Genetic Variability of Alnus cordata (Loisel.) Duby Populations and Introgressive Hybridization with A. glutinosa (L.) Gaertn. in Southern Italy: Implication for Conservation and Management of Genetic Resources
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Isolation, SSR Amplification and Genotyping
2.2. Genetic Validation and Assignment of Species and Hybrids
2.3. Genetic Diversity and Population Structure
3. Results
3.1. Genetic Validation and Assignment of Species and Hybrids
3.2. Genetic Diversity and Population Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cai, Y.; Wang, F.; Tan, G.; Hu, Z.; Wang, Y.; Ng, W.L.; Wu, W.; Liu, Y.; Zhou, R. Hybridization of Bornean Melastoma: Implications for Conservation of Endemic Plants in Southeast Asia. Bot. Lett. 2019, 166, 117–124. [Google Scholar] [CrossRef]
- Rhymer, J.M.; Simberloff, D. Extinction by Hybridization and Introgression. Annu. Rev. Ecol. Syst. 1996, 27, 83–109. [Google Scholar] [CrossRef]
- Huxel, G.R. Rapid Displacement of Native Species by Invasive Species: Effects of Hybridization. Biol. Conserv. 1999, 89, 143–152. [Google Scholar] [CrossRef] [Green Version]
- Todesco, M.; Pascual, M.A.; Owens, G.L.; Ostevik, K.L.; Moyers, B.T.; Hübner, S.; Heredia, S.M.; Hahn, M.A.; Caseys, C.; Bock, D.G.; et al. Hybridization and Extinction. Evol. Appl. 2016, 9, 892–908. [Google Scholar] [CrossRef] [PubMed]
- Brock, M.T. The Potential for Genetic Assimilation of a Native Dandelion Species, Taraxacum Ceratophorum (Asteraceae), by the Exotic Congener T. Officinale. Am. J. Bot. 2004, 91, 656–663. [Google Scholar] [CrossRef] [Green Version]
- Klips, R.A.; Culley, T.M. Natural Hybridization between Prairie Milkweeds, Asclepias Sullivantii and Asclepias Syriaca: Morphological, Isozyme, and Hand-Pollination Evidence. Int. J. Plant Sci. 2004, 165, 1027–1037. [Google Scholar] [CrossRef]
- Hardig, T.M.; Allison, J.R.; Schilling, E.E. Molecular Evidence of Hybridization between Liatris Oligocephala (Asteraceae) and More-Widespread Congener: A Preliminary Assessment of the Potential for Extinction. Castanea 2005, 70, 246–254. [Google Scholar] [CrossRef]
- Burgess, K.S.; Husband, B.C. Habitat Differentiation and the Ecological Costs of Hybridization: The Effects of Introduced Mulberry (Morus alba) on a Native Congener (M. rubra). J. Ecol. 2006, 94, 1061–1069. [Google Scholar] [CrossRef]
- Zika, P.F. The Status of Impatiens Capensis (Balsaminaceae) on the Pacific Northwest Coast. J. Torrey Bot. Soc. 2006, 133, 593–600. [Google Scholar] [CrossRef]
- Xie, C.-Y.; El-Kassaby, Y.A.; Ying, C.C. Genetics of Red Alder (Alnus rubra Bong.) Populations in British Columbia and Its Implications for Gene Resources Management. New For. 2002, 24, 97–112. [Google Scholar] [CrossRef]
- Hamann, A.; El-Kassaby, Y.A.; Koshy, M.P.; Namkoong, G. Multivariate Analysis of Allozymic and Quantitative Trait Variation in Alnus Rubra: Geographic Patterns and Evolutionary Implications. Can. J. For. Res. 2011. [Google Scholar] [CrossRef]
- Schrader, J.A.; Graves, W.R. Infraspecific Systematics of Alnus maritima (Betulaceae) from Three Widely Disjunct Provenances. Castanea 2002, 67, 380–401. [Google Scholar]
- Gibson, J.P.; Rice, S.A.; Stucke, C.M. Comparison of Population Genetic Diversity between a Rare, Narrowly Distributed Species and a Common, Widespread Species of Alnus (Betulaceae). Am. J. Bot. 2008, 95, 588–596. [Google Scholar] [CrossRef]
- Mejnartowicz, L. Genetic Variation within and among Naturally Regenerating Populations of Alder (Alnus glutinosa). Acta Soc. Bot. Pol. 2008, 77, 105–110. [Google Scholar] [CrossRef] [Green Version]
- Prat, D.; Leger, C.; Bojovic, S. Genetic Diversity among Alnus glutinosa (L.) Gaertn. Populations. Acta Oecol. 1992, 13, 469–477. [Google Scholar]
- King, R.A.; Ferris, C. Chloroplast DNA Phylogeography of Alnus glutinosa (L.) Gaertn. Mol. Ecol. 1998, 7, 1151–1161. [Google Scholar] [CrossRef]
- Havrdová, A.; Douda, J.; Krak, K.; Vít, P.; Hadincová, V.; Zákravský, P.; Mandák, B. Higher Genetic Diversity in Recolonized Areas than in Refugia of Alnus glutinosa Triggered by Continent-Wide Lineage Admixture. Mol. Ecol. 2015, 24, 4759–4777. [Google Scholar] [CrossRef] [PubMed]
- Huh, M.K. Genetic Diversity and Population Structure of Korean Alder (Alnus japonica; Betulaceae). Can. J. For. Res. 2011. [Google Scholar] [CrossRef]
- Bousquet, J.; Cheliak, W.M.; Lalonde, M. Genetic Differentiation among 22 Mature Populations of Green Alder (Alnus crispa) in Central Quebec. Can. J. For. Res. 2011. [Google Scholar] [CrossRef]
- Mandák, B.; Havrdová, A.; Krak, K.; Hadincová, V.; Vít, P.; Zákravský, P.; Douda, J. Recent Similarity in Distribution Ranges Does Not Mean a Similar Postglacial History: A Phylogeographical Study of the Boreal Tree Species Alnus incana Based on Microsatellite and Chloroplast DNA Variation. New Phytol. 2016, 210, 1395–1407. [Google Scholar] [CrossRef] [Green Version]
- Hylander, N. On Cut-Leaved and Small-Leaved Forms of Alnus Glutinosa and A. Incana. Sven. Bot Tidskr 1957, 51, 437–453. [Google Scholar]
- Parnell, J. Variation and Hybridisation of Alnus Miller in Ireland. Watsonia 1994, 20, 61–71. [Google Scholar]
- Vander Mijnsbrugge, K. Morphological Dissection of Leaf, Bud and Infructescence Traits of the Interfertile Native A. Glutinosa and Non-Native A. Incana in Flanders (Northern Part of Belgium). Trees 2015, 29, 1661–1672. [Google Scholar] [CrossRef]
- Poljak, I.; Idžojtić, M.; Šapić, I.; Vukelić, J.; Zebec, M. Population Variability of Grey (Alnus incana (L.) Moench) and Black Alder (A. glutinosa (L.) Gaertn.) in the Mura and Drava Region According to the Leaf Morphology. Šumar. List 2014, 1–2, 7–17. [Google Scholar]
- Furlow, J.J. The Systematics of the American Species of Alnus (Betulaceae). Rhodora 1979, 81, 1–121. [Google Scholar]
- Hall, R.B.; Burgess, D. Evaluation of Alnus Species and Hybrids. Biomass 1990, 22, 21–34. [Google Scholar] [CrossRef]
- Vít, P.; Douda, J.; Krak, K.; Havrdová, A.; Mandák, B. Two New Polyploid Species Closely Related to Alnus glutinosa in Europe and North Africa—An Analysis Based on Morphometry, Karyology, Flow Cytometry and Microsatellites. Taxon 2017, 66, 567–583. [Google Scholar] [CrossRef]
- Ducci, F.; Tani, A. Alnus Cordata—Technical Guidelines for Genetic Conservation and Use for Italian Alder; Bioversity International: Rome, Italy, 2009; ISBN 978-92-9043-786-4. [Google Scholar]
- San-Miguel-Ayanz, J.; de Rigo, D.; Caudullo, G.; Durrant, T.H.; Mauri, A. European Atlas of Forest Tree Species; Publication Office of the European Union: Luxembourg, 2016; ISBN 978-92-79-36740-3. [Google Scholar]
- Borghetti, M.; Cocco, S.; Lambardi, M.; Raddi, S. Response to Water Stress of Italian Alder Seedlings from Diverse Geographic Origins. Can. J. For. Res. 1989, 19, 1071–1076. [Google Scholar] [CrossRef]
- King, R.A.; Ferris, C. Chloroplast DNA and Nuclear DNA Variation in the Sympatric Alder Species, Alnus cordata (Lois.) Duby and A. glutinosa (L.) Gaertn. Biol. J. Linn. Soc. 2000, 70, 147–160. [Google Scholar] [CrossRef]
- Xie, C.-Y. Ten-Year Results from Red Alder (Alnus rubra Bong.) Provenance-Progeny Testing and Their Implications for Genetic Improvement. New For. 2008, 36, 273–284. [Google Scholar] [CrossRef]
- Malkoçoğlu, A.; Özdemir, T. The Machining Properties of Some Hardwoods and Softwoods Naturally Grown in Eastern Black Sea Region of Turkey. J. Mater. Process. Technol. 2006, 173, 315–320. [Google Scholar] [CrossRef]
- Bekhta, P.; Hiziroglu, S.; Shepelyuk, O. Properties of Plywood Manufactured from Compressed Veneer as Building Material. Mater. Des. 2009, 30, 947–953. [Google Scholar] [CrossRef]
- Klemmedson, J.O. Ecological Importance of Actinomycete-Nodulated Plants in the Western United States. Bot. Gaz. 1979, 140, S91–S96. [Google Scholar] [CrossRef]
- Mejnartowicz, L. Influence of nursery environment and pollution on alders. In Genetic Response of Forest Systems to Changing Environmental Conditions; Müller-Starck, G., Schubert, R., Eds.; Forestry Sciences; Springer: Dordrecht, The Netherlands, 2001; pp. 63–73. ISBN 978-94-015-9839-2. [Google Scholar]
- Innangi, M.; Danise, T.; d’Alessandro, F.; Curcio, E.; Fioretto, A. Dynamics of Organic Matter in Leaf Litter and Topsoil within an Italian Alder (Alnus cordata (Loisel.) Desf.) Ecosystem. Forests 2017, 8, 240. [Google Scholar] [CrossRef] [Green Version]
- Lepais, O.; Bacles, C.F.E. De Novo Discovery and Multiplexed Amplification of Microsatellite Markers for Black Alder (Alnus glutinosa) and Related Species Using SSR-Enriched Shotgun Pyrosequencing. J. Hered. 2011, 102, 627–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lance, S.L.; Jones, K.L.; Hagen, C.; Glenn, T.C.; Jones, J.M.; Gibson, J.P. Development and Characterization of Nineteen Polymorphic Microsatellite Loci from Seaside Alder, Alnus Maritima. Conserv. Genet. 2009, 10, 1907. [Google Scholar] [CrossRef]
- Paetkau, D.; Waits, L.P.; Clarkson, P.L.; Craighead, L.; Vyse, E.; Ward, R.; Strobeck, C. Variation in Genetic Diversity across the Range of North American Brown Bears. Conserv. Biol. 1998, 12, 418–429. [Google Scholar] [CrossRef]
- Chapuis, M.-P.; Estoup, A. Microsatellite Null Alleles and Estimation of Population Differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef] [Green Version]
- Anderson, E.C.; Thompson, E.A. A Model-Based Method for Identifying Species Hybrids Using Multilocus Genetic Data. Genetics 2002, 160, 1217–1229. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- QGIS Development Team. QGIS Geographic Information System; Open Source Geospatial Foundation Project. Available online: http://www.qgis.org (accessed on 4 May 2021).
- Kalinowski, S.T. Hp-Rare 1.0: A Computer Program for Performing Rarefaction on Measures of Allelic Richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (Version 3.0): An Integrated Software Package for Population Genetics Data Analysis. Evol. Bioinform. Online 2007, 1, 47–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pritchard, J.K.; Wen, X.; Falush, D. Documentation of Structure Software: Version 2.3. 2009. Available online: https://web.stanford.edu/group/pritchardlab/structure.html (accessed on 10 December 2021).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A Program for Identifying Clustering Modes and Packaging Population Structure Inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [Green Version]
- Banaev, E.V.; Bažant, V. Study of Natural Hybridization between Alnus incana (L.) Moench. and Alnus glutinosa (L.) Gaertn. J. For. Sci. 2008, 53, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Parfenov, V.I. Dependence of Distribution and Adaptation of Plant Species on the Area Borders; Nauka Tekhnika: Minsk, Belarus, 1980; p. 205. [Google Scholar]
- Rieseberg, L.H.; Blackman, B.K. Speciation Genes in Plants. Ann. Bot. 2010, 106, 439–455. [Google Scholar] [CrossRef] [Green Version]
- Harrison, R.G.; Larson, E.L. Hybridization, Introgression, and the Nature of Species Boundaries. J. Hered. 2014, 105, 795–809. [Google Scholar] [CrossRef] [Green Version]
- Dang, M.; Yue, M.; Zhang, M.; Zhao, G.; Zhao, P. Gene Introgression among Closely Related Species in Sympatric Populations: A Case Study of Three Walnut (Juglans) Species. Forests 2019, 10, 965. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.-Y.; Sun, Y.-W.; Bai, X.-R.; Dang, M.; Feng, X.-J.; Zulfiqar, S.; Zhao, P. Population Structure, Genetic Diversity, and Gene Introgression of Two Closely Related Walnuts (Juglans Regia and J. Sigillata) in Southwestern China Revealed by EST-SSR Markers. Forests 2018, 9, 646. [Google Scholar] [CrossRef] [Green Version]
- Chhatre, V.E.; Evans, L.M.; DiFazio, S.P.; Keller, S.R. Adaptive Introgression and Maintenance of a Trispecies Hybrid Complex in Range-Edge Populations of Populus. Mol. Ecol. 2018, 27, 4820–4838. [Google Scholar] [CrossRef] [PubMed]
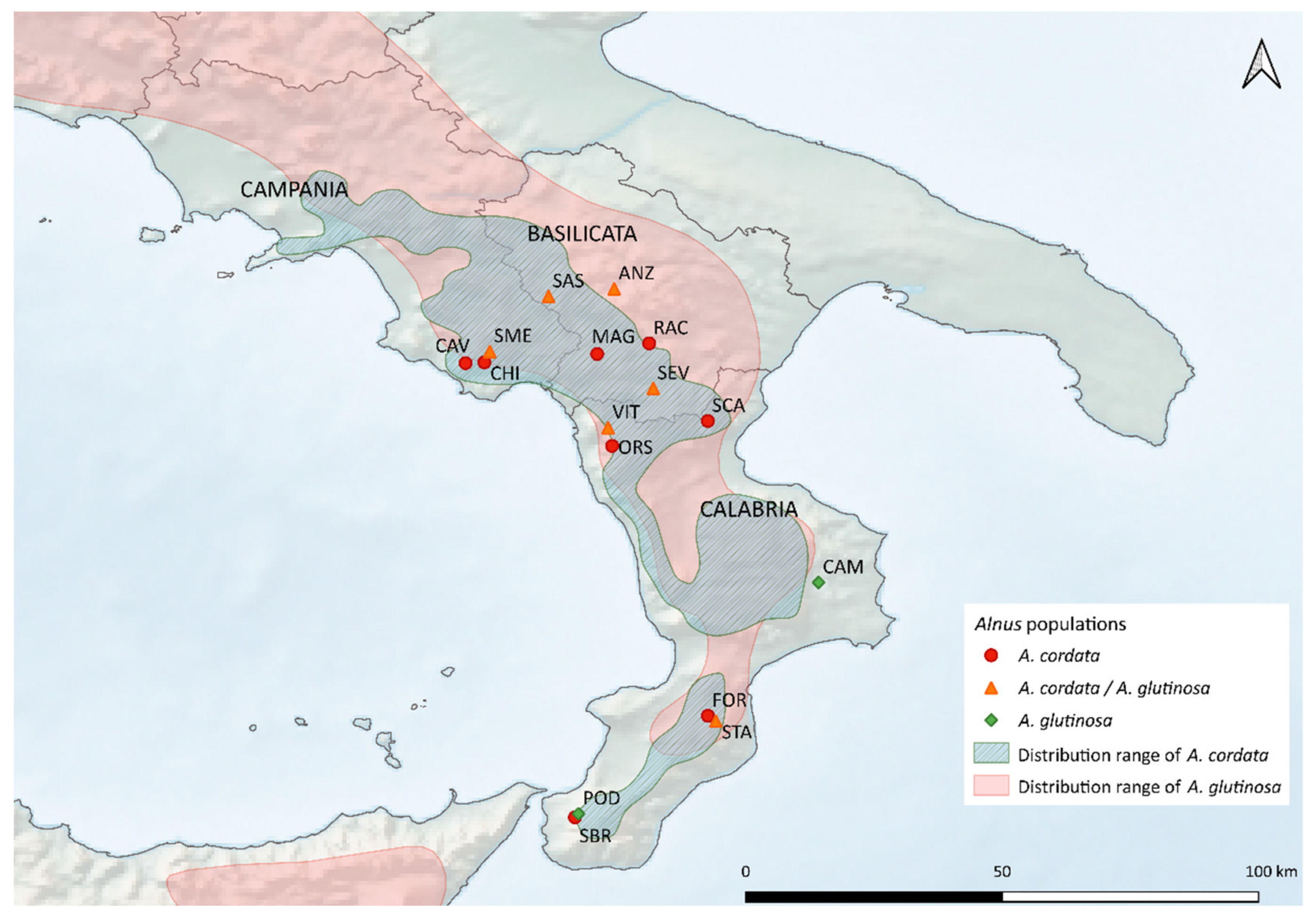





| Pop ID | Municipality | Pr | Lat. | Long. | Elev. (m) | Ni | A. cordata | A. glutinosa | Uncert. |
|---|---|---|---|---|---|---|---|---|---|
| Campania Region | |||||||||
| CAV | Cuccaro Vetere | SA | 40.1613 | 15.2946 | 680 | 19 | 19 | 0 | 0 |
| CHI | Montano Antilia | SA | 40.1645 | 15.3779 | 700 | 20 | 19 | 0 | 1 |
| SME | Rofrano | SA | 40.2130 | 15.4018 | 521 | 25 | 15 | 5 | 5 |
| Basilicata Region | |||||||||
| RAC | Gallicchio | PZ | 40.2488 | 16.1076 | 370 | 18 | 18 | 0 | 0 |
| SEV | Chiaromonte | PZ | 40.0491 | 16.1255 | 630 | 20 | 13 | 7 | 0 |
| MAG | Moliterno | PZ | 40.2017 | 15.8772 | 740 | 20 | 20 | 0 | 0 |
| ANZ | Anzi | PZ | 40.4913 | 15.9526 | 585 | 16 | 13 | 3 | 0 |
| SAS | Sasso di Castalda | PZ | 40.4570 | 15.6617 | 740 | 24 | 15 | 6 | 3 |
| Calabria Region | |||||||||
| CAM | Santa Severina | KR | 39.1889 | 16.8576 | 100 | 17 | 0 | 17 | 0 |
| SCA | Plataci | CS | 39.9044 | 16.3673 | 1156 | 18 | 18 | 0 | 0 |
| VIT | Mormanno | CS | 39.8745 | 15.9254 | 180 | 26 | 17 | 4 | 5 |
| ORS | Papasidero | CS | 39.7942 | 15.9435 | 220 | 19 | 19 | 0 | 0 |
| FOR | Brognaturo | VV | 38.5984 | 16.3674 | 950 | 19 | 19 | 0 | 0 |
| STA | Brognaturo | VV | 38.5751 | 16.4023 | 1018 | 22 | 6 | 2 | 14 |
| SBR | S. Stefano in Aspromonte | RC | 38.1487 | 15.7789 | 1000 | 20 | 20 | 0 | 0 |
| POD | S. Stefano in Aspromonte | RC | 38.1646 | 15.7941 | 616 | 16 | 0 | 16 | 0 |
| Pop | N | Na | Ne | I | Ho | He | uHe | Fis |
|---|---|---|---|---|---|---|---|---|
| Alnus cordata (Loisel.) Duby | 216 | 5.571 | 2.177 | 0.853 | 0.391 | 0.437 | 0.438 | 0.190 |
| Alnus glutinosa (L.) Gaertn. | 71 | 9.429 | 4.237 | 1.528 | 0.533 | 0.664 | 0.669 | 0.203 |
| Hybrids | 32 | 8.429 | 5.060 | 1.722 | 0.616 | 0.770 | 0.782 | 0.224 |
| Pop | Species | N | Na | Ne | Ho | He | uHe | Fis |
|---|---|---|---|---|---|---|---|---|
| CAV | Alnus cordata | 19 | 3.143 | 2.074 | 0.436 | 0.452 | 0.465 | 0.098 |
| CHI | 20 | 3.286 | 2.001 | 0.436 | 0.439 | 0.450 | 0.035 | |
| RAC | 18 | 3.143 | 2.060 | 0.436 | 0.421 | 0.434 | −0.034 | |
| MAG | 20 | 3.000 | 2.050 | 0.430 | 0.418 | 0.429 | 0.006 | |
| SCA | 18 | 3.429 | 1.997 | 0.395 | 0.368 | 0.379 | −0.088 | |
| ORS | 19 | 3.000 | 2.014 | 0.308 | 0.379 | 0.389 | 0.243 | |
| FOR | 19 | 2.857 | 2.064 | 0.391 | 0.378 | 0.389 | −0.061 | |
| SBRU | 20 | 3.000 | 1.930 | 0.343 | 0.358 | 0.368 | 0.188 | |
| SME | Mixed A. cordata/A. glutinosa | 25 | 7.571 | 4.199 | 0.576 | 0.724 | 0.739 | 0.237 ** |
| SEV | 20 | 6.857 | 3.811 | 0.479 | 0.729 | 0.748 | 0.359 ** | |
| ANZ | 16 | 6.143 | 3.365 | 0.509 | 0.653 | 0.674 | 0.263 ** | |
| SAS | 24 | 7.286 | 3.949 | 0.523 | 0.699 | 0.714 | 0.290 ** | |
| VIT | 25 | 6.714 | 3.587 | 0.391 | 0.688 | 0.702 | 0.461 ** | |
| STA | 21 | 6.857 | 4.141 | 0.459 | 0.709 | 0.726 | 0.400 ** | |
| CAM | Alnus glutinosa | 17 | 6.286 | 3.281 | 0.483 | 0.593 | 0.612 | 0.187 ** |
| POD | 16 | 4.286 | 2.793 | 0.514 | 0.546 | 0.565 | 0.039 |
| Locus | Fis | Fit | Fst | Nm |
|---|---|---|---|---|
| alma1 | 0.470 | 0.639 | 0.320 | 0.532 |
| alma7 | 0.113 | 0.168 | 0.062 | 3.789 |
| alma11 | 0.073 | 0.181 | 0.116 | 1.903 |
| alng4 | 0.283 | 0.434 | 0.211 | 0.937 |
| AG10 | 0.023 | 0.165 | 0.146 | 1.466 |
| AG20 | 0.508 | 0.761 | 0.515 | 0.236 |
| AG13 | 0.098 | 0.177 | 0.088 | 2.600 |
| Mean | 0.224 | 0.361 | 0.208 | 1.638 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villani, F.; Castellana, S.; Beritognolo, I.; Cherubini, M.; Chiocchini, F.; Battistelli, A.; Mattioni, C. Genetic Variability of Alnus cordata (Loisel.) Duby Populations and Introgressive Hybridization with A. glutinosa (L.) Gaertn. in Southern Italy: Implication for Conservation and Management of Genetic Resources. Forests 2021, 12, 655. https://doi.org/10.3390/f12060655
Villani F, Castellana S, Beritognolo I, Cherubini M, Chiocchini F, Battistelli A, Mattioni C. Genetic Variability of Alnus cordata (Loisel.) Duby Populations and Introgressive Hybridization with A. glutinosa (L.) Gaertn. in Southern Italy: Implication for Conservation and Management of Genetic Resources. Forests. 2021; 12(6):655. https://doi.org/10.3390/f12060655
Chicago/Turabian StyleVillani, Fiorella, Simone Castellana, Isacco Beritognolo, Marcello Cherubini, Francesca Chiocchini, Alberto Battistelli, and Claudia Mattioni. 2021. "Genetic Variability of Alnus cordata (Loisel.) Duby Populations and Introgressive Hybridization with A. glutinosa (L.) Gaertn. in Southern Italy: Implication for Conservation and Management of Genetic Resources" Forests 12, no. 6: 655. https://doi.org/10.3390/f12060655






