A Circular Chloroplast Genome of Fagus sylvatica Reveals High Conservation between Two Individuals from Germany and One Individual from Poland and an Alternate Direction of the Small Single-Copy Region
Abstract
1. Introduction
2. Materials and Methods
2.1. Initial Draft Assembly
2.2. Manual Curation
2.3. Annotation of Genic Regions
2.4. Comparison of Chloroplast Assemblies
3. Results
3.1. General Features of the Chloroplast Assembly of Bhaga
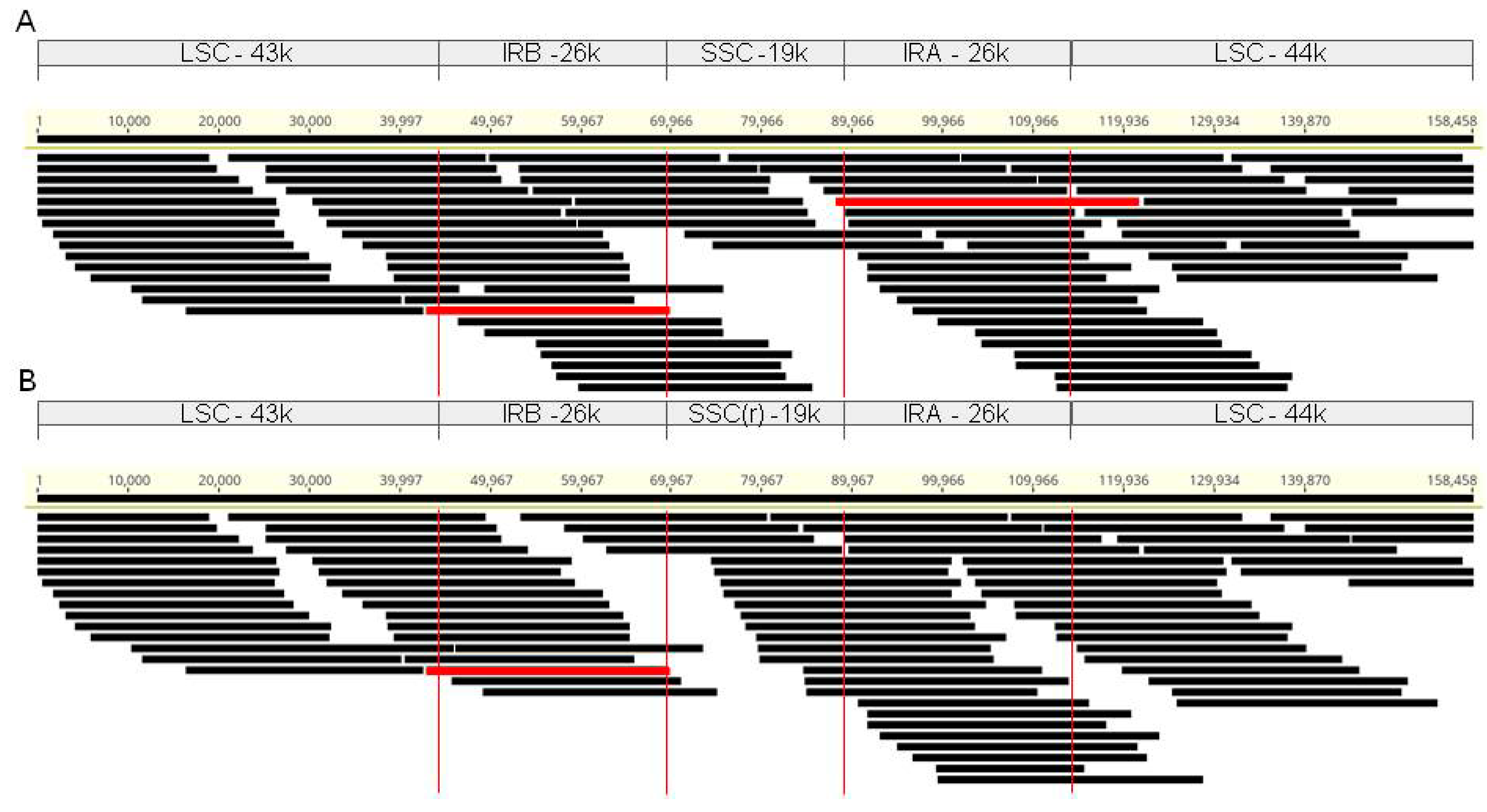
3.2. Comparison of Chloroplast Assemblies
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giesecke, T.; Hickler, T.; Kunkel, T.; Sykes, M.T.; Bradshaw, R.H. Towards an understanding of the Holocene distribution of Fagus sylvatica L. J. Biogeogr. 2007, 34, 118–131. [Google Scholar] [CrossRef]
- Brunet, J.; Fritz, Ö.; Richnau, G. Biodiversity in European beech forests-a review with recommendations for sustainable forest management. Ecol. Bull. 2010, 53, 77–94. [Google Scholar]
- Jump, A.S.; Hunt, J.M.; Peñuelas, J. Rapid climate change-related growth decline at the southern range edge of Fagus sylvatica. Glob. Chang. Biol. 2006, 12, 2163–2174. [Google Scholar] [CrossRef]
- Gessler, A.; Keitel, C.; Kreuzwieser, J.; Matyssek, R.; Seiler, W.; Rennenberg, H. Potential risks for European beech (Fagus sylvatica L.) in a changing climate. Trees 2007, 21, 1–11. [Google Scholar] [CrossRef]
- Albert, R.E.I.F.; Xystrakis, F.; Gaertner, S.; Sayer, U. Floristic change at the drought limit of European beech (Fagus sylvatica L.) to downy oak (Quercus pubescens) forest in the temperate climate of central Europe. Not. Bot. Horti Agrobot. Cluj-Napoca 2017, 45, 646–654. [Google Scholar]
- Sandurska, E.; Ulaszewski, B.; Burczyk, J. Genetic Insights into Ecological Succession from Oak- (Quercus robur L.) to Beech- (Fagus sylvatica L.) Dominated Forest Stands. Acta Biol. Crac. Bot. 2017, 59, 23–33. [Google Scholar] [CrossRef]
- Comps, B.; Thiébaut, B.; Paule, L.; Merzeau, D.; Letouzey, J. Allozymic variability in beechwoods (Fagus sylvatica L.) over central Europe: Spatial differentiation among and within populations. Heredity 1990, 65, 407–417. [Google Scholar] [CrossRef]
- Leonardi, S.; Menozzi, P. Genetic variability of Fagus sylvatica L. in Italy: The role of postglacial recolonization. Heredity 1995, 75, 35–44. [Google Scholar] [CrossRef][Green Version]
- Demesure, B.; Comps, B.; Petit, R.J. Chloroplast DNA Phylogeography of the Common Beech (Fagus sylvatica L.) in Europe. Evolution 1996, 50, 2515. [Google Scholar] [CrossRef]
- Sjölund, M.J.; González-Díaz, P.; Moreno-Villena, J.J.; Jump, A.S. Understanding the legacy of widespread population translocations on the post-glacial genetic structure of the European beech, Fagus sylvatica L. J. Biogeogr. 2017, 44, 2475–2487. [Google Scholar] [CrossRef]
- Cuervo-Alarcon, L.; Arend, M.; Müller, M.; Sperisen, C.; Finkeldey, R.; Krutovsky, K.V. Genetic variation and signatures of natural selection in populations of European beech (Fagus sylvatica L.) along precipitation gradients. Tree Genet. Genomes 2018, 14, 84. [Google Scholar] [CrossRef]
- Müller, M.; Lopez, P.A.; Papageorgiou, A.C.; Tsiripidis, I.; Gailing, O. Indications of Genetic Admixture in the Transition Zone between Fagus sylvatica L. and Fagus sylvatica ssp. orientalis Greut. & Burd. Diversity 2019, 11, 90. [Google Scholar] [CrossRef]
- Sebastiani, F.; Carnevale, S.; Vendramin, G.G. A new set of mono-and dinucleotide chloroplast microsatellites in Fagaceae. Mol. Ecol. Notes 2004, 4, 259–261. [Google Scholar] [CrossRef]
- Vettori, C.; Vendramin, G.G.; Anzidei, M.; Pastorelli, R.; Paffetti, D.; Giannini, R. Geographic distribution of chloroplast variation in Italian populations of beech (Fagus sylvatica L.). Theor. Appl. Genet. 2004, 109, 1–9. [Google Scholar] [CrossRef]
- Hatziskakis, S.; Papageorgiou, A.C.; Gailing, O.; Finkeldey, R. High chloroplast haplotype diversity in Greek populations of beech (Fagus sylvatica L.). Plant Biol. 2009, 11, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Meger, J.; Ulaszewski, B.; Vendramin, G.G.; Burczyk, J. Using reduced representation libraries sequencing methods to identify cpDNA polymorphisms in European beech (Fagus sylvatica L). Tree Genet. Genomes 2019, 15, 7. [Google Scholar] [CrossRef]
- McLachlan, J.S.; Clark, J.S.; Manos, P.S. Molecular Indicators of Tree Migration Capacity Under Rapid Climate Change. Ecology 2005, 86, 2088–2098. [Google Scholar] [CrossRef]
- Magri, D.; Vendramin, G.G.; Comps, B.; Dupanloup, I.; Geburek, T.; Gomory, D.; Latalowa, M.; Litt, T.; Paule, L.; Roure, J.M.; et al. A new scenario for the quaternary history of European beech populations: Palaeobotanical evidence and genetic conse-quences. New Phytol. 2006, 171, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Mader, M.; Liesebach, H.; Liesebach, M.; Kersten, B. The complete chloroplast genome sequence of Fagus sylvatica L. (Fagaceae). Mitochondrial DNA Part B 2019, 4, 1818–1819. [Google Scholar] [CrossRef]
- Yang, J.; Takayama, K.; Youn, J.-S.; Pak, J.-H.; Kim, S.-C. Plastome characterization and phylogenomics of East Asian beeches with a special emphasis on Fagus multinervis on Ulleung Island, Korea. Genes 2020, 11, 1338. [Google Scholar] [CrossRef]
- Mishra, B.; Gupta, D.K.; Pfenninger, M.; Hickler, T.; Langer, E.; Nam, B.; Paule, J.; Sharma, R.; Ulaszewski, B.; Warmbier, J.; et al. A reference genome of the European beech (Fagus sylvatica L.). GigaScience 2018, 7, giy063. [Google Scholar] [CrossRef] [PubMed]
- Hackl, T.; Hedrich, R.; Schultz, J.; Förster, F. proovread: Large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 2014, 30, 3004–3011. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Stadermann, K.B.; Holtgräwe, D.; Weisshaar, B. Chloroplast genome sequence of Arabidopsis thaliana accession Landsberg erecta, assembled from single-molecule, real-time sequencing data. Genome Announc. 2016, 4, e00975-16. [Google Scholar] [CrossRef] [PubMed]
- Pinard, D.; Myburg, A.A.; Mizrachi, E. The plastid and mitochondrial genomes of Eucalyptus grandis. BMC Genom. 2019, 20, 132. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Manos, P.S.; Stanford, A.M. The historical biogeography of Fagaceae: Tracking the Tertiary history of temperate and subtropical forests of the northern hemisphere. Int. J. Plant Sci. 2001, 162, S77–S93. [Google Scholar] [CrossRef]
- Chaisson, M.J.; Tesler, G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinform. 2012, 13, 238. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A.V. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Barna, M. Adaptation of European beech (Fagus sylvatica L.) to different ecological conditions: Leaf size variation. Pol. J. Ecol. 2004, 52, 35–45. [Google Scholar]
- Maringer, J.; Ascoli, D.; Küffer, N.; Schmidtlein, S.; Conedera, M. What drives European beech (Fagus sylvatica L.) mortality after forest fires of varying severity? For. Ecol. Manag. 2016, 368, 81–93. [Google Scholar] [CrossRef]
- Rey, F.; Gobet, E.; Schwörer, C.; Wey, O.; Hafner, A.; Tinner, W. Causes and mechanisms of synchronous succession trajectories in primeval Central European mixed Fagus sylvatica forests. J. Ecol. 2019, 107, 1392–1408. [Google Scholar] [CrossRef]
- Gauzere, J.; Delzon, S.; Davi, H.; Bonhomme, M.; De Cortazar-Atauri, I.G.; Chuine, I. Integrating interactive effects of chilling and photoperiod in phenological process-based models. A case study with two European tree species: Fagus sylvatica and Quercus petraea. Agric. For. Meteorol. 2017, 244, 9–20. [Google Scholar] [CrossRef]
- Krajmerová, D.; Hrivnák, M.; Ditmarová, Ľ.; Jamnická, G.; Kmeť, J.; Kurjak, D.; Gömöry, D. Nucleotide polymorphisms associated with climate, phenology and physiological traits in European beech (Fagus sylvatica L.). New For. 2017, 48, 463–477. [Google Scholar] [CrossRef]
- Sjölund, M.J.; González-Díaz, P.; Moreno-Villena, J.J.; Jump, A.S. Gene flow at the leading range edge: The long-term consequences of isolation in European beech (Fagus sylvatica L. Kuhn). J. Biogeogr. 2019, 46, 2787–2799. [Google Scholar] [CrossRef]
- Capblancq, T.; Morin, X.; Gueguen, M.; Renaud, J.; Lobreaux, S.; Bazin, E. Climate-associated genetic variation in Fagus sylvatica and potential responses to climate change in the French Alps. J. Evol. Biol. 2020, 33, 783–796. [Google Scholar] [CrossRef]
- Tsukamoto, M.; Akada, S.; Matsuda, S.; Jouyu, H.; Kisanuki, H.; Tomaru, N.; Torimaru, T. Assessments of fine-scale spatial patterns of SNPs in an old-growth beech forest. Heredity 2020, 125, 240–252. [Google Scholar] [CrossRef]
- Ali, T.; Schmuker, A.; Runge, F.; Solovyeva, I.; Nigrelli, L.; Paule, J.; Buch, A.-K.; Xia, X.; Ploch, S.; Orren, O.; et al. Morphology, phylogeny, and taxonomy of Microthlaspi (Brassicaceae: Coluteocarpeae) and related genera. Taxon 2016, 65, 79–98. [Google Scholar] [CrossRef]
- Pluess, A.R.; Frank, A.; Heiri, C.; Lalagüe, H.; Vendramin, G.G.; Oddou-Muratorio, S. Genome–environment association study suggests local adaptation to climate at the regional scale in Fagus sylvatica. New Phytol. 2016, 210, 589–601. [Google Scholar] [CrossRef]
- Comps, B.; Gömöry, D.; Letouzey, J.; Thiébaut, B.; Petit, R.J. Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics 2001, 157, 389–397. [Google Scholar]
- Martínez, I.; González-Taboada, F. Seed dispersal patterns in a temperate forest during a mast event: Performance of alternative dispersal kernels. Oecologia 2009, 159, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Bialozyt, R.; Bradley, L.R.; Bradshaw, R.H. Modelling the spread of Fagus sylvatica and Picea abies in southern Scandinavia during the late Holocene. J. Biogeogr. 2012, 39, 665–675. [Google Scholar] [CrossRef]
- Oddou-Muratorio, S.; Bontemps, A.; Klein, E.K.; Chybicki, I.J.; Vendramin, G.G.; Suyama, Y. Comparison of direct and indirect genetic methods for estimating seed and pollen dispersal in Fagus sylvatica and Fagus crenata. For. Ecol. Manag. 2010, 259, 2151–2159. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, B.; Ulaszewski, B.; Ploch, S.; Burczyk, J.; Thines, M. A Circular Chloroplast Genome of Fagus sylvatica Reveals High Conservation between Two Individuals from Germany and One Individual from Poland and an Alternate Direction of the Small Single-Copy Region. Forests 2021, 12, 180. https://doi.org/10.3390/f12020180
Mishra B, Ulaszewski B, Ploch S, Burczyk J, Thines M. A Circular Chloroplast Genome of Fagus sylvatica Reveals High Conservation between Two Individuals from Germany and One Individual from Poland and an Alternate Direction of the Small Single-Copy Region. Forests. 2021; 12(2):180. https://doi.org/10.3390/f12020180
Chicago/Turabian StyleMishra, Bagdevi, Bartosz Ulaszewski, Sebastian Ploch, Jaroslaw Burczyk, and Marco Thines. 2021. "A Circular Chloroplast Genome of Fagus sylvatica Reveals High Conservation between Two Individuals from Germany and One Individual from Poland and an Alternate Direction of the Small Single-Copy Region" Forests 12, no. 2: 180. https://doi.org/10.3390/f12020180
APA StyleMishra, B., Ulaszewski, B., Ploch, S., Burczyk, J., & Thines, M. (2021). A Circular Chloroplast Genome of Fagus sylvatica Reveals High Conservation between Two Individuals from Germany and One Individual from Poland and an Alternate Direction of the Small Single-Copy Region. Forests, 12(2), 180. https://doi.org/10.3390/f12020180





