Transcriptome Analysis for Fraxinus mandshurica Rupr. Seedlings from Different Carbon Sequestration Provenances in Response to Nitrogen Deficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Cultivation, Nitrogen Treatment, Harvesting and Determination of Dry Weight
2.2. cDNA Library Construction and Sequencing
2.3. De Novo cDNA Assembly and Functional Annotation
2.4. Differential Gene Expression Analysis
2.5. Quantitative Real-Time PCR Analysis
2.6. Determination of Phytohormone and LigninContent
2.7. Determination of the Content and Amount of C and N
2.8. Determination of Physiological Traits
2.9. Determination of Enzyme Activities
2.10. Statistical Analysis and Graphic Production
3. Results
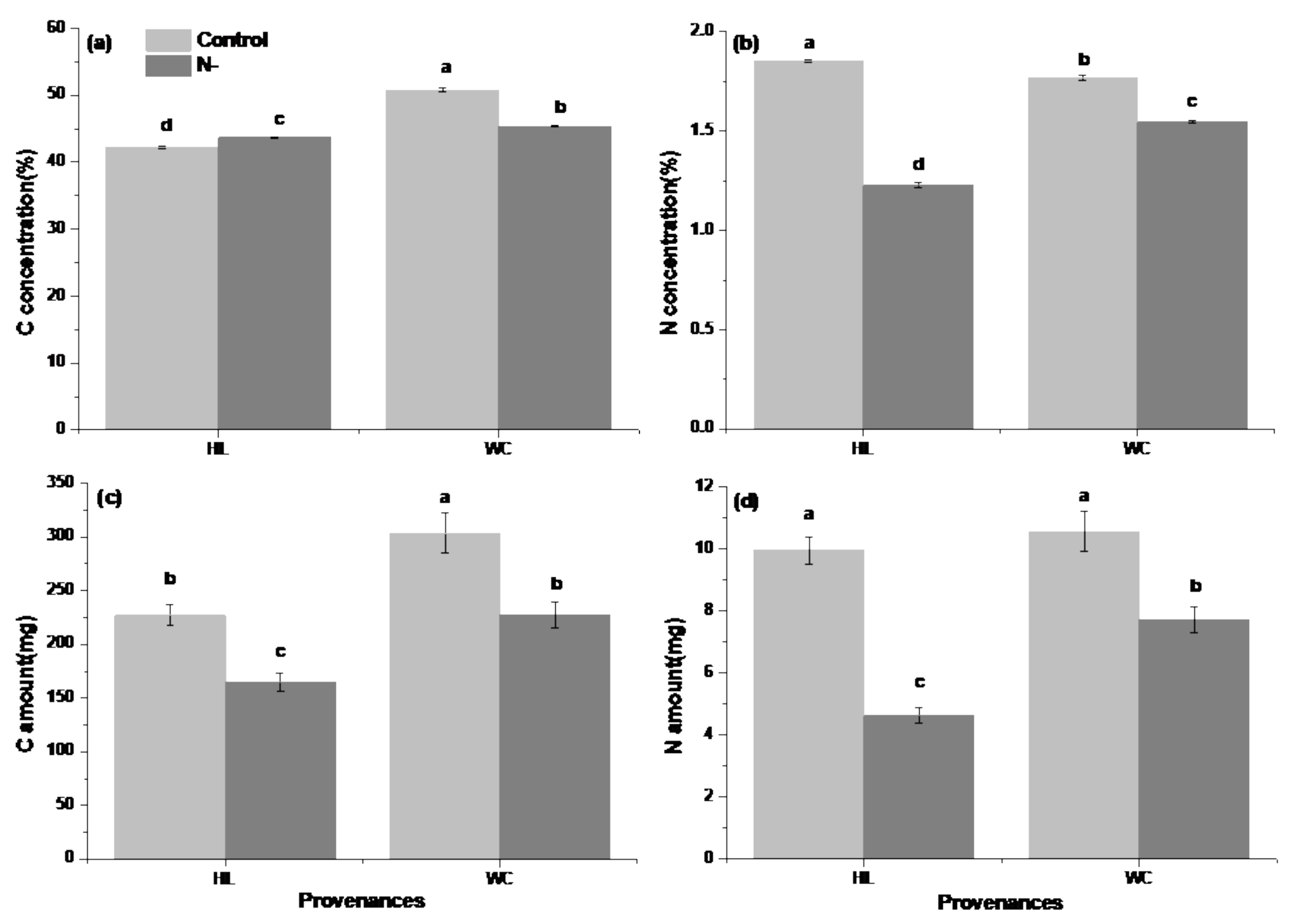
3.1. Analysis of Growth and Physiological Differences between Provenances
3.2. RNA-Seq and de Novo Transcriptome Assembly
3.3. Annotation and Functional Classification of Unigenes
3.4. Identification of Differentially Expressed Genes (DEGs)
3.5. KEGG Pathway Analysis for DEGs between the Two F. mandshurica Provenances
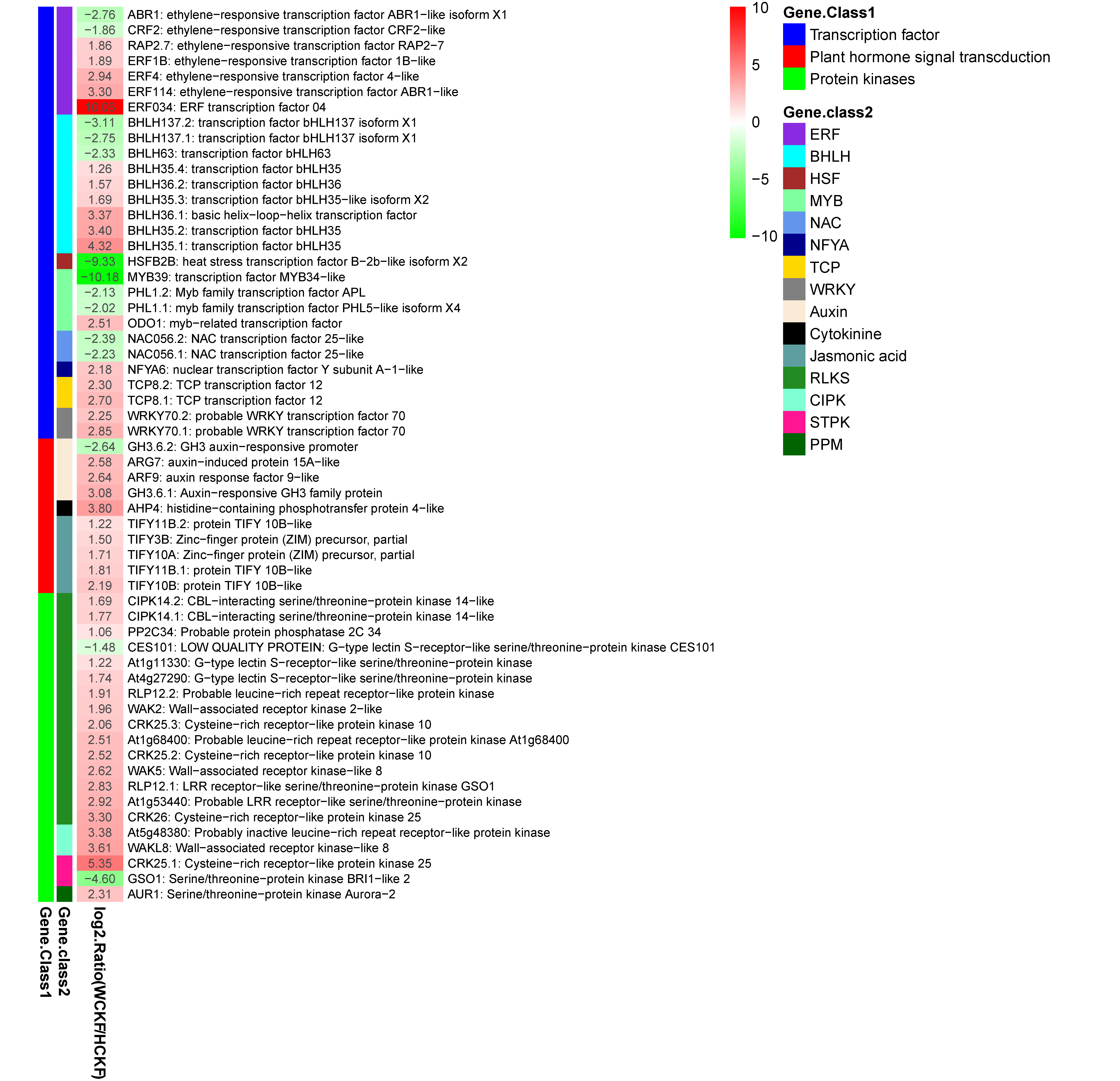
3.6. KEGG Pathway Analysis for DEGs Between the Two F. mandshurica Provenances in Response to Nitrogen Deficiency
4. Discussion
4.1. Growth and Physiological Differences between Provenances
4.2. DEGs Related to C Sequestration
4.3. Identification of DEGs Related to Nitrogen Deficiency
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nieto, V.; Giraldo-Charria, D.; Sarmiento, M.; Borralho, N. Effects of provenance and genetic variation on the growth and stem formation of Eucalyptus pellita in Colombia. J. Trop. For. Sci. 2016, 28, 227–234. [Google Scholar]
- Chhin, S.; Zalesny, R.S.; Parker, W.C.; Brissette, J. Dendroclimatic analysis of white pine (Pinus strobus L.) using long-term provenance test sites across eastern North America. For. Ecosyst. 2018, 5, 18–32. [Google Scholar] [CrossRef]
- Skulason, B.; Hansen, O.K.; Nielsen, U.B. Provenance variation in Christmas tree characteristics in subalpine fir (Abies lasiocarpa) planted in Denmark and Iceland. Scand. J. For. Res. 2018, 33, 222–232. [Google Scholar] [CrossRef]
- Pan, W.; Sun, J.; Xia, X.; Liu, R.; Wu, X.; Li, Y. Provenance variation in growth, stem-form, and wood basic density of 24-year-old Liriodendron. Austrian J. For. Sci. 2018, 343–362. [Google Scholar]
- Václav, B.; Petr, N.; Jaroslav, D. Results of Czech ash provenance experiment. J. For. Sci. 2017, 63, 263–274. [Google Scholar] [CrossRef]
- Zhou, C.B.; Gong, W. Effect of provenance and climate on xylem anatomy of Haloxylon ammodendron (C. A. mey) bunge in the gurbantunggut desert, China. Appl. Ecol. Environ. Res. 2017, 15, 1309–1321. [Google Scholar] [CrossRef]
- Lu, P.; Parker, W.C.; Colombo, S.J.; Man, R. Restructuring tree provenance test data to conform to reciprocal transplant experiments for detecting local adaptation. J. Appl. Ecol. 2016, 53, 1088–1097. [Google Scholar] [CrossRef]
- Thomson, A.M.; Riddell, C.L.; Parker, W.H. Boreal forest provenance tests used to predict optimal growth and response to climate change: 2. Black spruce. Can. J. Res. 2009, 39, 143–153. [Google Scholar] [CrossRef]
- Hebel, I.; Haas, R.; Dounavi, A. Genetic Variation of Common Ash (Fraxinus excelsior L.) Populations From Provenance Regions in Southern Germany by Using Nuclear and Chloroplast Microsatellites. Silvae Genetica 2006, 55, 38–44. [Google Scholar] [CrossRef]
- Calvo, L.; Hernández, V.; Valbuena, L.; Taboada, A. Provenance and seed mass determine seed tolerance to high temperatures associated to forest fires in Pinus pinaster. Ann. For. Sci. 2015, 73, 381–391. [Google Scholar] [CrossRef]
- Santoni, I.; Callone, E.; Sandak, A.; Sandak, J.; Dire, S. Solid state NMR and IR characterization of wood polymer structure in relation to tree provenance. Carbohydr. Polym. 2015, 117, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Spinnler, D.; Egli, P.; Körner, C. Provenance effects and allometry in beech and spruce under elevated CO2 and nitrogen on two different forest soils. Basic Appl. Ecol. 2003, 4, 467–478. [Google Scholar] [CrossRef]
- AmusAnt, N.; Digeon, A.; Descroix, L.; BruneAu, O.; BezArD, V.; BeAuchène, J. Planting rosewood for the sustainable essential oil productionInfluence of surrounding forest and seed provenance on tree growth and essential oil yield. Bois Forets des Tropiques 2015, 326, 57–65. [Google Scholar] [CrossRef]
- Klisz, M.; Ukalska, J.; Koprowski, M.; Tereba, A.; Puchałka, R.; Przybylski, P.; Jastrzębowski, S.; Nabais, C. Effect of provenance and climate on intra-annual density fluctuations of Norway spruce Picea abies (L.) Karst. in Poland. Agric. For. Meteorol. 2019, 269–270, 145–156. [Google Scholar] [CrossRef]
- Montwé, D.; Isaac-Renton, M.; Hamann, A.; Spiecker, H. Cold adaptation recorded in tree rings highlights risks associated with climate change and assisted migration. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Santiso, X.; Retuerto, R. Low among-provenance differences in structural and functional plasticity in response to nutrients in saplings of the circum-Mediterranean tree Arbutus unedo L. Tree Physiol. 2015, 35, 1118–1128. [Google Scholar] [CrossRef]
- Jiang, L.; Ball, G.; Hodgman, C.; Coules, A.; Zhao, H.; Lu, C. Analysis of Gene Regulatory Networks of Maize in Response to Nitrogen. Genes 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Nian, J.; Xie, X.; Yu, H.; Zhang, J.; Bai, J.; Dong, G.; Hu, J.; Bai, B.; Chen, L.; et al. Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice. Nat. Commun. 2018, 9, 735–744. [Google Scholar] [CrossRef]
- Bi, Y.; Wang, R.; Zhu, T.; Rothstein, S.J. Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genom. 2007, 8, 281–298. [Google Scholar] [CrossRef]
- Hao, Q.N.; Zhou, X.A.; Sha, A.H.; Wang, C.; Zhou, R.; Chen, S.L. Identification of genes associated with nitrogen-use efficiency by genome-wide transcriptional analysis of two soybean genotypes. BMC Genom. 2011, 12, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liang, N.; He, L.; Yu, L.; Cao, Y.; Xia, D.; Xiao, Y.; Zhang, G.; Zeng, F.; Zhan, Y. Variation in carbon traits among Fraxinus mandshurica populations and allometric equations between carbon traits and growth traits. New For. 2021. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Yang, W.; Yoon, J.; Choi, H.; Fan, Y.; Chen, R.; An, G. Transcriptome analysis of nitrogen-starvation-responsive genes in rice. BMC Plant Biol. 2015, 15. [Google Scholar] [CrossRef]
- He, Z.; Zhan, Y.; Zeng, F.; Zhao, X.; Wang, X. Drought physiology and gene expression characteristics of Fraxinus interspecific hybrids. Plant Growth Regul. 2016, 78, 179–193. [Google Scholar] [CrossRef]
- Wu, Y.; Jun, R.; Chen, W.; Liu, X.; Kong, F.; Ke, Y.; Shin, H.; Yuan, J. Effect of low-nitrogen stress on photosynthesis and chlorophyll fluorescence characteristics of maize cultivars with different lownitrogen tolerances. J. Integr. Agric. 2019, 18, 1246–1256. [Google Scholar] [CrossRef]
- Hao, Y.-Q.; Lu, G.-Q.; Wang, L.-H.; Wang, C.-L.; Guo, H.-M.; Li, Y.-F.; Cheng, H.-M. Overexpression of AmDUF1517 enhanced tolerance to salinity, drought, and cold stress in transgenic cotton. J. Integr. Agric. 2018, 17, 2204–2214. [Google Scholar] [CrossRef]
- Rosen, H. A Modfied Ninhydrin Calorimetric Analysis for Amino Acids. Arch. Biochem. Biophys. 1957, 67, 10–15. [Google Scholar] [CrossRef]
- Turano, F.J.; Muhitch, M.J. Differential accumulation of ferredoxin- and NADH-dependent glutamate synthase activities, peptides, and transcripts in developing soybean seedlings in response to light, nitrogen, and nodulation. Physiol. Plant. 1999, 107, 407–418. [Google Scholar] [CrossRef]
- Brugière, N.; Dubois, F.; Limami, A.M.; Lelandais, M.; Roux, Y.; Sangwan, R.S.; Hirel, B. Glutamine Synthetase in the Phloem Plays a Major Role in Controlling Proline Production. Plant Cell 1999, 11, 1995–2011. [Google Scholar] [CrossRef]
- Guiz, C.; Hirel, B.; Shedlofsky, G.; Gadal, P. Occurrence and influence of light on the relative proportions of two glutamine sythetases in rice leaves. Plant Sci. Lett. 1979, 15, 271–277. [Google Scholar] [CrossRef]
- Weber, J.C.; Larwanou, M.; Abasse, T.A.; Kalinganire, A. Growth and survival of Prosopis africana provenances tested in Niger and related to rainfall gradients in the West African Sahel. For. Ecol. Manag. 2008, 256, 585–592. [Google Scholar] [CrossRef]
- Saenger, P.; West, P.W. Phenotypic variation of the mangrove species Avicennia marina (Forssk.) Vierh. from seven provenances around Australia. Aquat. Bot. 2018, 149, 28–32. [Google Scholar] [CrossRef]
- Ginwal, H.S. Provenance and family variation in growth performance of Eucalyptus tereticornis (Sm.) in a provenance cum progeny trial in Midnapore, India. For. Ecol. Manag. 2009, 258, 2529–2534. [Google Scholar] [CrossRef]
- Miranda, I.; Almeida, M.H.; Pereira, P. Provenance and site variation of wood density in Eucalyptus globulus labill. At harvest age and its relation to a non-destructive early assessment. For. Ecol. Manag. 2001, 149, 235–240. [Google Scholar] [CrossRef]
- Bergin, D.O.; Kimberley, M.O.; Low, C.B. Provenance variation in Podocarpus totara (D. Don): Growth, tree form and wood density on a coastal site in the north of the natural range, New Zealand. For. Ecol. Manag. 2008, 253, 1367–1378. [Google Scholar] [CrossRef]
- Fkiri, S.; Guibal, F.; Fady, B.; Khorchani, A.E.; Khaldi, A.; Khouja, M.L.; Nasr, Z. Tree-rings to climate relationships in nineteen provenances of four black pines sub-species (Pinus nigra Arn.) growing in a common garden from Northwest Tunisia. Dendrochronologia 2018, 50, 44–51. [Google Scholar] [CrossRef]
- Luo, J.; Li, H.; Liu, T.; Polle, A.; Peng, C.; Luo, Z.-B. Nitrogen metabolism of two contrasting poplar species during acclimation to limiting nitrogen availability. J. Exp. Bot. 2013, 64, 4207–4224. [Google Scholar] [CrossRef]
- Yan, B.; Wu, B.; Gao, Y.; Wu, J.; Niu, J.; Xie, Y.; Cui, Z.; Zhang, Z. Effects of nitrogen and phosphorus on the regulation of nonstructural carbohydrate accumulation, translocation and the yield formation of oilseed flax. Field Crop. Res. 2018, 2019, 229–241. [Google Scholar] [CrossRef]
- Hause, B.; Vörös, K.; Kogel, K.; Besser, K.; Wasternack, C. A Jasmonate-responsive Lipoxygenase of Barley Leaves is Induced by Plant Activators but not by Pathogens. J. Plant Physiol. 1999, 154, 459–462. [Google Scholar] [CrossRef]
- Vargas, J.M.M.; Castillo, K.C.; Gallardo, N.M.; Ortiz, J.J.O.; Frier, J.P.D.; Winkler, R. Modulation of steroidal glycoalkaloid biosynthesis in tomato (Solanum lycopersicum) by jasmonic acid. Plant Sci. 2018, 277, 155–165. [Google Scholar] [CrossRef]
- Alsoufi, A.S.M.; Pączkowski, C.; Szakiel, A.; Długosz, M. Effect of jasmonic acid and chitosan on triterpenoid production in Calendula officinalis hairy root cultures. Phytochem. Lett. 2019, 31, 5–11. [Google Scholar] [CrossRef]
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The tify family previously known as ZIM. Trends Plant Sci. 2007, 12, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Zarattini, M.; Farjad, M.; Launay, A.; Cannella, D.; Soulié, M.C.; Bernacchia, G.; Fagard, M. Every cloud has a silver lining how abiotic stresses affect gene expression in plant pathogen interactions. J. Exp. Bot. 2020. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, C.M.; Lunn, J.E. A Tale of Two Sugars Trehalose 6-Phosphate and Sucrose. Plant Physiol. 2016, 172, 7–27. [Google Scholar] [CrossRef]
- Baena-González, E.; Lunn, J.E. SnRK1 and trehalose 6-phosphate—Two ancient pathways converge to regulate plant metabolism and growth. Curr. Opin. Plant Biol. 2020, 55, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zhao, X.; Qi, G.; Bai, Z.; Wang, Y.; Wang, S.; Ma, Y.; Liu, Q.; Hu, R.; Zhou, G. Integrated analysis of transcriptome and metabolites reveals an essential role of metabolic flux in starch accumulation under nitrogen starvation in duckweed. Biotechnol. Biofuels 2017, 10, 167–180. [Google Scholar] [CrossRef]
- Oh, Y.; Robertson, S.L.; Parker, J.; Muddiman, D.C.; Dean, R.A. Comparative proteomic analysis between nitrogen supplemented and starved conditions in Magnaporthe oryzae. Proteome Sci. 2017, 15, 20. [Google Scholar] [CrossRef]
- Quan, X.; Zeng, J.; Ye, L.; Chen, G.; Han, Z.; Shah, J.M.; Zhang, G. Transcriptome profiling analysis for two Tibetan wild barley genotypes in responses to low nitrogen. BMC Plant Biol. 2016, 16, 30. [Google Scholar] [CrossRef]
- Bi, Y.; Meyer, A.; Downs, G.S.; Shi, X.; El-kereamy, A.; Lukens, L.; Rothstein, S.J. High throughput RNA sequencing of a hybrid maize and its parents shows different mechanisms responsive to nitrogen limitation. BMC Genom. 2014, 15, 77–88. [Google Scholar] [CrossRef]
- Gutiérrez, R.A.; Lejay, L.V.; Dean, A.; Chiaromonte, F.; Shasha, D.E.; Coruzzi, G.M. Qualitative network models and genome-wide expression data define carbonnitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007, 8, R7. [Google Scholar] [CrossRef] [PubMed]
- Amiour, N.; Imbaud, S.; Clément, G.; Agier, N.; Zivy, M.; Valot, B.; Balliau, T.; Armengaud, P.; Quilleré, I.; Cañas, R.; et al. The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize. J. Exp. Bot. 2012, 63, 5017–5033. [Google Scholar] [CrossRef]
- Guo, X.; Yu, C.; Luo, L.; Wan, H.; Zhen, N.; Xu, T.; Tan, J.; Pan, H.; Zhang, Q. Transcriptome of the floral transition in Rosa chinensis ‘Old Blush’. BMC Genom. 2017, 18, 199–216. [Google Scholar] [CrossRef]
- Ning, P.; Yang, L.; Li, C.; Fritschi, F.B. Post-silking carbon partitioning under nitrogen deficiency revealed sink limitation of grain yield in maize. J. Exp. Bot. 2018, 69, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- PlavcovÁ, L.; Hacke, U.G.; Almeida-Rodriguez, A.M.; Li, E.; Douglas, C.J. Gene expression patterns underlying changes in xylem structure and function in response to increased nitrogen availability in hybrid poplar. Plant Cell Environ. 2013, 36, 186–199. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, X.; Yang, J.; Liu, W.; Du, Q.; Wang, H.; Fu, C.; Li, W.X. MicroRNA528 Affects Lodging Resistance of Maize by Regulating Lignin Biosynthesis under Nitrogen-Luxury Conditions. Mol. Plant 2018, 11, 806–814. [Google Scholar] [CrossRef]
- Camargo, E.L.O.; Nascimento, L.C.; Soler, M.; Salazar, M.M.; Lepikson-Neto, J.; Marques, W.L.; Alves, A.; Teixeira, P.J.P.L.; Mieczkowsk, P.; Carazzolle, M.F.; et al. Contrasting nitrogen fertilization treatments impact xylem gene expression and secondary cell wall lignification in Eucalyptus. BMC Plant Biol. 2014, 14, 256–272. [Google Scholar] [CrossRef] [PubMed]
- Pascual, M.B.; El-Azaz, J.; de la Torre, F.N.; Cañas, R.A.; Avila, C.; Cánovas, F.M. Biosynthesis and Metabolic Fate of Phenylalanine in Conifers. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Donaldson, L.; Kim, H.; Phillips, L.; Flint, H.; Steward, D.; Torr, K.; Koch, G.; Schmitt, U.; Ralph, J. Suppression of 4-Coumarate-CoA Ligase in the Coniferous Gymnosperm Pinus radiata. Plant Physiol. 2009, 149, 370–383. [Google Scholar] [CrossRef]
- Moura, J.C.M.S.; Bonine, C.A.V.; Viana, J.O.F.; Dornelas, M.C.; Mazzafera, P. Abiotic and Biotic Stresses and Changes in the Lignin Content and Composition in Plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Liu, Q.; Zheng, L.; He, F.; Zhao, F.-J.; Shen, Z.; Zheng, L. Transcriptional and physiological analyses identify a regulatory role for hydrogen peroxide in the lignin biosynthesis of copper-stressed rice roots. Plant Soil 2014, 387, 323–336. [Google Scholar] [CrossRef]
- García-Rojas, M.; Meneses, M.; Oviedo, K.; Carrasco, C.; Defilippi, B.; González-Agüero, M.; León, G.; Hinrichsen, P. Exogenous gibberellic acid application induces the overexpression of key genes for pedicel lignification and an increase in berry drop in table grape. Plant Physiol. Biochem. 2018, 126, 32–38. [Google Scholar] [CrossRef]
- Luo, J.; Wang, X.; Feng, L.; Li, Y.; He, J. The mitogen-activated protein kinase kinase 9 (MKK9) modulates nitrogen acquisition and anthocyanin accumulation under nitrogen-limiting condition in Arabidopsis. Biochem. Biophys. Res. Commun. 2017, 487, 539–544. [Google Scholar] [CrossRef]
- Albacete, A.A.; Martínez-Andújar, C.; Pérez-Alfocea, F. Hormonal and metabolic regulation of source–sink relations under salinity and drought: From plant survival to crop yield stability. Biotechnol. Adv. 2014, 32, 12–30. [Google Scholar] [CrossRef]
- Hisano, H.; Matsuura, T.; Mori, I.C.; Yamane, M.; Sato, K. Endogenous hormone levels affect the regeneration ability of callus derived from different organs in barley. Plant Physiol. Biochem. 2016, 99, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Verbon, E.H.; Liberman, L.M. Beneficial Microbes Affect Endogenous Mechanisms Controlling Root Development. Trends Plant Sci. 2016, 21, 218–229. [Google Scholar] [CrossRef]
- Xiong, Q.; Tang, G.; Zhong, L.; He, H.; Chen, X. Response to Nitrogen Deficiency and Compensation on Physiological Characteristics, Yield Formation, and Nitrogen Utilization of Rice. Front. Plant Sci. 2018, 9, 1075. [Google Scholar] [CrossRef] [PubMed]
- Grossman, A.; Takahashi, H. Macronutrient Utilization by Photosynthetic Eukaryotes and the Fabric of Interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 163–210. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Diaz, C.; Colombani, V.C.; Loudet, O.; Belluomo, P.; Moreau, L.; Vedele, F.D.; Morot-Gaudry, J.F.; Daubresse, C.M. Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 74–83. [Google Scholar] [CrossRef]








| Code | Stage Comparison | Log2 Ratio () | Total DEGs | Upregulated | Downregulated |
|---|---|---|---|---|---|
| CP1 | HCKF vs. WCKF | WCKF/HCKF | 669 | 384 | 285 |
| CP2 | HCKR vs. WCKR | WCKR/HCKR | 149 | 72 | 77 |
| CP3 | WCKF vs. WCTF | WCTF/WCKF | 305 | 165 | 140 |
| CP4 | WCKR vs. WTR | WTR/WCKR | 4225 | 1907 | 2318 |
| CP5 | HCKR vs. HTR | HTR/HCKR | 4531 | 2386 | 2145 |
| CP6 | HCKF vs. HTF | HTF/HCKF | 1414 | 991 | 423 |
| CP7 | HTF vs. WTF | WTF/HTF | 141 | 74 | 67 |
| CP8 | HTR vs. WTR | WTR/HTR | 178 | 134 | 44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Yu, L.; Liu, Z.; Liu, J.; Ji, X.; Zhang, X.; Liu, M.; Mei, Y.; Zeng, F.; Zhan, Y. Transcriptome Analysis for Fraxinus mandshurica Rupr. Seedlings from Different Carbon Sequestration Provenances in Response to Nitrogen Deficiency. Forests 2021, 12, 257. https://doi.org/10.3390/f12020257
Zhao X, Yu L, Liu Z, Liu J, Ji X, Zhang X, Liu M, Mei Y, Zeng F, Zhan Y. Transcriptome Analysis for Fraxinus mandshurica Rupr. Seedlings from Different Carbon Sequestration Provenances in Response to Nitrogen Deficiency. Forests. 2021; 12(2):257. https://doi.org/10.3390/f12020257
Chicago/Turabian StyleZhao, Xingtang, Lei Yu, Zhang Liu, Jianfei Liu, Xintong Ji, Xu Zhang, Mengqi Liu, Yushuo Mei, Fansuo Zeng, and Yaguang Zhan. 2021. "Transcriptome Analysis for Fraxinus mandshurica Rupr. Seedlings from Different Carbon Sequestration Provenances in Response to Nitrogen Deficiency" Forests 12, no. 2: 257. https://doi.org/10.3390/f12020257
APA StyleZhao, X., Yu, L., Liu, Z., Liu, J., Ji, X., Zhang, X., Liu, M., Mei, Y., Zeng, F., & Zhan, Y. (2021). Transcriptome Analysis for Fraxinus mandshurica Rupr. Seedlings from Different Carbon Sequestration Provenances in Response to Nitrogen Deficiency. Forests, 12(2), 257. https://doi.org/10.3390/f12020257





