Analysis on Characteristics of Vegetation and Soil Bacterial Community under 20 Years’ Restoration of Different Tree Species: A Case Study of the Qinling Mountains
Abstract
1. Introduction
2. Material and Methods
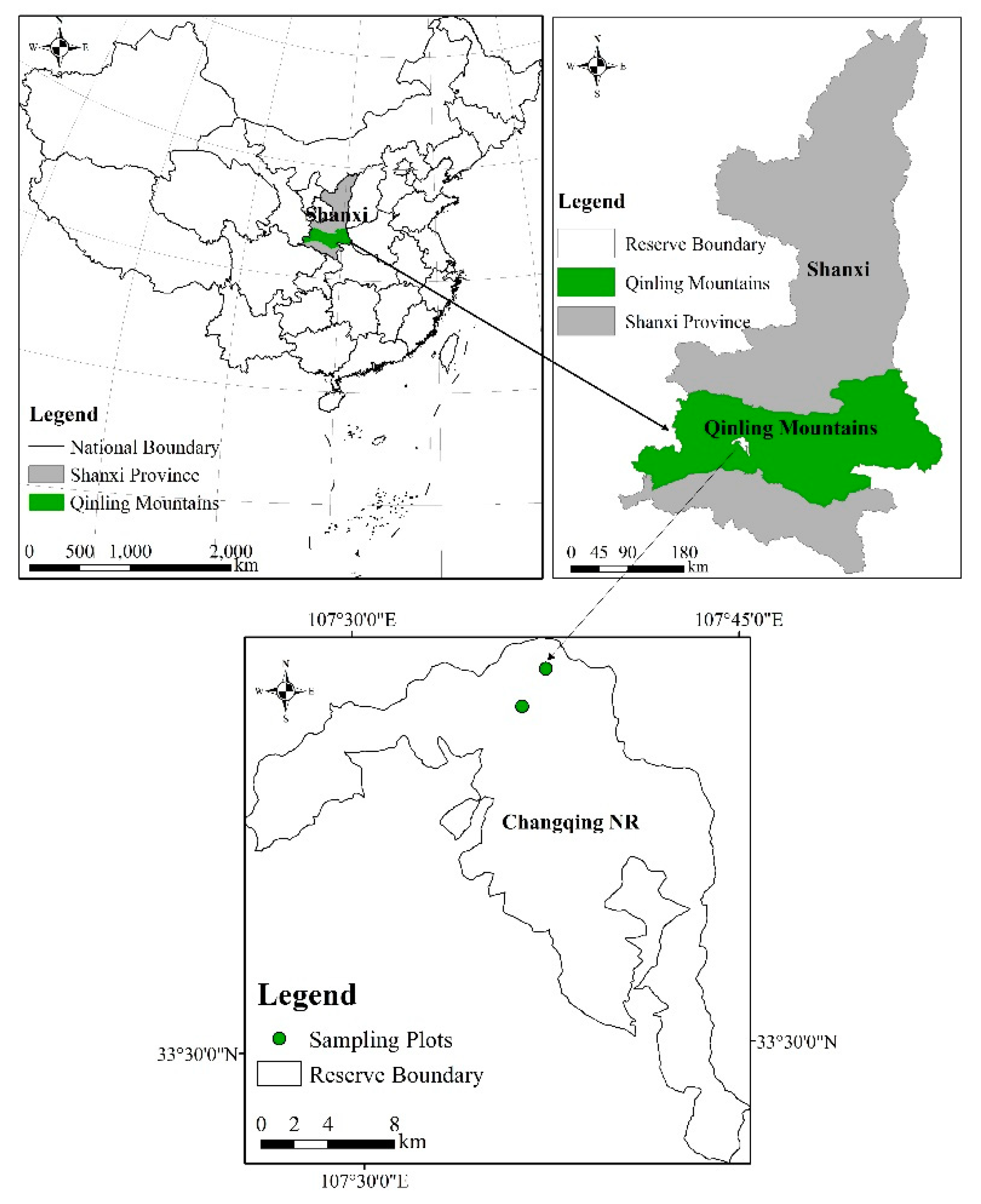
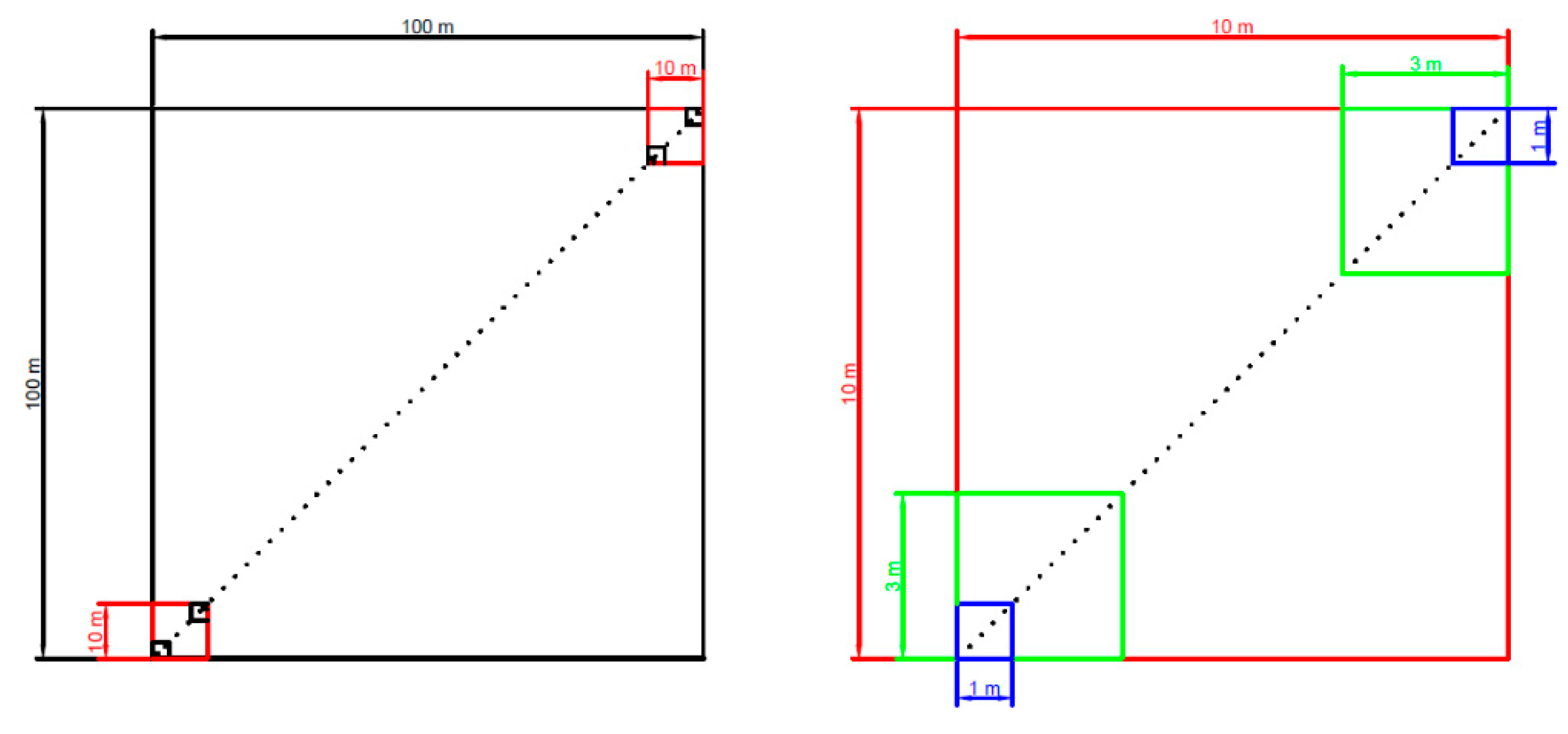
2.1. Field Site and Sampling
2.2. Sample Analysis
2.2.1. Biomass Estimation of Plant
2.2.2. Physico–Chemical Analysis
2.2.3. Soil Microbial DNA Extraction, PCR Amplification and Sequencing
2.3. Pyrosequencing Data Treatment
2.4. Data Processing and Statistical Analysis
3. Results
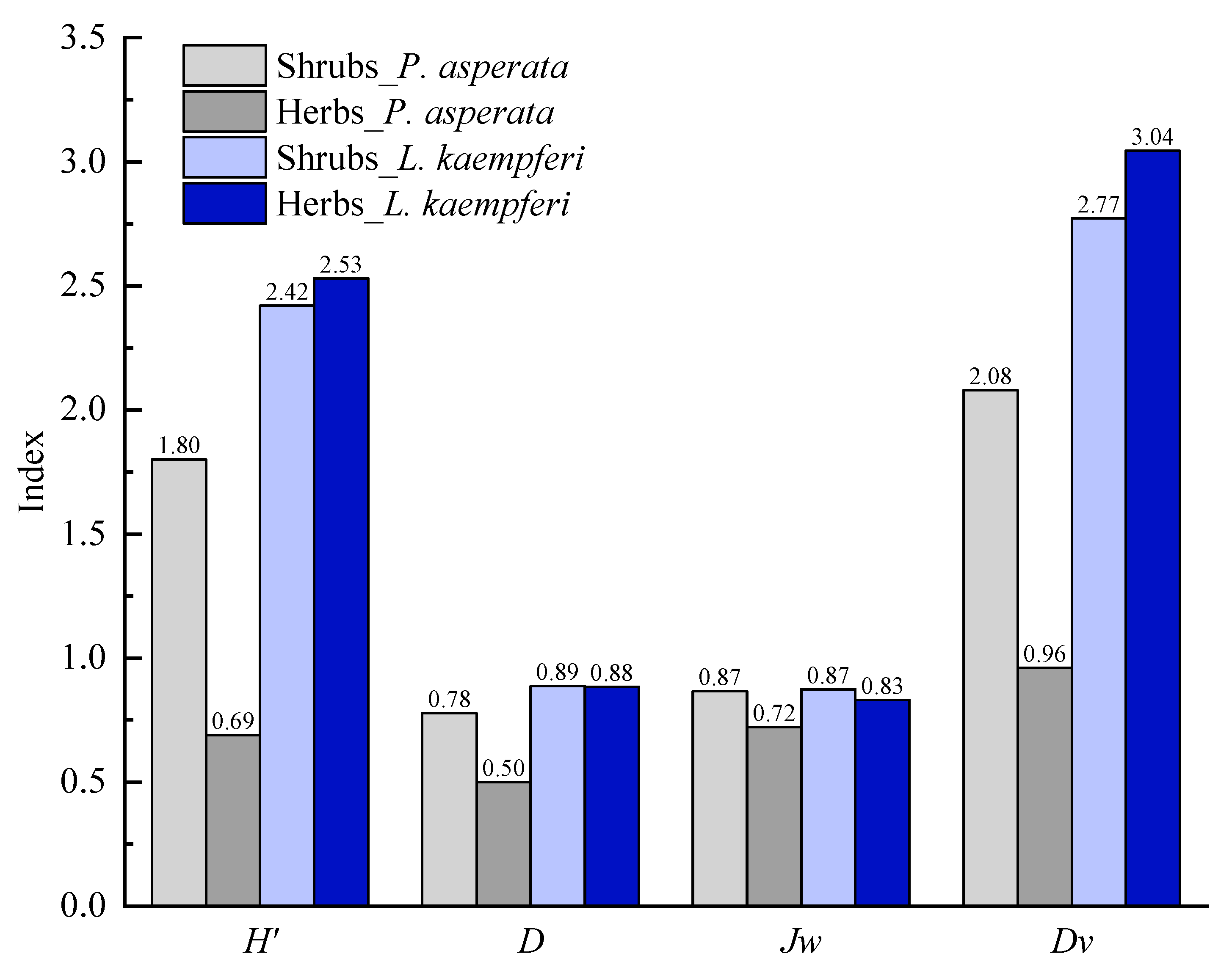
3.1. Diversities and Biomass of Vegetation
3.1.1. Alpha Diversity of P. asperata Forest
3.1.2. Alpha Diversity of L. kaempferi Forest
3.1.3. Vegetation Biomass of P. asperata and L. kaempferi Forest
3.2. Physico–Chemical Properties of Soils
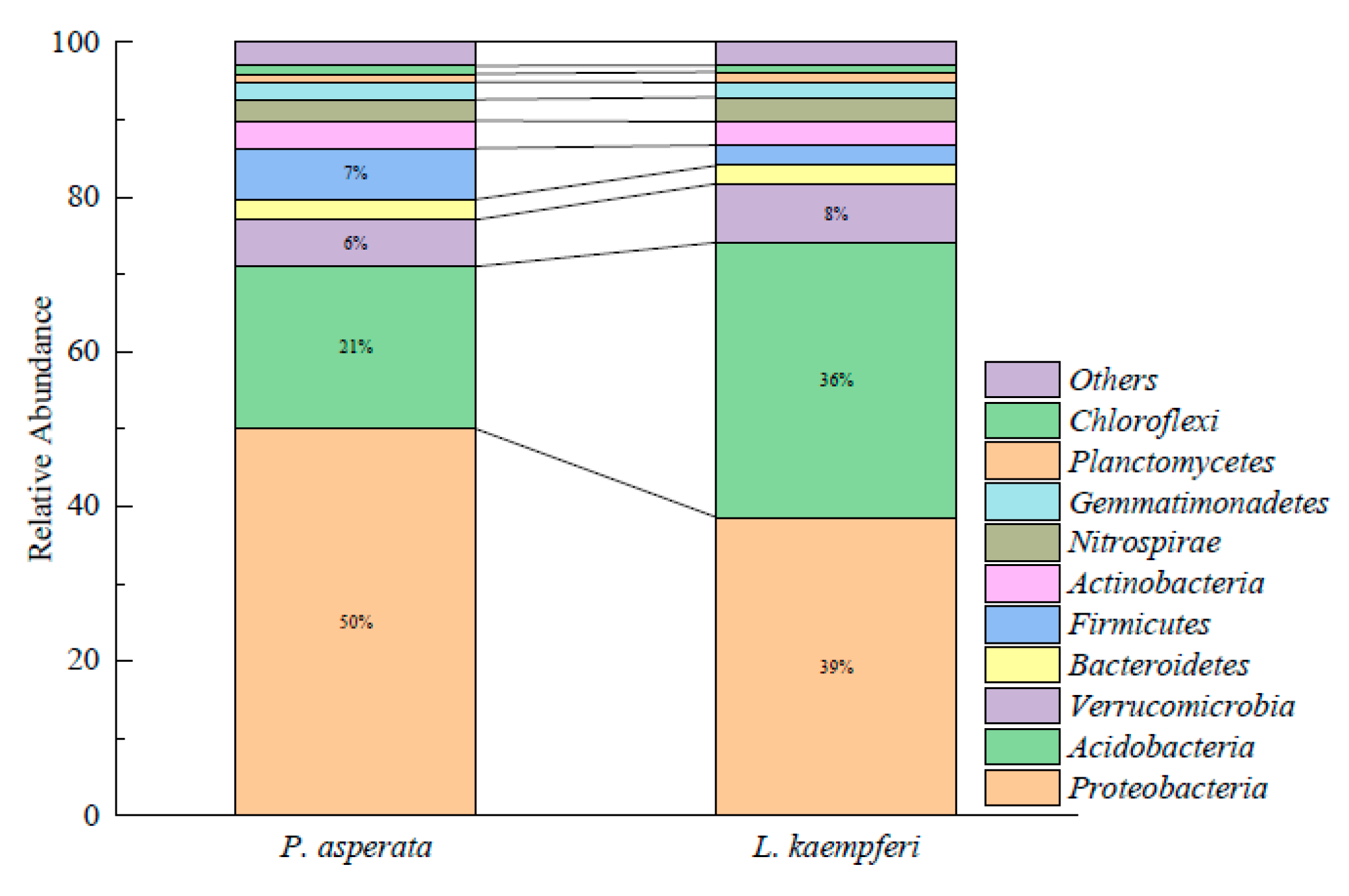
3.3. Diversity and Composition of Soil Bacterial Community
4. Discussion
4.1. Distribution Characteristics of Vegetation
4.2. Factors That Effected Soil Physico–Chemical Properties
4.3. Effects of Tree Species on Soil Bacterial Community Composition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stephens, S.S.; Wagner, M.R. Forest Plantations and Biodiversity: A Fresh Perspective. J. For. 2007, 105, 307–313. [Google Scholar] [CrossRef]
- Carnus, J.-M.; Parrotta, J.; Brockerhoff, E.; Arbez, M.; Jactel, H.; Kremer, A.; Lamb, D.; O’Hara, K.; Walters, B. Planted Forests and Biodiversity. J. For. 2006, 104, 65–77. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, J.; Zhao, N.; Liu, Y.; Wang, Y.; Wilson, J.P.; Yue, T. Estimation of China’s forest stand biomass carbon sequestration based on the continuous biomass expansion factor model and seven forest inventories from 1977 to 2013. For. Ecol. Manag. 2019, 448, 528–534. [Google Scholar] [CrossRef]
- Liu, J.; Dang, P.; Gao, Y.; Zhu, H.; Zhu, H.; Zhao, F.; Zhao, Z. Effects of tree species and soil properties on the composition and diversity of the soil bacterial community following afforestation. For. Ecol. Manag. 2018, 427, 342–349. [Google Scholar] [CrossRef]
- FAO. Global Forest Resources Assessment 2015: How Are the World’s Forests Changing? The Food and Agriculture Organization: Rome, Italy, 2016. [Google Scholar]
- Song, X.C.; Wang, H.L.; Qin, W.D.; Deng, X.J.; Tian, H.D.; Tan, Y.B.; Wang, S.H.; Cao, J.Z. Effects of stand of artificial forests on soil microbila functhional diversity. Chin. J. Appl. Ecol. 2019, 30, 841–848. [Google Scholar]
- He, Y.J.; Liang, X.Y.; Qin, L.; Li, Z.Y.; Shao, M.X.; Tan, L. Community characteristics and soil properties of coniferous plantation forest monocultures in the early stages after close-to-nature transformation management in southern subtropical China. Acta Ecol. Sin. 2013, 33, 2484–2495. [Google Scholar]
- Chodak, M.; Klimek, B.; Azarbad, H.; Jaźwa, M. Functional diversity of soil microbial communities under Scots pine, Norway spruce, silver birch and mixed boreal forests. Pedobiologia 2015, 58, 81–88. [Google Scholar] [CrossRef]
- Zheng, H.; Ouyang, Z.Y.; Wang, X.K.; Fang, Z.G.; Zhao, T.Q.; Miao, H. Effects of regenerating forest cover on soil microbial communities: A case study in hilly red soil region, Southern China. For. Ecol. Manag. 2005, 217, 244–254. [Google Scholar] [CrossRef]
- Xu, Q.; Jiang, P.; Xu, Z. Soil microbial functional diversity under intensively managed bamboo plantations in southern China. J. Soils Sediments 2008, 8, 177–183. [Google Scholar] [CrossRef]
- Chan, O.C.; Yang, X.; Fu, Y.; Feng, Z.; Sha, L.; Casper, P.; Zou, X. 16S rRNA gene analyses of bacterial community structures in the soils of evergreen broad-leaved forests in south-west China. FEMS Microbiol Ecol. 2006, 58, 13. [Google Scholar] [CrossRef]
- Wheeler, C.E.; Omeja, P.A.; Chapman, C.A.; Glipin, M.; Tumwesigye, C.; Lewis, S.L. Carbon sequestration and biodiversity following 18 years of active tropical forest restoration. For. Ecol. Manag. 2016, 373, 44–55. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, Y. Biomass, Carbon and Nutrient Storage in a 30-Year-Old Chinese Cork Oak (Quercus Variabilis) Forest on the South Slope of the Qinling Mountains, China. Forests 2015, 6, 1239–1255. [Google Scholar] [CrossRef]
- Zheng, H.; Chen, F.; Ouyang, Z.; Tu, N.; Xu, W.; Wang, X.; Miao, H.; Li, X.; Tian, Y. Impacts of reforestation approaches on runoff control in the hilly red soil region of Southern China. J. Hydrol. 2008, 356, 174–184. [Google Scholar] [CrossRef]
- Wang, H.H.; Chu, H.L.; Dou, Q.; Xie, Q.Z.; Tang, M.; Sung, C.K.; Wang, C.Y. Phosphorus and Nitrogen Drive the Seasonal Dynamics of Bacterial Communities in Pinus Forest Rhizospheric Soil of the Qinling Mountains. Front. Microbiol. 2018, 9, 1930. [Google Scholar] [CrossRef]
- Liu, X.; Wu, P.; Shao, X.; Songer, M.; Cai, Q.; Zhu, Y.; He, X. Spatiotemporally monitoring forest landscape for giant panda habitat through a high learning-sensitive neural network in Guanyinshan Nature Reserve in the Qinling Mountains, China. Environ. Earth Sci. 2017, 76, 1–12. [Google Scholar] [CrossRef]
- Jiang, S.G. Review on soil bulk density determination method. Hubei Agric. S 2019, 58, 82–91. [Google Scholar]
- De Feudis, M.; Falsone, G.; Vianello, G.; Vittori Antisari, L. The Conversion of Abandoned Chestnut Forests to Managed Ones Does Not Affect the Soil Chemical Properties and Improves the Soil Microbial Biomass Activity. Forests 2020, 11, 786. [Google Scholar] [CrossRef]
- Bao, S.D. Soil Agrochemical Analysis, 3rd ed.; China Agriculture Press: Beijing, China, 2000. [Google Scholar]
- Xue, W.P. Characteristics of Soil Organic Carbon Density in Pinus Armandii and Picea Asperata Forest in Qinling Mountains. Master’s Thesis, Northwest Agriculture & Forestry University, Xi’an, China, 2016. [Google Scholar]
- Bremner, J.; Mulvaney, C. Nitrogen-total. Methods Soil Anal. 1996, 5, 1085–1121. [Google Scholar]
- Zhang, C.; Xue, S.; Liu, G.B.; Song, Z.L. A comparison of soil qualities of different revegetation types in the Loess Plateau, China. Plant. Soil 2011, 347, 163–178. [Google Scholar] [CrossRef]
- Editorial committee of Chinese flora, Chinese Academy of Sciences. Flora of China; Editorial committee of Chinese flora, Science Press: Bejing, China, 1993. [Google Scholar]
- Ji, L. Biomass and Diversity of Shrubs and Herbs under Three Type different Canopy Forests in Jingouling Forestry Station. Master’s Thesis, Beijing Forestry University, Beijing, China, 2016. [Google Scholar]
- Guangquan, L.; Shidong, Z.; Xiaoning, T. Distributional characteristice on biomass and nutrient elements of Pine-oak forest belt in MT. Qingling. Sci. Silvae Sin. 2001, 1, 28–36. [Google Scholar]
- Xu, L.; Yu, G.; He, N.; Wang, Q.; Gao, Y.; Wen, D.; Ge, J. Carbon storage in China’s terrestrial ecosystems: A synthesis. Sci. Rep. 2018, 8, 2806. [Google Scholar] [CrossRef]
- Liu, H. Changes in Landscapes Pattern and Carbon Storage of Main Forest Tyes at Huoditang Forest Region in the Qinling Mountains. Ph.D. Thesis, Northwest Agriculture & Forestry University, Xi’an, China, 2005. [Google Scholar]
- Lin, H.; Ruide, L.; Dexiang, W.; Lianbin, S.; Hui, Z. Carbon density of arbor layer in Pinus tabulaeformis community in Huoditang forest region in Qinling Mountains. J. Northeast For. Univ. 2009, 37, 23–25. [Google Scholar]
- Kang, L. Research on Arborous Layer Aboveground Biomass and Gross Productivity of the Typical Forest Types on the Southern Slope of Qinling Mountains. Master’s Thesis, Northwest Agriculture & Forestry University, Xi’an, China, 2012. [Google Scholar]
- Ren, Y.H. Carbon Sequestration of Main Forest Tyoes at Huoditang Forest Region in the Qinling Mountains. Master’s Thesis, Northwest Agriculture & Forestry University, Xi’an, China, 2012. [Google Scholar]
- Zhao, Y.; Wang, P.; Fan, W.; Zhu, Y. Nutrient cycling in Quercus varlabilis plantations of different ages classes in hilly region of Taihang Mountain. Sci. Soil Water Conserv. 2009, 7, 66–71. [Google Scholar]
- Wen, D.; He, N.P. Spatial patterns of litter density and their controlling factors in forests and grasslands of China. Acta Ecol. Sin. 2016, 36, 2876–2884. [Google Scholar]
- Niu, X.Y.; Sun, X.M.; Chen, D.S.; Zhan, S.G. Soil microorganisms, nutrients and enzyme activity of Larix kaempferi plantation under different ages in mountainous region of eastern Liaoning Province, China. Chin. J. Appl. Ecol. 2015, 26, 2663–2672. [Google Scholar]
- Zhang, J.; Peng, C.; Zhu, Q.; Xue, W.; Shen, Y.; Yang, Y.; Wang, M. Temperature sensitivity of soil carbon dioxide and nitrous oxide emissions in mountain forest and meadow ecosystems in China. Atmos. Environ. 2016, 142, 340–350. [Google Scholar] [CrossRef]
- Wang, D.; Geng, Z.C.; She, D.; He, W.X.; Hou, L. Soil organic carbon storage and vertical distribution of carbon and nitrogen across different forest types in the Qinling Mountains. Acta Ecol. Sin. 2015, 35, 9. [Google Scholar]
- Don, A.; Schumacher, J.; Scherer-Lorenzen, M.; Scholten, T.; Schulze, E.D. Spatial and vertical variation of soil carbon at two grassland sites—Implications for measuring soil carbon stocks. Geoderma 2007, 141, 272–282. [Google Scholar] [CrossRef]
- Jia, P.; Hao, W.; Li, G.F. Study on P Content Changement in Larch Plantation. J. Northeast Forest Univ. 1998, 68–70. [Google Scholar]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
- Shi, L.-L.; Mortimer, P.E.; Ferry Slik, J.W.; Zou, X.-M.; Xu, J.; Feng, W.-T.; Qiao, L. Variation in forest soil fungal diversity along a latitudinal gradient. Fungal Divers. 2013, 64, 305–315. [Google Scholar] [CrossRef]
- Cheng, D.; Tian, Z.; Feng, L.; Xu, L.; Wang, H. Diversity analysis of the rhizospheric and endophytic bacterial communities of Senecio vulgaris L. (Asteraceae) in an invasive range. Peerj 2019, 6, e6162. [Google Scholar] [CrossRef] [PubMed]
- Kembel, S.W.; Eisen, J.A.; Pollard, K.S.; Green, J.L. The phylogenetic diversity of metagenomes. PLoS ONE 2011, 6, e23214. [Google Scholar] [CrossRef] [PubMed]
- Horton, M.W.; Bodenhausen, N.; Beilsmith, K.; Meng, D.; Muegge, B.D.; Subramanian, S.; Vetter, M.M.; Vilhjalmsson, B.J.; Nordborg, M.; Gordon, J.I.; et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun 2014, 5, 5320. [Google Scholar] [CrossRef]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina del Rio, T.; Jones, C.D.; Tringe, S.G.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860. [Google Scholar] [CrossRef]
- Lauber, C.L.; Ramirez, K.S.; Aanderud, Z.; Lennon, J.; Fierer, N. Temporal variability in soil microbial communities across land-use types. ISME J. 2013, 7, 1641–1650. [Google Scholar] [CrossRef]
- Placella, S.A.; Brodie, E.L.; Firestone, M.K. Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc. Natl. Acad. Sci. USA 2012, 109, 10931–10936. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Ren, C.; Zhang, W.; Zhong, Z.; Han, X.; Yang, G.; Feng, Y.; Ren, G. Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics. Sci. Total Environ. 2018, 610–611, 750–758. [Google Scholar] [CrossRef]
- Reynolds, H.L.; Packer, A.; Bever, J.D.; Clay, K. Grassroots Ecology: Plant–Microbe–Soil Interactions As Drivers Of Plant Community Structure And Dynamics. Ecology 2003, 84, 2281–2291. [Google Scholar] [CrossRef]
- Thakur, M.P.; Milcu, A.; Manning, P.; Niklaus, P.A.; Roscher, C.; Power, S.; Reich, P.B.; Scheu, S.; Tilman, D.; Ai, F.; et al. Plant diversity drives soil microbial biomass carbon in grasslands irrespective of global environmental change factors. Glob. Chang. Biol. 2015, 21, 4076–4085. [Google Scholar] [CrossRef]
- Miki, T. Microbe-mediated plant–soil feedback and its roles in a changing world. Ecol. Res. 2012, 27, 509–520. [Google Scholar] [CrossRef]
- Wang, N.F.; Zhang, T.; Zhang, F.; Wang, E.T.; He, J.F.; Ding, H.; Zhang, B.T.; Liu, J.; Ran, X.B.; Zang, J.Y. Diversity and structure of soil bacterial communities in the Fildes Region (maritime Antarctica) as revealed by 454 pyrosequencing. Front. Microbiol 2015, 6, 1188. [Google Scholar] [CrossRef]




| Site | Forest Types | Age | Temperature (°C) | Precipitation (mm) | Elevation (m) | Latitude | Longitude |
|---|---|---|---|---|---|---|---|
| A | P. asperata Mast. | 20 | 6.27 | 696.72 | 2271 | 33.703773° | 107.624529° |
| B | L.kaempferi (Lamb.) Carr. | 20 | 7.07 | 675.53 | 2039 | 33.683742° | 107.608682° |
| Sites | Tree Species | Occurrence Frequency (%) | Density (Trees·ha−1) | Average DBH (cm) | Average TH (m) |
|---|---|---|---|---|---|
| A | P. asperata Mast. | 88.37 | 1900 | 6.85 | 8.92 |
| Pinus armandi Franch. | 6.98 | 150 | 33.00 | 12.00 | |
| Acer davidii Franch. | 4.65 | 100 | 2.39 | 5.50 | |
| B | L. kaempferi (Lamb.) Carr. | 97.78 | 2200 | 15.37 | 11.06 |
| Prunus armandii Franch. | 2.22 | 50 | 2.55 | 5 |
| Forest Types | pH | SOC (g·kg−1) | TN (g·kg−1) | TP (g·kg−1) | C/N Ratio |
|---|---|---|---|---|---|
| P. asperata | 4.08 ± 0.02 | 26.53 ± 0.14 | 5.19 ± 0.07 | 0.92 ± 0.01 | 5.12 |
| L. kaempferi | 3.93 ± 0.08 | 21.36 ± 0.39 | 4.18 ± 0.09 | 1.13 ± 0.01 | 5.11 |
| Plant Biomass | α_Plant | Bacterial Composition | α_Bacterial | SOC | pH | TN | TP | |
|---|---|---|---|---|---|---|---|---|
| Plant Biomass | 1.000 | 0.964 ** | 0.856 * | 0.946 ** | 0.487 | 0.622 | 0.484 | −0.669 |
| α_plant | 1.000 | 0.779 | 0.983 ** | 0.546 | 0.214 | 0.497 | −0.673 | |
| Bacterial Composition | 1.000 | 0.698 | 0.703 | 0.717 | 0.759 | −0.875 * | ||
| α_bacterial | 1.000 | 0.414 | 0.475 | 0.342 | −0.545 | |||
| SOC | 1.000 | 0.860 * | 0.964 ** | −0.956 ** | ||||
| pH | 1.000 | 0.863* | −0.884 ** | |||||
| TN | 1.000 | −0.967 ** | ||||||
| TP | 1.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Liu, X.; Tian, Z.; Shao, X. Analysis on Characteristics of Vegetation and Soil Bacterial Community under 20 Years’ Restoration of Different Tree Species: A Case Study of the Qinling Mountains. Forests 2021, 12, 562. https://doi.org/10.3390/f12050562
Sun W, Liu X, Tian Z, Shao X. Analysis on Characteristics of Vegetation and Soil Bacterial Community under 20 Years’ Restoration of Different Tree Species: A Case Study of the Qinling Mountains. Forests. 2021; 12(5):562. https://doi.org/10.3390/f12050562
Chicago/Turabian StyleSun, Wanlong, Xuehua Liu, Zhaoxue Tian, and Xiaoming Shao. 2021. "Analysis on Characteristics of Vegetation and Soil Bacterial Community under 20 Years’ Restoration of Different Tree Species: A Case Study of the Qinling Mountains" Forests 12, no. 5: 562. https://doi.org/10.3390/f12050562
APA StyleSun, W., Liu, X., Tian, Z., & Shao, X. (2021). Analysis on Characteristics of Vegetation and Soil Bacterial Community under 20 Years’ Restoration of Different Tree Species: A Case Study of the Qinling Mountains. Forests, 12(5), 562. https://doi.org/10.3390/f12050562





