sucrose-phosphate synthase [2.4.1.14] | Unigene23727_All | |
| Fructokinase [2.7.1.4] | CL11976.Contig1_All | |
| Trehalose 6-phosphate synthase [2.4.1.15] | Unigene12426_All | |
| Trehalose 6-phosphate phosphatase [3.1.3.12] | Unigene12426_All | |
| II | Sucrose synthetase [2.4.1.13] | Unigene23727_All |
| Sucrose-phosphate synthase [2.4.1.14] | Unigene23727_All | |
| beta-glucosidase [3.2.1.21] | CL7021.Contig36_All | |
| Trehalose 6-phosphate synthase [2.4.1.15] | Unigene12426_All | |
| Trehalose 6-phosphate phosphatase [3.1.3.12] | Unigene12426_All | |
| III | Sucrose synthetase [2.4.1.13] | CL6258.Contig2_All, Unigene49637_All, CL11646.Contig2_All |
| beta-glucosidase [3.2.1.21] | CL7021.Contig3_All | |
| UTP--glucose-1-phosphate uridylyltransferase [2.7.7.9] | Unigene95924_All | |
| beta-amylase [3.2.1.2] | Unigene54154_All | |
| Fructokinase [2.7.1.4] | CL2613.Contig2_All | |
| Trehalose 6-phosphate synthase [2.4.1.15] | Unigene12426_All | |
| Trehalose 6-phosphate phosphatase [3.1.3.12] | Unigene12426_All |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Liang, H.; Tao, L.; Xiong, L.; Liang, W.; Shi, Z.; Zhao, Z. Transcriptome Sequencing and Differential Expression Analysis Reveal Molecular Mechanisms for Starch Accumulation in Chestnut. Forests 2020, 11, 388. https://doi.org/10.3390/f11040388
Li S, Liang H, Tao L, Xiong L, Liang W, Shi Z, Zhao Z. Transcriptome Sequencing and Differential Expression Analysis Reveal Molecular Mechanisms for Starch Accumulation in Chestnut. Forests. 2020; 11(4):388. https://doi.org/10.3390/f11040388
Chicago/Turabian StyleLi, Shengxing, Haiying Liang, Liang Tao, Liquan Xiong, Wenhui Liang, Zhuogong Shi, and Zhiheng Zhao. 2020. "Transcriptome Sequencing and Differential Expression Analysis Reveal Molecular Mechanisms for Starch Accumulation in Chestnut" Forests 11, no. 4: 388. https://doi.org/10.3390/f11040388




