Ecostoichiometry Reveals the Separation of Microbial Adaptation Strategies in a Bamboo Forest in an Urban Wetland under Simulated Nitrogen Deposition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Overview of the Study Area
2.2. Field Experiment and Soil Sampling
2.3. Measurement of Soil Properties
2.4. DNA Extraction, Polymerase Chain Reaction, and Illumina HiSeq Sequencing
2.5. Data Processing and Statistical Analysis
3. Results
3.1. The Effects of SND on Soil Physicochemical Properties and Ecostoichiometry
3.2. Effect of SND on the Alpha Diversity of Bacteria and Fungi
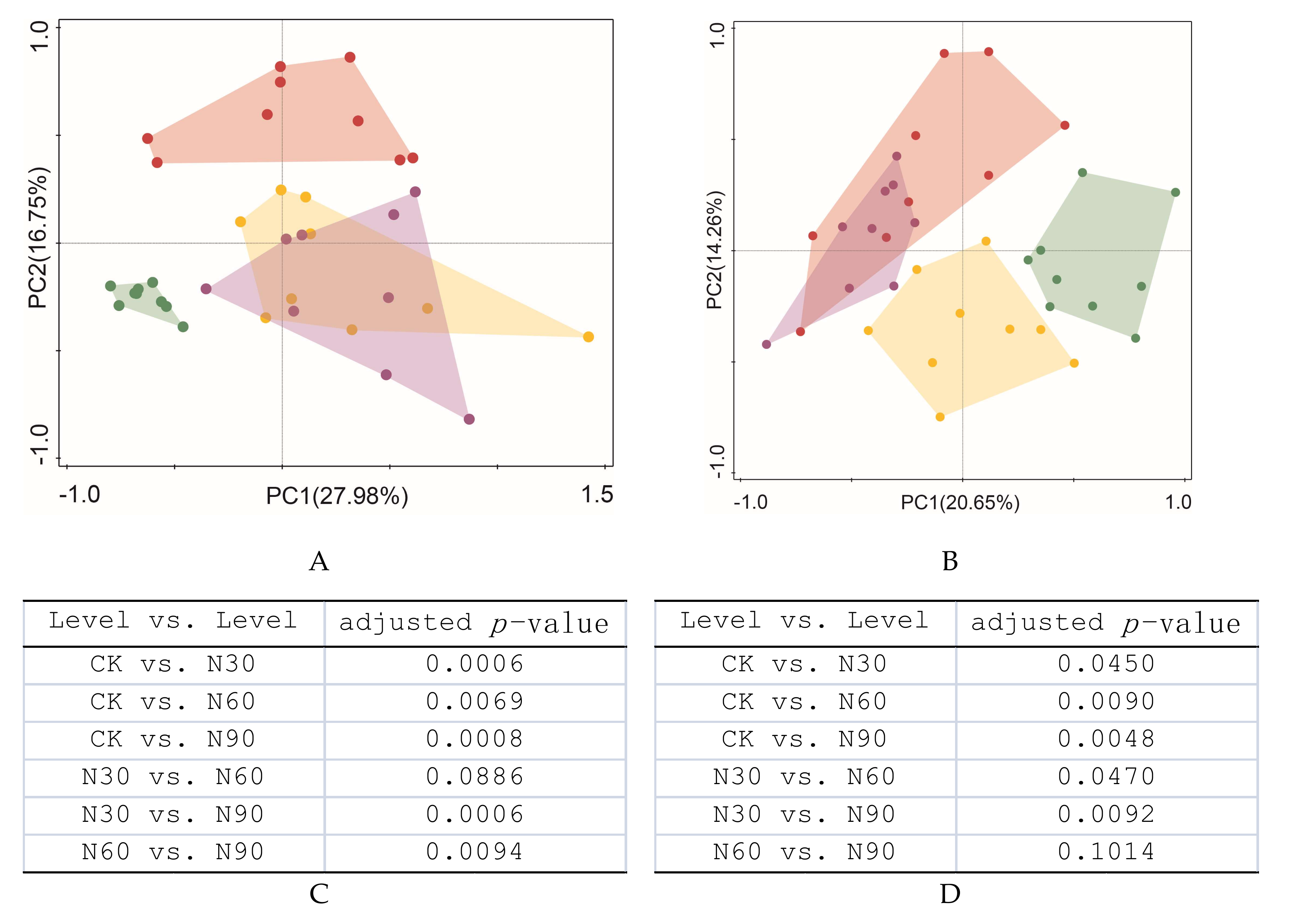
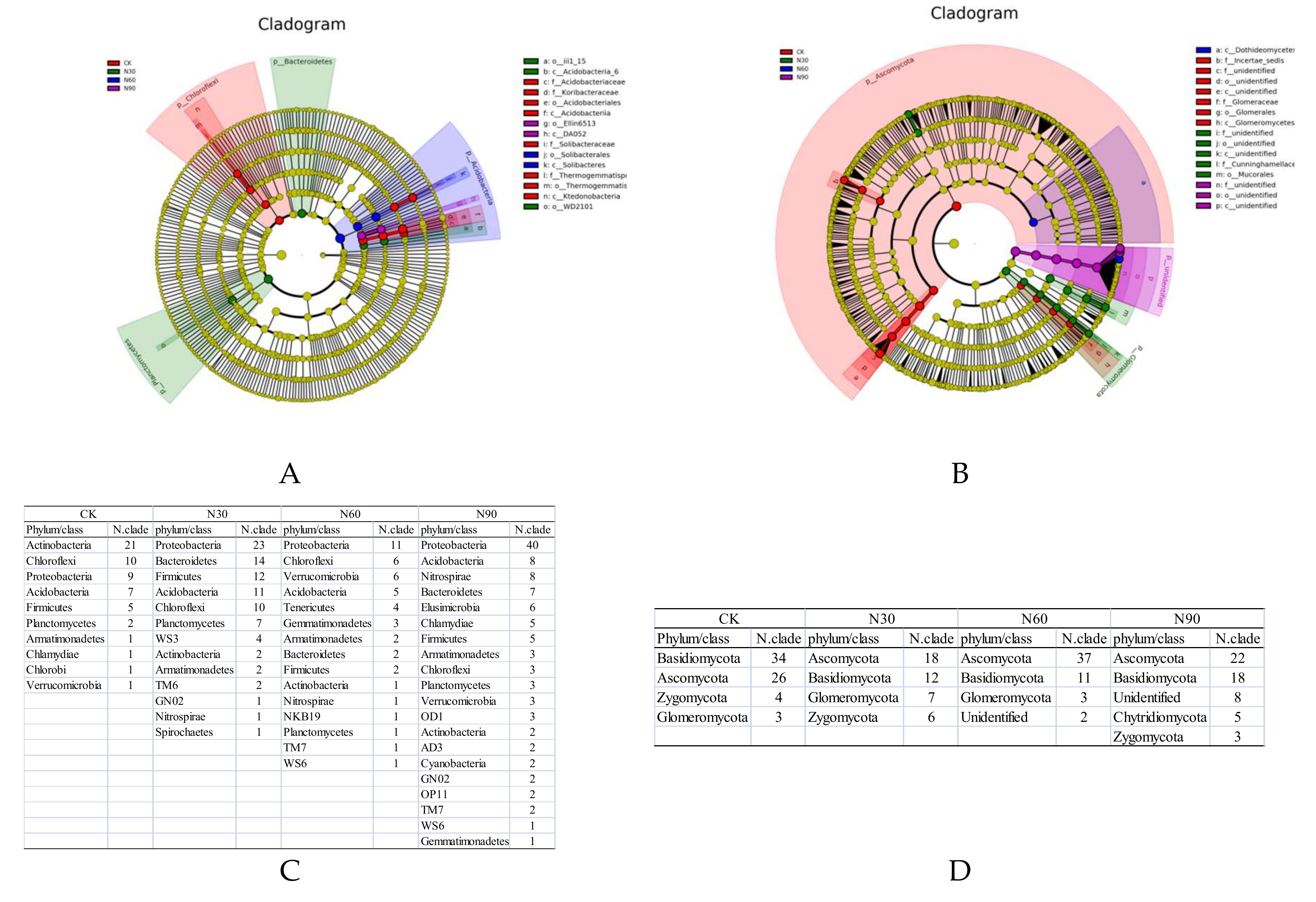
3.3. Effects of SND on the Beta Diversity of Soil Bacteria and Fungi
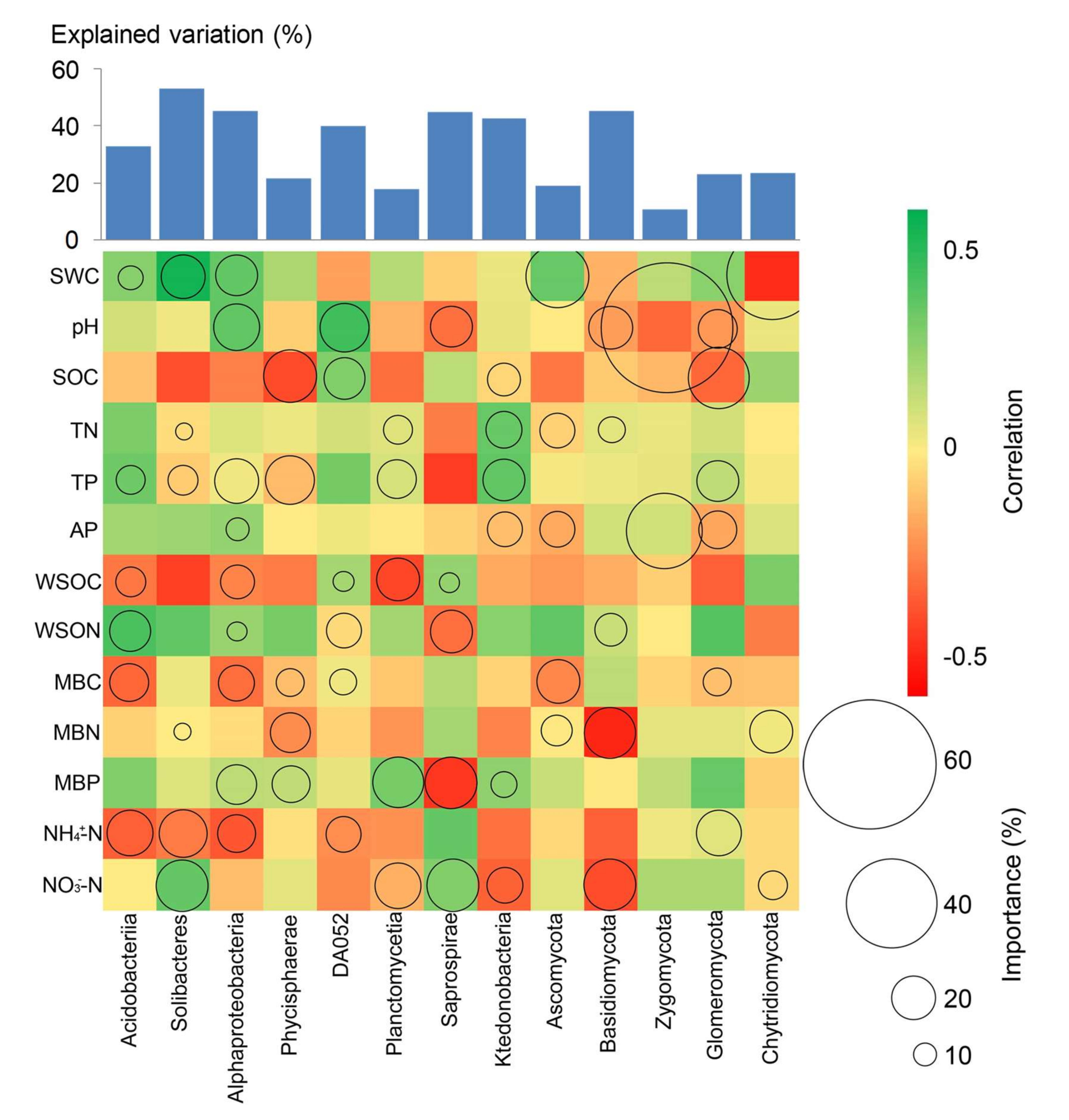
3.4. Association between Soil Properties and the Microbial Community Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Houlton, B.Z.; Wang, Y.-P.; Vitousek, P.; Field, C.B. A unifying framework for dinitrogen fixation in the terrestrial biosphere. Nature 2008, 454, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Galloway, J.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.; Martinelli, L.; Seitzinger, S.P.; Sutton, M.A. Transformation of the Nitrogen Cycle: Recent Trends, Questions, and Potential Solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [Green Version]
- Fanin, N.; Fromin, N.; Buatois, B.; Hättenschwiler, S. An experimental test of the hypothesis of non-homeostatic consumer stoichiometry in a plant litter-microbe system. Ecol. Lett. 2013, 16, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Mooshammer, M.; Wanek, W.; Zechmeister-Boltenstern, S.; Richter, A. Stoichiometric imbalances between terrestrial decomposer communities and their resources: Mechanisms and implications of microbial adaptations to their resources. Front. Microbiol. 2014, 5, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, K.S.; Craine, J.M.; Fierer, N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Chang. Boil. 2012, 18, 1918–1927. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, S.; Razavi, B.S.; Zhran, M.; Wang, J.; Zhu, Z.; Wu, J.; Ge, T. Differentiated response of plant and microbial C: N: P stoichiometries to phosphorus application in phosphorus-limited paddy soil. Eur. J. Soil Boil. 2019, 95, 103122. [Google Scholar] [CrossRef]
- Zhang, J.; Elser, J.J. Carbon:Nitrogen:Phosphorus Stoichiometry in Fungi: A Meta-Analysis. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Schimel, J.P.; Bennett, J. Nitrogen mineralization: Challenges of a changing paradigm. Ecology 2004, 85, 591–602. [Google Scholar] [CrossRef]
- Frost, P.C.; Stelzer, R.S.; Lamberti, G.A.; Elser, J. ROSEMARY MACKAY FUND ARTICLE: Ecological stoichiometry of trophic interactions in the benthos: Understanding the role of C:N:P ratios in lentic and lotic habitats. J. North Am. Benthol. Soc. 2002, 21, 515–528. [Google Scholar] [CrossRef] [Green Version]
- Elser, J.; Acharya, K.; Kyle, M.; Cotner, J.B.; Makino, W.; Markow, T.; Hobbie, S.E.; Fagan, W.; Schade, J.; Hood, J.; et al. Growth rate-stoichiometry couplings in diverse biota. Ecol. Lett. 2003, 6, 936–943. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Zhang, Y.; Duan, C.; Wang, X.; Zhang, X.; Ju, W.; Chen, H.; Yue, S.; Wang, Y.; Li, S.; et al. Ecoenzymatic stoichiometry reveals microbial phosphorus limitation decreases the nitrogen cycling potential of soils in semi-arid agricultural ecosystems. Soil Tillage Res. 2020, 197, 104463. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Liptzin, D. C:N:P stoichiometry in soil: Is there a ‘Redfield ratio’ for themicrobial biomass? Biogeochemistry 2007, 85, 235–252. [Google Scholar] [CrossRef]
- Chen, X.; Daniell, T.; Neilson, R.; O’Flaherty, V.; Griffiths, B. Microbial and microfaunal communities in phosphorus limited, grazed grassland change composition but maintain homeostatic nutrient stoichiometry. Soil Boil. Biochem. 2014, 75, 94–101. [Google Scholar] [CrossRef]
- Ren, C.; Zhao, F.; Kang, D.; Yang, G.; Han, X.; Tong, X.; Feng, Y.; Ren, G. Linkages of C:N:P stoichiometry and bacterial community in soil following afforestation of former farmland. For. Ecol. Manag. 2016, 376, 59–66. [Google Scholar] [CrossRef]
- González-Chávez, M.D.C.A.; Aitkenhead-Peterson, J.A.; Gentry, T.J.; Zuberer, D.; Hons, F.; Loeppert, R. Soil microbial community, C, N, and P responses to long-term tillage and crop rotation. Soil Tillage Res. 2010, 106, 285–293. [Google Scholar] [CrossRef]
- Peng, Y.; Thomas, S. Influence of Non-nitrogenous Soil Amendments on Soil CO2 Efflux and Fine Root Production in an N-Saturated Northern Hardwood Forest. Ecosystem 2010, 13, 1145–1156. [Google Scholar] [CrossRef]
- Crowther, T.W.; Thomas, S.M.; Maynard, D.S.; Baldrián, P.; Covey, K.; Frey, S.D.; Van Diepen, L.T.A.; Bradford, M.A. Biotic interactions mediate soil microbial feedbacks to climate change. Proc. Natl. Acad. Sci. USA 2015, 112, 7033–7038. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.; Leake, J.R.; Lee, J.; Campbell, C. Changes in soil microbial biomass and microbial activities in response to 7 years simulated pollutant nitrogen deposition on a heathland and two grasslands. Environ. Pollut. 1998, 103, 239–250. [Google Scholar] [CrossRef]
- Rinnan, R.; Michelsen, A.; Bååth, E.; Jonasson, S. Fifteen years of climate change manipulations alter soil microbial communities in a subarctic heath ecosystem. Glob. Chang. Boil. 2007, 13, 28–39. [Google Scholar] [CrossRef]
- Compton, J.E.; Watrud, L.S.; Porteous, L.A.; DeGrood, S. Response of soil microbial biomass and community composition to chronic nitrogen additions at Harvard forest. For. Ecol. Manag. 2004, 196, 143–158. [Google Scholar] [CrossRef]
- Hart, S.C.; Perry, D.A. Transferring soils from high- to low-elevation forests increases nitrogen cycling rates: Climate change implications. Glob. Chang. Boil. 1999, 5, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Heuck, C.; Weig, A.R.; Spohn, M. Soil microbial biomass C:N:P stoichiometry and microbial use of organic phosphorus. Soil Boil. Biochem. 2015, 85, 119–129. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Hill, B.H.; Shah, J.J.F. Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 2009, 462, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Peñuelas, J.; Poulter, B.; Sardans, J.; Ciais, P.; Van Der Velde, M.; Bopp, L.; Boucher, O.; Godderis, Y.; Hinsinger, P.; Llusià, J.; et al. Human-induced nitrogen–phosphorus imbalances alter natural and managed ecosystems across the globe. Nat. Commun. 2013, 4, 2934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Z.Y.; Chen, H.Y.H. Decoupling of nitrogen and phosphorus in terrestrial plants associated with global changes. Nat. Clim. Chang. 2015, 5, 465–469. [Google Scholar] [CrossRef]
- Hemkemeyer, M.; Christensen, B.T.; Martens, R.; Tebbe, C.C. Soil particle size fractions harbour distinct microbial communities and differ in potential for microbial mineralisation of organic pollutants. Soil Boil. Biochem. 2015, 90, 255–265. [Google Scholar] [CrossRef]
- Sessitsch, A.; Weilharter, A.; Gerzabek, M.H.; Kirchmann, H.; Kandeler, E. Microbial Population Structures in Soil Particle Size Fractions of a Long-Term Fertilizer Field Experiment. Appl. Environ. Microbiol. 2001, 67, 4215–4224. [Google Scholar] [CrossRef] [Green Version]
- Neumann, D.; Heuer, A.; Hemkemeyer, M.; Martens, R.; Tebbe, C.C. Response of microbial communities to long-term fertilization depends on their microhabitat. FEMS Microbiol. Ecol. 2013, 86, 71–84. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Zhou, J.; Peng, Y.; He, Y.; Yang, H.; Mao, J.; Zhang, M.; Wang, Y.; Wang, S. Atmospheric wet deposition of nitrogen and sulfur in the agroecosystem in developing and developed areas of Southeastern China. Atmos. Environ. 2014, 89, 102–108. [Google Scholar] [CrossRef]
- Li, W.C.; Sheng, H.Y.; Jiang, Y.P.; Wen, X. Soil CO2 flux and its influence factors of different bamboo plantations in the dike-pond ecosystem. Sci. Silvae Sin. 2018, 54, 13–22. [Google Scholar]
- Fang, Y.; Yoh, M.; Koba, K.; Zhu, W.; Takebayashi, Y.; Xiao, Y.; Lei, C.; Mo, J.; Zhang, W.; Lu, X. Nitrogen deposition and forest nitrogen cycling along an urban-rural transect in southern China. Glob. Chang. Boil. 2011, 17, 872–885. [Google Scholar] [CrossRef]
- Li, W.; Sheng, H.; Ekawati, D.; Jiang, Y.; Yang, H. Variations in the Compositions of Soil Bacterial and Fungal Communities Due to Microhabitat Effects Induced by Simulated Nitrogen Deposition of a Bamboo Forest in Wetland. Forest 2019, 10, 1098. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Joergensen, R.G.; Pommerening, B.; Chaussod, R.; Brookes, P.C. Measurement of soil microbial biomass C by fumigation-extraction—An automated procedure. Soil Boil. Biochem. 1990, 22, 1167–1169. [Google Scholar] [CrossRef]
- Brookes, P.; Powlson, D.; Jenkinson, D. Phosphorus in the soil microbial biomass. Soil Boil. Biochem. 1984, 16, 169–175. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Lindahl, B. Fungal identification biases in microbiome projects. Environ. Microbiol. Rep. 2016, 8, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.-Y.; Zhang, C.-X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2012, 20, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R. Vagan: Community Ecology Package. R Package Version 22-1. 2015. Available online: http://CRAN.R-project.org/package=vegan (accessed on 12 November 2019).
- Wickham, H. Ggplots2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Boil. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Field, A.; Miles, J.; Field, Z. Discovering Statistics Using R; Sage Publications: London, UK, 2012. [Google Scholar]
- Wei, C.; Yu, Q.; Bai, E.; Lü, X.-T.; Li, Q.; Xia, J.; Kardol, P.; Liang, W.; Wang, Z.; Han, X. Nitrogen deposition weakens plant-microbe interactions in grassland ecosystems. Glob. Chang. Boil. 2013, 19, 3688–3697. [Google Scholar] [CrossRef]
- Aber, J.D.; Magill, A.H. Chronic nitrogen additions at the Harvard Forest (USA): The first 15 years of a nitrogen saturation experiment. For. Ecol. Manag. 2004, 196, 1–5. [Google Scholar] [CrossRef]
- Sirulnik, A.G.; Allen, E.; Meixner, T.; Fenn, M.E.; Allen, M.F. Changes in N cycling and microbial N with elevated N in exotic annual grasslands of southern California. Appl. Soil Ecol. 2007, 36, 1–9. [Google Scholar] [CrossRef]
- Yang, K.; Zhu, J.; Gu, J.; Yu, L.; Wang, Z. Changes in soil phosphorus fractions after 9 years of continuous nitrogen addition in a Larix gmelinii plantation. Ann. For. Sci. 2014, 72, 435–442. [Google Scholar] [CrossRef]
- Hall, S.J.; Matson, P.A. Nutrient status of tropical rain forests influences soil n dynamics after n additions. Ecol. Monogr. 2003, 73, 107–129. [Google Scholar] [CrossRef]
- Liu, X.-R.; Ren, J.-Q.; Li, S.-G.; Zhang, Q.-W. Effects of Simulated Nitrogen Deposition on Soil Net Nitrogen Mineralization in the Meadow Steppe of Inner Mongolia, China. PLoS ONE 2015, 10, 0134039. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sheng, H.; Liu, Y.; Chen, W. Responses of soil bacterial compositions to concentrations of nitrogen forms in the process of Moso bamboo invasion. Ecol. Res. 2019, 34, 743–752. [Google Scholar] [CrossRef]
- Deng, Q.; Hui, D.F.; Dennis, S.; Reddy, K.C. Responses of terrestrial ecosystem phosphorus cycling tonitrogen addition: A meta-analysis. Glob. Ecol. Biogeogr. 2017, 26, 713–728. [Google Scholar] [CrossRef]
- Ma, B.B.; Zhou, X.L.; Zhang, Q.; Qin, M.S.; Hu, L.G.; Yang, K.N.; Xie, Z.; Ma, W.B.; Chen, B.B.; Feng, H.Y.; et al. How do soil micro-organisms respond to N, P and NPadditions? Application of the ecological framework of (co-) limitation by multiple resources. J. Ecol. 2019, 107, 2329–2345. [Google Scholar] [CrossRef]
- Van Diepen, L.; Lilleskov, E.; Pregitzer, K.; Miller, R. Simulated nitrogen deposition causes a decline of intra- and extraradieal abundance of arbuseular mycorhizal fungi and changes in microbial community structure in northem hardwood forests. Ecosystems 2010, 13, 683–695. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhang, X.Y.; Wen, X.F.; Wang, S.Q.; Wang, H.M. The effect of nitrogen deposition on forest soil organic matter and litter deeompostion and the microbial mechanism. Acta Ecol. Sin. 2013, 33, 1337–1346. [Google Scholar] [CrossRef]
- Gundersen, P.; Callesen, I.; De Vries, W. Nitrate leaching in forest ecosystems is related to forest floor C/N ratios. Nitrogen, the Confer-N-s 1998, 102, 403–407. [Google Scholar]
- Moorhead, D.L.; Sinsabaugh, R.L. A theoretical model of litter decay and microbial interaction. Ecol. Monogr. 2006, 76, 151–174. [Google Scholar] [CrossRef]
- Walbridge, M.; Richardson, C.; Swank, W. Vertical distribution of biological and geochemical phosphorus subcycles in two southern Appalachian forest soils. Biogeochemistry 1991, 13, 61–85. [Google Scholar] [CrossRef]
- Che, R.; Qin, J.; Tahmasbian, I.; Wang, F.; Zhou, S.; Xu, Z.; Cui, X. Litter amendment rather than phosphorus can dramatically change inorganic nitrogen pools in a degraded grassland soil by affecting nitrogen-cycling microbes. Soil Boil. Biochem. 2018, 120, 145–152. [Google Scholar] [CrossRef]
- Fierer, N.; Strickland, M.S.; Liptzin, D.; Bradford, M.A.; Cleveland, C.C. Global patterns in belowground communities. Ecol. Lett. 2009, 12, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, S.; Xu, Z.; Yan, Q.; Li, X.; Van Nostrand, J.D.; He, Z.; Yao, F.; Han, X.; Zhou, J.; et al. Responses of soil microbial functional genes to global changes are indirectly influenced by aboveground plant biomass variation. Soil Boil. Biochem. 2017, 104, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Phillips, R.P.; Finzi, A.; Bernhardt, E.S. Enhanced root exudation induces microbial feedbacks to N cycling in a pine forest under long-term CO2 fumigation. Ecol. Lett. 2010, 14, 187–194. [Google Scholar] [CrossRef]
- Wassen, M.J.; Venterink, H.O.; Lapshina, E.; Tanneberger, F. Endangered plants persist under phosphorus limitation. Nature 2005, 437, 547–550. [Google Scholar] [CrossRef]


| SWC (wt.%) | pH | SOC (g·kg−1) | TN (g·kg−1) | TP (mg·kg−1) | AP (mg·kg−1) | WSOC (mg·kg−1) | WSON (mg·kg−1) | MBC (mg·kg−1) | MBN (mg·kg−1) | MBP (mg·kg−1) | NH4+−N (mg·kg−1) | NO3−−N (mg·kg−1) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CK | 15.3 ± 2.3b | 5.2 ± 0.3a | 44.5 ± 4.6a | 1.9 ± 0.2a | 51.3 ± 6.5a | 5.0 ± 1.9a | 26.3 ± 7.0a | 2.9 ± 0.5b | 432.8 ± 194.9a | 87.8 ± 24.7a | 5.4 ± 2.2b | 20.3 ± 6.0a | 4.5 ± 0.7b |
| N30 | 18.8 ± 2.6ab | 5.2 ± 0.2a | 40.2 ± 3.0ab | 2.0 ± 3.5a | 57.8 ± 10.2a | 5.6 ± 2.4a | 15.9 ± 3.4bc | 5.1 ± 1.6ab | 354.2 ± 132.7a | 72.5 ± 15.1a | 10.9 ± 4.8b | 17.6 ± 3.1ab | 5.0 ± 2.0b |
| N60 | 17.1 ± 3.2b | 4.8 ± 0.1b | 42.4 ± 5.8ab | 2.5 ± 0.7a | 50.2 ± 10.3a | 4.3 ± 2.4a | 19.9 ± 4.5b | 5.3 ± 2.5ab | 324.0 ± 163.6a | 69.6 ± 16.4a | 12.2 ± 5.8b | 16.7 ± 3.9ab | 6.6 ± 2.45ab |
| N90 | 21.8 ± 4.3a | 4.8 ± 0.2b | 39.1 ± 2.1b | 2.4 ± 0.4a | 63.8 ± 23.6a | 5.0 ± 2.2a | 11.1 ± 2.0c | 7.3 ± 2.6a | 377.1 ± 153.1a | 79.1 ± 34.6a | 34.4 ± 24.7a | 14.4 ± 1.4b | 8.3 ± 3.1a |
| Kingdom | Index | CK | N30 | N60 | N90 |
|---|---|---|---|---|---|
| Bacteria | Chao1 | 2033.5 ± 292.7b | 3146.7 ± 147.6a | 2512.0 ± 607.5ab | 2463.7 ± 399.5ab |
| Observed species (OTUs) | 1217 ± 148b | 1561 ± 332a | 1427 ± 258ab | 1473 ± 218ab | |
| Shannon | 8.36 ± 0.24a | 8.82 ± 0.50a | 8.69 ± 0.47a | 8.79 ± 0.38a | |
| Simpson | 0.9911 ± 0.0017a | 0.9931 ± 0.0025a | 0.9927 ± 0.0026a | 0.9936 ± 0.0021a | |
| Fungi | Chao1 | 171.2 ± 21.5a | 166.9 ± 21.4a | 153.8 ± 18.9a | 163.6 ± 20.9a |
| Observed species (OTUs) | 136 ± 16a | 133 ± 13a | 131 ± 13a | 127 ± 16a | |
| Shannon | 5.31 ± 0.52a | 5.21 ± 0.43a | 5.29 ± 0.35a | 4.87 ± 0.78a | |
| Simpson | 0.9343 ± 0.0421a | 0.9263 ± 0.0496a | 0.9382 ± 0.0282a | 0.8985 ± 0.09920a | |
| F/B | 0.1133 ± 0.0205a | 0.0887 ± 0.0183b | 0.0947 ± 0.0199ab | 0.0882 ± 0.0175b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Sheng, H.; Liu, Y.; Zhang, R.; Ekawati, D.; Qian, Y.; Lou, Y. Ecostoichiometry Reveals the Separation of Microbial Adaptation Strategies in a Bamboo Forest in an Urban Wetland under Simulated Nitrogen Deposition. Forests 2020, 11, 428. https://doi.org/10.3390/f11040428
Li W, Sheng H, Liu Y, Zhang R, Ekawati D, Qian Y, Lou Y. Ecostoichiometry Reveals the Separation of Microbial Adaptation Strategies in a Bamboo Forest in an Urban Wetland under Simulated Nitrogen Deposition. Forests. 2020; 11(4):428. https://doi.org/10.3390/f11040428
Chicago/Turabian StyleLi, Weicheng, Haiyan Sheng, Yaoyao Liu, Rui Zhang, Desy Ekawati, Yifan Qian, and Yi Lou. 2020. "Ecostoichiometry Reveals the Separation of Microbial Adaptation Strategies in a Bamboo Forest in an Urban Wetland under Simulated Nitrogen Deposition" Forests 11, no. 4: 428. https://doi.org/10.3390/f11040428





